Open Access Article
Open Access ArticleUltrasensitive melanoma biomarker detection using a microchip SERS immunoassay with anisotropic Au–Ag alloy nanoboxes†
Aswin Raj Kumar‡
 a,
Karthik Balaji Shanmugasundaram‡
a,
Karthik Balaji Shanmugasundaram‡ a,
Junrong Lia,
Zhen Zhanga,
Abu Ali Ibn Sina
a,
Junrong Lia,
Zhen Zhanga,
Abu Ali Ibn Sina *a,
Alain Wuethrich
*a,
Alain Wuethrich *a and
Matt Trau
*a and
Matt Trau *ab
*ab
aCentre for Personalized Nanomedicine, Australian Institute for Bioengineering and Nanotechnology (AIBN), The University of Queensland, Brisbane, QLD 4072, Australia. E-mail: a.wuethrich@uq.edu.au; a.sina@uq.edu.au; m.trau@uq.edu.au
bSchool of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, QLD 4072, Australia
First published on 4th August 2020
Abstract
The detection of circulating biomarkers in liquid biopsies has the potential to provide a non-invasive route for earlier cancer diagnosis and treatment management. Melanoma chondroitin sulfate proteoglycan (MCSP) is a membrane protein characteristic for melanoma cell migration and tissue invasion with its soluble form (sMCSP) serving as a potential promising diagnostic surrogate. However, at the initial disease stage, the detection of sMCSP is challenging because of its low abundance and the required high specificity to analyze sMCSP in complex bodily fluids. Herein, we report a highly sensitive and high-throughput microchip that enables Surface Enhanced Raman Spectroscopy (SERS) immunoassay for parallel detection of up to 28 samples. Key to assay speed and sensitivity is the stimulation of an alternating current-induced nanofluidic mixing that improves target-sensor collision and displacement of non-specific molecules. Anisotropic Au–Ag alloy nanoboxes (NB's) with strong plasmonic hot spots provide single SERS particle sensitivity that enables ultrasensitive sMCSP detection of as low as 0.79 pM (200 pg ml−1). As a proof of concept study, we investigate the assay performance in simulated melanoma patient samples.
Introduction
Molecular alterations are the main driving process of abnormal cell proliferation which give rise to a malignant tumor in cancer.1,2 Cancer cells release altered molecular materials such as soluble proteins, cell-free DNA, cell-free RNA, and exosomes into blood circulation which can be interrogated to study the dynamic evolution of cancer.3,4 The analysis of these circulating biomarkers is promising for minimally invasive diagnostics, monitoring therapy response, and to estimate the risk of metastatic progression and prognosis.5,6 Among the multitude of biomarkers present in liquid biopsy samples (e.g., blood, saliva, urine), soluble proteins belong to the most informative biomarkers as they reflect directly aberrations to molecular pathways.7The precise determination of low abundance soluble proteins, typically in the range of low pg ml−1 challenges many detection methods.8,9 In addition to the required high detection sensitivity, most liquid biopsy samples consist of a complex sample matrix, which further aggravates the specific detection of soluble proteins and increases the risk of false-positive results. In recent years, innovative detection strategies have been developed which include fluorescence nanoparticle probes, nanostructured devices, and surface enhanced Raman spectroscopy (SERS).9–11 SERS is a sensitive spectroscopic technique with the capability to detect single molecules.12 To enhance the relatively weak Raman signal of the Raman reporter, SERS makes use of metal structures to generate plasmonic “hot spots”, which increase the Raman signal by multiple orders of magnitude.13 SERS technique has been used for the detection of numerous biomolecules and also for the characterization of circulating tumor cell, soluble proteins and extracellular vesicles.14–17 The high sensitivity, molecular specificity, photostability and multiplexing capability of SERS nanotags conceive an attractive detection strategy for soluble proteins.17
The domain of microfabrication offers various platform devices suitable for detecting soluble proteins.17 Microfabrication technology for the production of biosensing chips enable the fabrication of ultrasensitive nanoscale devices with dimensions ranging from micrometer to millimeter.18 Advances in microfabrication and molding techniques such as photolithography have played a major role in the development of enhanced biosensing platforms that enable sensitive biomarker detection.18 Microdevices allow the integration of automated whole sample-to-answer workflow with controlled sample handling, which improves assay reproducibility. The micrometer to nanometer dimensions of the structures are suitable for handling minute volumes of patient samples and accelerate assay time by allowing high throughput analysis in a single device.18,19 The integration of a SERS immunoassay to a microchip could, therefore, be promising to achieve highly sensitive and specific soluble biomarker detection.
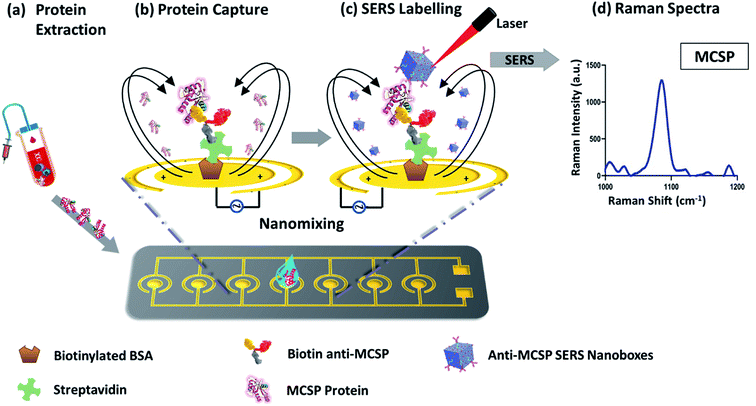
In this study, we present a microchip biosensor that utilizes SERS immunoassay to achieve highly sensitive and specific detection of a melanoma specific soluble protein from a complex biological system. The microchip biosensor utilizes alternating current-electrohydrodynamics (ac-EHD) to generate nanofluidic mixing of the sample solution within the nanometer scale of the chip surface. This nanomixing significantly increases target antigen interaction with antibody and SERS nanotags. Moreover, these ac-EHD nanomixing conditions are finely tunable to remove nonspecific attachment of the non-target molecules towards the sensor surface enabling highly specific detection of a soluble protein. To achieve ultrasensitive detection, we synthesize anisotropic core–shell nanoboxes of silver and gold to enhance the Raman scattering and achieve a detectable signal from single nanoparticles.20 The detection of single SERS particle sensitivity with Au–Ag alloy nanoboxes were determined in our previous study.20 Key to the strong signal intensity is the generation of strong electromagnetic fields on tips and corners of the Ag–Ag alloy nanoboxes used as SERS nanotags for enhanced sensitivity.20
As an exemplary demonstration of highly sensitive soluble protein detection in liquid biopsy, we select the melanoma marker MCSP (melanoma-associated chondroitin sulphate proteoglycan) which is overexpressed on melanoma cells.14 Proteolytic cleavage of MCSP and cell apoptosis are the main mechanisms for the release of soluble MCSP (sMCSP) into circulation. sMCSP has been found as a potential candidate for early melanoma detection and treatment monitoring. We study and optimize the performance of the microchip-SERS immunoassay in detecting sMCSP and evaluate its clinical applicability on simulated melanoma plasma samples. We believe that the microchip-SERS immunoassay could be useful to monitor disease progression in cancer immunotherapy owing to its highly sensitive, specific, and potential multiplexing capabilities.
Experimental
Cell culture
Melanoma cell line (MEL-28) from Ludwig Institute for Cancer Research (Melbourne, Australia) were characterized and used in their early passages. These cells were maintained and cultured in RPMI 1640 (Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, USA), 2 mM Glutamax (Thermo Fisher Scientific, USA) and 1% penicillin/streptomycin (Thermo Fisher Scientific, USA). Mycoplasma tests were conducted before and after the experiments. The cells were retained in the flask until the confluency level reaches to ∼90% and later scraped off with trypsin. The collected cells were washed with phosphate-buffered saline (PBS) and subjected to protein extraction.Immunoprecipitation
To extract the pure MCSP, we used Pierce Classic Magnetic Immunoprecipitation/Co-IP kit to perform magnetic immunopurification. Initially, the cultured melanoma cells were incubated on ice-cold lysis buffer (25 mM Tris HCl pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol) along with the addition of protease and phosphatase inhibitors (Thermo Fisher Scientific, Australia), with periodic mixing for 5 min. Later, the lysis buffer containing the cells were centrifuged (Allegra X-22 series benchtop centrifuge, Beckman Coulter) at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min to remove the cell debris and isolate the total protein from the cell pellet. The total protein quantification was completed using the Bradford assay. The total protein concentration 1000 μg of the cell lysate was combined with affinity-purified MCSP antibody and was incubated for 1 h at room temperature (RT) to obtain the antigen–antibody complex. Subsequently, immunoprecipitation was performed on the addition of pre-washed protein A/G magnetic beads along with the antigen–antibody mixture and incubated for 1 h at RT with constant mixing. Further, the magnetic beads were collected using a magnetic stand and the unbound samples were removed and discarded. Next, 100 μl of elution buffer (0.1 M glycine HCl, pH 2.5) was added to the magnetic beads and was incubated for 10 min at RT with continuous mixing to remove the target antigen from the magnetic beads. The final supernatant containing the MCSP protein was mixed with 10 μl of neutralization buffer (1 M Tris, pH 8.5) to every 100 μl of eluate to neutralize the elution buffer. The immunoprecipitation was concluded by determining the MCSP protein concentration using protein A280 assay in the nanodrop and the obtained samples were stored at −20 °C until further use.
000 × g for 10 min to remove the cell debris and isolate the total protein from the cell pellet. The total protein quantification was completed using the Bradford assay. The total protein concentration 1000 μg of the cell lysate was combined with affinity-purified MCSP antibody and was incubated for 1 h at room temperature (RT) to obtain the antigen–antibody complex. Subsequently, immunoprecipitation was performed on the addition of pre-washed protein A/G magnetic beads along with the antigen–antibody mixture and incubated for 1 h at RT with constant mixing. Further, the magnetic beads were collected using a magnetic stand and the unbound samples were removed and discarded. Next, 100 μl of elution buffer (0.1 M glycine HCl, pH 2.5) was added to the magnetic beads and was incubated for 10 min at RT with continuous mixing to remove the target antigen from the magnetic beads. The final supernatant containing the MCSP protein was mixed with 10 μl of neutralization buffer (1 M Tris, pH 8.5) to every 100 μl of eluate to neutralize the elution buffer. The immunoprecipitation was concluded by determining the MCSP protein concentration using protein A280 assay in the nanodrop and the obtained samples were stored at −20 °C until further use.
Western blot validation
Initially, the cultured MEL 28 cells were lysed, a protein ladder (P7712S, New England Biolabs, Australia) was taken along with 20 μg of the total protein lysate mixed with NuPAGE LDS sample buffer (4×), (Novex Life Technologies, Australia) and were heated at 95 °C for 5 min and loaded separately on an SDS-PAGE gel (NuPAGE 4–12% Bis–Tris gel, Novex Life Technologies, Australia). The gel containing the ladder and protein samples were added with an adequate amount of MOPS running buffer (1×) and were set to run at 200 V for 65 min. Subsequently, the proteins were transferred to the PVDF membrane using the NuPAGE transfer buffer (1×) when running under 30 V for 7 min. Then the membrane was given a quick wash using TBST (TBS (25 mM Tris, 0.15 M NaCl, pH 7.4) + 0.1% Tween 20) followed by blocking the membrane for 1 h using blocking buffer, (5% non-fat dry milk and TBS (25 mM Tris, 0.15 M NaCl pH 7.4)). Later, the membrane was incubated for 2 h in the selected MCSP monoclonal antibody (R&D systems, China) solution containing 5% non-fat dry milk and TBST. Following the incubation period, the membrane was washed (3 × 10 min) with TBST and then blocked again for 30 min. Further, the blocked membrane was immersed for 1 h in the infrared dye (IRDye 800CW Goat) tagged anti-mouse secondary antibody solution (Odyssey LI-COR, USA) and then rewashed (3 × 10 min) with the TBST solution. Finally, a fluorescence-based infrared imager (Odyssey LI-COR, USA) was used to capture the image of the PVDF membrane.Synthesis of SERS nanotags
Preparation of SERS nanotags involves the synthesis of Au–Ag alloy NB's followed by conjugation with Raman reporters and anti-MCSP antibody through dithiobis (succinimidyl propionate) DSP was prepared according to our previous publication.20 The binding properties and the polycrystalline structure of the nanoboxes were characterized and analyzed in our previous work.21 The crystalline structure of the nanoboxes was determined from the SEM and TEM images shown in ESI Fig. S4.† For the NB's synthesis, 45 μl of 25.4 mM HAuCl4 was added to 10 ml of ultrapure water under magnetic stirring followed by simultaneous addition of 170 μl of 6 mM AgNO3 and 30 μl of 0.1 M ascorbic acid. NB's formation began immediately which can be visually confirmed by the appearance of blue color in solution. After stirring for 1 min, 1 ml of the mixture was centrifuged at 600 × g for 15 min. The final product was resuspended in 300 μl of H2O.For the conjugation of Raman reporter and antibody on NB's, 8 μl of 1 mM MBA (4-mercaptobenzoic acid), 2 μl of 1 mM DSP were simultaneously introduced to 300 μl of Au–Ag alloy NB's concentrated from 1 ml of the prepared solution. To form intact monolayers, the mixture was incubated at 25 °C, 350 rpm for 6 h followed by centrifugation at 600 × g for 15 min to remove residual reactants and resuspended in 300 μl of 0.1 mM PBS. Then, 2 μl of pure MCSP antibody was added and the solution was incubated overnight at 4 °C to immobilize the antibodies onto the surface of Au–Ag alloy NB's. Finally, the product was centrifuged at 600 × g for 15 min and resuspended into 200 μl of 0.1% BSA to relieve free antibodies and prevent non-specific adsorption and detaching of Raman reporters. The final product was stored at 4 °C until further use.
Biosensor fabrication
Microchip biosensor was fabricated according to previous literature.22 The fabrication and device design are shown in ESI Fig. S2 and S3.† Briefly, the electrodes layout and a second soda-lime mask that carried the pattern was designed using Layout L-Edit V15 (Tanner Research, Monrovia, USA) and written to a 5 × 5-inch soda lime chrome mask (Shenzhen Qingyi Precision Mask Making, Shenzhen, Singapore) using a direct laser writer μPG 101 (Heidelberg Instruments, Heidelberg, Austria) with 5-micron head attached. Standard photolithography was used in the fabrication of the electrodes. 1.1 mm thick, 4-inch Borofloat glass wafers (Bonda Technology Pte Ltd, Downtown Core, Singapore) were spin coated with a negative photoresist AZnLOF 2070 (Microchemicals GmbH, Ulm, Germany) at 3000 rpm for 30 s. The wafers were UV exposed at a constant rate of 250 mJ cm−1 using a mask aligner (EVG 620, EV Group, St Florian am Inn, Austria) after a softbake at 110 °C for 2 min followed by another bake at 110 °C for 1 min. Subsequently, the wafers were developed in AZ726 MIF Developer (Microchemicals GmbH, Ulm, Germany) for 45 s. A thin layer of Ti (10 nm) and Au (200 nm) was deposited with Temescal FC-2000 electron beam evaporator (Ferrotec, Livermore, USA) before over lifting to reveal the inner and outer circular electrodes connected to large gold connecting pads. PDMS (polydimethylsiloxane) channels to perform immunoassay was prepared by curing activated silicon elastomer solution (Sylgard® 184, Dow, Midland, USA) at 80 °C for 20 min. PDMS structures were thermally bonded on the fabricated device for 2 h at 65 °C after punching wells of 6 mm in diameter.Biosensor functionalisation
Device functionalization onto the gold electrode for protein biomarker detection follows the standard biotin-avidin chemistry. First, a solution of 250 μg ml−1 biotinylated bovine serum albumin (Thermo Fisher Scientific, Scoresby, Australia) in phosphate buffered saline (PBS 10 mM, pH 7.4) was incubated for 2 h, followed by 1 h incubation with 100 μg ml−1 of Streptavidin (Thermo Fisher Scientific, Scoresby, Australia). Next, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 dilution of Biotin-MCSP (Miltenyi Biotech, Australia) in PBS was added to the electrode wells and incubated for 1 h. The reactions were carried out at room temperature. The electrodes were washed with PBS one time in between each step to remove unbound biomolecules. Finally, the device was incubated with 1% BSA in PBS (Thermo Fisher Scientific, Scoresby, Australia) for 1 h at room temperature as a blocking step to complete biosensor functionalization.
50 dilution of Biotin-MCSP (Miltenyi Biotech, Australia) in PBS was added to the electrode wells and incubated for 1 h. The reactions were carried out at room temperature. The electrodes were washed with PBS one time in between each step to remove unbound biomolecules. Finally, the device was incubated with 1% BSA in PBS (Thermo Fisher Scientific, Scoresby, Australia) for 1 h at room temperature as a blocking step to complete biosensor functionalization.
Detection of target protein biomarker
Initially, 50 μl of protein lysate containing pure MCSP with desired concentration was incubated on the wells of the functionalized biosensor under ac-EHD conditions of f = 500 Hz, Vpp = 800 mV for t = 30 min using signal generator 33510B (Agilent Technologies, Mulgrave, Australia). The wells were washed with PBS three times to remove nonspecific biomolecules. Next, pure MCSP antibody conjugated with SERS nanotags was incubated in the wells with an applied electric field of f = 500 Hz, Vpp = 800 mV for t = 20 min to complete the assay. This was followed by 3 times wash with wash buffer (0.1% BSA, 0.01% Tween-20 in PBS) to remove unbound SERS nanotags. The process was carried out with the healthy human serum samples obtained from Ventyx Wesley Research Institute Tissue Bank (Auchenflower, Australia) and diluted to 5-fold with PBS.SERS mapping and data analysis
SERS spectra and images were recorded on WITec alpha 300 R confocal microscope. The sample was excited for Raman scattering using a HeNe laser with a power of 35.0 mW at 633 nm wavelength. The integration time was set to 0.1 s with a step size of 1 μm and a mapping area of 60 μm × 60 μm (60-pixel × 60-pixel) using 20× microscopic objective. A silicon wafer that produces a first-order photon peak at ∼520 cm−1 was used for calibration of the instrument based on Raman intensity. Fluorescence and background removal from the raw spectra were done with Vancouver Raman algorithm based on the fifth-order polynomial fitting method. Multiple positions from each sample was scanned to improve precision and reduce any possible errors during the experiment. Triplicates were obtained for each sample and the average of peak height used to plot the intensity graph corresponding to the concentration of protein biomarkers in liquid biopsies.Results and discussion
Principle of microchip biosensing platform
The microchip platform consists of an array of 28 electrodes in 4 rows which has the potential to simultaneously analyze different biomarkers or run 28 samples in parallel. Individual electrodes are made up of inner and outer circular ring structure, on which PDMS wells are embedded. We have previously studied the effect of biosensing platform geometry on target capture and EHD fluid flow and optimized it for optimal assay performance for biomarker detection.22 Here, we investigated the effect of ac-EHD induced nanomixing and SERS nanotags with anisotropic Au–Ag alloy NB's as substrates for sensitive and specific detection of a soluble protein. The application of optimized ac-EHD conditions (f = 500 Hz, Vpp = 800 mV) during sample and SERS nanotags incubation facilitated maximum target capture and maximizes the disposition of weakly bound molecules (non-specific). Upon application of the electric field, a lateral variation arose in the number and distribution of charges across each electrode pair. The non-uniform forces generated on each electrode pair varied as the lateral force on the larger electrode was stronger than that of the smaller electrode. This difference in force generated as a result of asymmetric electrode pairs gave rise to bulk fluid flow towards the larger electrodes which increased the target sensor interactions and resulted in increased specificity.The detail workflow of the assay has been demonstrated in Scheme 1. Briefly, the target MSCP protein was extracted and purified by standard immunoprecipitation method from Mel-28 melanoma cell line. The presence of MCSP protein in these cells was determined by western blot study, shown in ESI Fig. S1.† The purified MCSP was then added to the anti-MCSP functionalized microchip and specifically captured with ac-EHD nanofluidic mixing. The captured MCSP protein molecules were detected by Au–Ag alloy SERS nanotags with their characteristic peaks of MBA (4-mercaptobenzoic acid) reporter molecule at an intensity of 1075 cm−1. The peak at 1075 cm−1 was selected because it provides the highest sensitivity amongst the other peaks of MBA's Raman spectrum. The entire Raman spectrum for the MBA reporter with its characteristic peak (1075 cm−1) are shown in ESI Fig. S2.5.†
Study of ac-EHD nanomixing
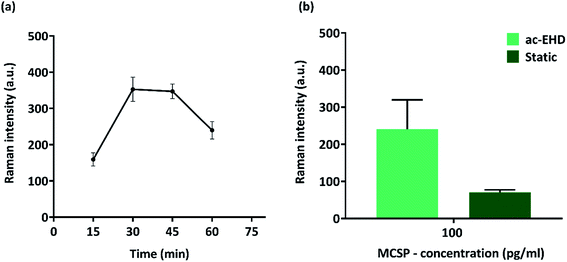
To investigate the effect of ac-EHD induced nanomixing time on the performance of the immunoassay, we conducted experiments with IP-purified MCSP (100 pg ml−1) at time intervals of 15, 30, 45, and 60 min for antigen incubation under an applied electric field of f = 500 Hz, Vpp = 800 mV. The signal intensity was minimum after 15 min incubation due to reduced nanomixing time which resulted in a limited number of the target antigen – primary antibody interaction. Fig. 1(a) shows that the maximum Raman signal was observed at 30 min after which it tends to stabilize till 45 min. The signal intensity dropped drastically when the time interval reached 60 min which might be due to excessive nanoshearing of the target antigen. 30 min was selected for the upcoming experiments and compared to a static incubation time of 1 h Fig. 1(b) shows an approx. 4-fold increase (100 pg ml−1) in detection sensitivity upon application of electric field when compared to static incubation with a significant reduction in overall assay time.Biosensor selectivity
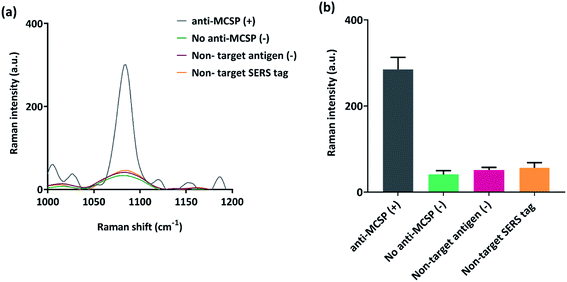
To investigate the specificity of our capture immunoassay, experiments were run with the positive sample of MCSP protein (100 pg ml−1) and non-target protein PD-L1 (100 pg ml−1) was taken as the negative control. Additional control experiments without primary antibody and with non-target PD-L1 SERS nanotags were run to assess the detection bias due to the presence of potential targets in complex serum samples. The samples were run through the microchip platform under optimized ac-EHD conditions. Fig. 2(a) shows the typical overlaid SERS spectra where a strong Raman signal is observed for anti-MCSP positive control (+) sample. The negative controls (−) were without primary antibody (no anti-MCSP), non-target protein (PD-L1), and non-target conjugated with SERS tags (anti-PD-L1 SERS) showed a negligible Raman signal. The data suggest that the capture immunoassay is highly specific for MCSP detection using the ac-EHD microchip platform.Assay sensitivity detection
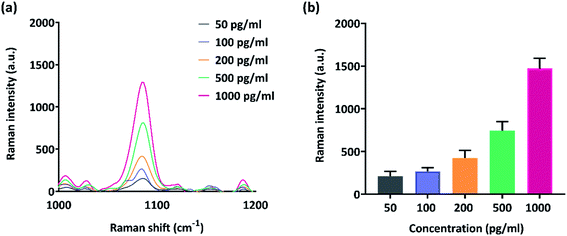
Biosensors with high sensitivity are vital and mandatory for prognostic applications and to observe post-treatment drug resistance including metastatic relapses. The dynamic range and sensitivity of the microchip-SERS immunoassay was studied with MCSP protein serially diluted in concentrations ranging from 1 pg ml−1 to 1000 pg ml−1. The samples were driven through the device under applied electric field conditions of f = 500 Hz, Vpp = 800 mV, t = 30 min. Fig. 3(a) shows the representative overlaid Raman spectra obtained from different concentrations. An increase in Raman signal is observed with increasing protein concentration and the platform can detect protein as low as 50 pg ml−1. The coefficient of variation was 1.15% and the coefficient of determination as (R2) = 0.9878. These parameters are very well suited to detect trace levels of biomarkers in blood biopsy with negligible false positive results during cancer immunotherapy.Simulated melanoma patient plasma analysis
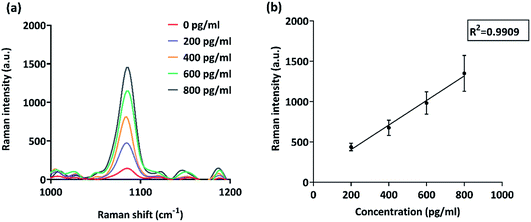
Unfavorable prognosis is correlated with overexpression of MCSP as it promotes growth and tissue invasion by tumor cells with the help of certain signaling molecules such as focal adhesion kinase (FAK)23 and mitogenic ERK.24 Thus, sensitive detection of soluble MCSP is vital to aid the above-mentioned strategies. We explored the potential of our microchip platform in capturing and quantifying subtle changes in MCSP levels from a complex biological matrix.Pure MCSP was spiked in human serum to obtain different concentrations ranging from 200 pg ml−1 to 800 pg ml−1. The simulated samples were run on functionalized gold electrodes under ac-EHD conditions of f = 500 Hz, Vpp = 800 mV, and t = 30 min. Blank (0 pg ml−1) was taken as the negative control to assess any significant noise observed in the assay due to the presence of other potential targets and non-specific molecules in human serum. It is evident from Fig. 4(a) that there is a significant difference between the 200 pg ml−1 and blank (0 pg ml−1) which determines the potential of the microchip platform in sensitive and specific detection of protein biomarkers from liquid biopsies.
Serological tumor biomarkers can act as a predictive and prognostic tool in determining the state of metastasis owing to their increased expression in advanced stage cancer.25 These biomarker levels can be used to identify patients that benefited from targeted treatment which are specific and provides molecular endpoints that predict and monitor treatment efficacy. Numerous melanoma biomarkers have been associated with overall-survival, tumor progression, recurrence, and stages of metastasis.25 An ideal diagnostic tool should be capable of quantifying biomarkers present in blood and other body fluids non-invasively. The tool should exhibit enough sensitivity without interference from non-specific molecules to predict therapy response and reduce false-positive results. Common commercial ELISA kits provide MCSP detection sensitivity in the range of pg ml−1, but have drawbacks on laborious and time-consuming workflows, requiring relatively large volumes of reagents and samples. With our microchip-SERS immunoassay the analysis time is reduced to 5 h with minimal sample handling steps involved and the total volume of sample reduced to 50 μl (ESI Table S1†). Our microchip-SERS immunoassay provided a comparative sensitivity with the previously published studies on the detection of melanoma-associated biomarkers.26–28 The comparative studies referred for the sensitivity detection are added as a table in the ESI Table S2.† We believe our microchip-SERS immunoassay achieved all the essential criteria for clinical application and may potentially be applicable in clinical settings.
Conclusion
We have demonstrated a microchip-SERS immunoassay for highly sensitive analysis of circulating sMCSP. The integration of a SERS nanotag system into a microchip platform equipped with an array of ac-EHD electrodes was achieved successfully. Instrumental for sensitivity enhancement was the deployment of anisotropic Au–Ag alloy nanoparticles which enables single-particle SERS spectra. The ac-EHD electrodes engendered a nanoscopic fluid flow that increased specificity by increasing collisions of sMCSP with the anti-MSCP functionalized electrodes and simultaneously minimized the binding of non-target species. Evaluated on simulated melanoma patient plasma samples, the SERS immunoassay could detect as low as 200 pg μl−1 of sMCSP and process 28 samples simultaneously. We envision that the platform could be extended towards detection of multiple clinically relevant biomarkers such as DNA/RNA, exosome, circulating free DNA (cfDNA), etc. This would enable comprehensive profiling by monitoring a panel of biomarkers simultaneously from liquid biopsies to support personalized cancer therapy.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by NHMRC Investigator Grant (APP1175047 for AAIS and APP1173669 for AW), UQ ECR Grant (UQECR1945613 for AAIS and AW) and ARC DP (DP180102836 for MT). These grants have significantly contributed to the environment to stimulate the research described here. The fabrication works were conducted at the Queensland node of the Australian National Fabrication Facility (Q-ANFF).References
- S. Hanash and A. Taguchi, Nat. Rev. Cancer, 2010, 10, 652–660 CrossRef CAS PubMed.
- W. Li, J. M.-K. Ng, C. C. Wong, E. K. W. Ng and J. Yu, Oncogene, 2018, 37, 4903–4920 CrossRef CAS PubMed.
- E. Crowley, F. Di Nicolantonio, F. Loupakis and A. Bardelli, Nat. Rev. Clin. Oncol., 2013, 10, 472 CrossRef CAS PubMed.
- G. Siravegna, S. Marsoni, S. Siena and A. J. Bardelli, Nat. Rev. Clin. Oncol., 2017, 14, 531 CrossRef CAS PubMed.
- E. Heitzer, I. S. Haque, C. E. Roberts and M. R. Speicher, Nat. Rev. Genet., 2019, 20, 71–88 CrossRef CAS PubMed.
- K. Pantel and C. Alix-Panabières, Nat. Rev. Clin. Oncol., 2019, 16, 409–424 CrossRef CAS PubMed.
- R. Harpio and R. J. Einarsson, Clin. Biochem., 2004, 37, 512–518 CrossRef CAS PubMed.
- S. K. Arya and P. Estrela, Biosens. Bioelectron., 2018, 117, 620–627 CrossRef CAS PubMed.
- Y. Wu, W. Guo, W. Peng, Q. Zhao, J. Piao, B. Zhang, X. Wu, H. Wang, X. Gong and J. Chang, ACS Appl. Mater. Interfaces, 2017, 9, 9369–9377 CrossRef CAS PubMed.
- J. Li, A. Wuethrich, S. Dey, R. E. Lane, A. A. Sina, J. Wang, Y. Wang, S. Puttick, K. M. Koo and M. Trau, Adv. Funct. Mater., 2020, 1909306 CrossRef.
- A. Wuethrich, A. R. Rajkumar, K. B. Shanmugasundaram, K. R. Khondakar, S. Dey, C. B. Howard, A. A. I. Sina and M. Trau, Analyst, 2019, 144, 6914–6921 RSC.
- P. Mao, C. Liu, G. Favraud, Q. Chen, M. Han, A. Fratalocchi and S. Zhang, Nat. Commun., 2018, 9, 1–8 CrossRef CAS PubMed.
- T. Chen, H. Wang, G. Chen, Y. Wang, Y. Feng, W. S. Teo, T. Wu and H. Chen, ACS Nano, 2010, 4, 3087–3094 CrossRef CAS PubMed.
- S. C.-H. Tsao, J. Wang, Y. Wang, A. Behren, J. Cebon and M. Trau, Nat. Commun., 2018, 9, 1–10 CrossRef CAS PubMed.
- J. Wang, A. Wuethrich, A. A. I. Sina, R. E. Lane, L. L. Lin, Y. Wang, J. Cebon, A. Behren and M. Trau, Sci. Adv., 2020, 6, eaax3223 CrossRef PubMed.
- I. Khalil, W. A. Yehye, N. M. Julkapli, S. Rahmati, A. A. I. Sina, W. J. Basirun and M. R. Johan, Biosens. Bioelectron., 2019, 131, 214–223 CrossRef CAS PubMed.
- K. K. Reza, A. A. I. Sina, A. Wuethrich, Y. S. Grewal, C. B. Howard, D. Korbie and M. Trau, Biosens. Bioelectron., 2019, 126, 178–186 CrossRef CAS PubMed.
- K. R. Khondakar, S. Dey, A. Wuethrich, A. A. I. Sina and M. Trau, Acc. Chem. Res., 2019, 52, 2113–2123 CrossRef CAS PubMed.
- S. Dey, K. M. Koo, Z. Wang, A. A. Sina, A. Wuethrich and M. J. Trau, Lab Chip, 2019, 19, 738–748 RSC.
- J. Li, G. Zhang, J. Wang, I. S. Maksymov, A. D. Greentree, J. Hu, A. Shen, Y. Wang and M. Trau, ACS Appl. Mater. Interfaces, 2018, 10, 32526–32535 CrossRef CAS PubMed.
- J. Li, J. Wang, Y. S. Grewal, C. B. Howard, L. J. Raftery, S. Mahler, Y. Wang and M. Trau, Anal. Chem., 2018, 90, 10377–10384 CrossRef CAS PubMed.
- A. Wuethrich, A. A. I. Sina, M. Ahmed, T.-Y. Lin, L. G. Carrascosa and M. Trau, Nanoscale, 2018, 10, 10884–10890 RSC.
- J. Yang, M. A. Price, C. L. Neudauer, C. Wilson, S. Ferrone, H. Xia, J. Iida, M. A. Simpson and J. B. J. McCarthy, J. Cell Biol., 2004, 165, 881–891 CrossRef CAS PubMed.
- J. Yang, M. A. Price, G. Y. Li, M. Bar-Eli, R. Salgia, R. Jagedeeswaran, J. H. Carlson, S. Ferrone, E. A. Turley and J. B. J. McCarthy, Cancer Res., 2009, 69, 7538–7547 CrossRef CAS PubMed.
- L. Brochez and J. M. Naeyaert, Br. J. Dermatol., 2000, 143, 256–268 CrossRef CAS PubMed.
- J. Song, S. L. Merbs, L. J. Sokoll, D. W. Chan and Z. Zhang, Clin. Proteomics, 2019, 16(1), 1–10 CrossRef CAS PubMed.
- C. Huillet, A. Adrait, D. Lebert, G. Picard, M. Trauchessec, M. Louwagie, A. Dupuis, L. Hittinger, B. Ghaleh, P. Le Corvoisier, M. Jaquinod, J. Garin, C. Bruley and V. Brun, Mol. Cell. Proteomics, 2012, 11(2) CrossRef CAS PubMed.
- E. P. Kartalov, J. F. Zhong, A. Scherer, S. R. Quake, C. R. Taylor and W. French Anderson, BioTechniques, 2006, 40(1), 85–90 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra05032f |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2020 |