Ligand functionalized copper nanoclusters for versatile applications in catalysis, sensing, bioimaging, and optoelectronics
Shayan
Shahsavari†
 ab,
Samaneh
Hadian-Ghazvini†
c,
Fahimeh
Hooriabad Saboor†
d,
Iman
Menbari Oskouie
be,
Masoud
Hasany
ab,
Samaneh
Hadian-Ghazvini†
c,
Fahimeh
Hooriabad Saboor†
d,
Iman
Menbari Oskouie
be,
Masoud
Hasany
 f,
Abdolreza
Simchi
f,
Abdolreza
Simchi
 *gh and
Andrey L.
Rogach
*gh and
Andrey L.
Rogach
 *ij
*ij
aDepartment of Chemistry, Sharif University of Technology, Tehran, Iran
bNanoclub Elites Association, Tehran, Iran
cInstitute of Biochemistry and Biophysics, University of Tehran, Tehran, Iran
dDepartment of Chemical Engineering, University of Mohaghegh Ardabili, Ardabil, Iran
eSchool of Medicine, Tehran University of Medical Sciences, Tehran, Iran
fDepartment of Chemical and Petroleum Engineering, Sharif University of Technology, Tehran, Iran
gDepartment of Materials Science and Engineering, Sharif University of Technology, P.O. Box 11365-9466, 14588 Tehran, Iran. E-mail: simchi@sharif.edu; Fax: +98 (21) 6600 5717; Tel: +98 (21) 6616 5226
hInstitute for Nanoscience and Nanotechnology, Sharif University of Technology, P.O. Box 11365-9466, 14588 Tehran, Iran
iDepartment of Materials Science and Engineering, and Centre for Functional Photonics, City University of Hong Kong, 83 Tat Chee Avenue, Kowloon, Hong Kong S.A.R. E-mail: andrey.rogach@cityu.edu.hk
jShenzhen Research Institute, City University of Hong Kong, Shenzhen, 518057, China
First published on 20th September 2019
Abstract
Copper nanoclusters (Cu NCs) have emerged as a valuable member of the family of ligand-protected few-atomic metal nanoparticles and show fascinating properties of color-controlled light emission, combined with the advantages of versatile solution-based chemical synthesis at low cost. Synthetic methods of Cu NCs using various types of functional ligands and scaffolds allow tuning their emission wavelength and improving their environmental stability. Depending on the method of preparation and the ligands used, Cu NCs have already been applied for a wide variety of applications in catalysis, sensing, bioimaging, theranostics, and optoelectronics. This review highlights the potential of Cu NCs and links synthetic procedures and functionalization with different ligands with their properties and applications.
1. Introduction
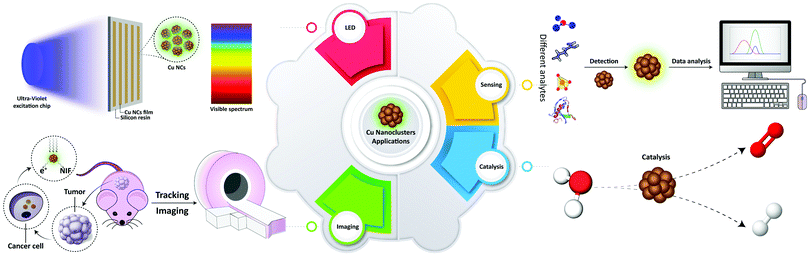
Functional nanomaterials have undergone an impressive development and are now starting to impact diverse aspects of human life. An important class of nanomaterials is metal nanoparticles (NPs) with chemical, electrical, magnetic, and optical properties often different from their bulk phases,1 which have made them applicable for a wide range of applications. A recent development in the synthesis of metal NPs is the fabrication of sub-nanometer structures which are composed of a few or few tens of atoms. These so-called metal nanoclusters (MNCs) provide a link between larger, plasmonic metal NPs and molecular-like compounds. Their sizes are close to the Fermi wavelength of electrons, so that they exhibit discrete electronic states and show fluorescence emission.1–3 In recent years, many studies have been devoted to gold and silver NCs.4 In spite of the lower cost and ready availability of copper, Cu NCs were more difficult to synthesize, while their photoluminescence quantum yields (PLQY) tended to be lower.2 At the same time, the photostability of Cu NCs is better than that of organic dyes. In comparison with many II–VI and IV–VI semiconductor quantum dots with high PLQY, toxicity issues of Cu NCs are fewer,4 while their cellular uptake could be easier, due to the smaller size.5 Due to the useful chemical, optical and electrical properties of Cu NCs, efforts on broadening their applicability in various fields such as catalysts, chemical sensors, biological imaging agents, and electronic devices have increased in recent years.2,3,6–8The aim of this article is to present a comprehensive review on the application-related aspects of Cu NCs (summarized in Table 1), which were not so much in the focus of other reviews. Recent advances in the synthetic protocols with an emphasis on their advantages and shortcomings are considered, and properties of Cu NCs related to several possible applications (Scheme 1) such as catalysis, detection and sensing, biological imaging, theranostics, and light emitting devices (LEDs) are discussed. The review concludes with the future trends and outlooks for the further development of the Cu NC field.
| Application | Synthesis method | Cu NCs | Cluster formula | Type of reaction | Reactant | Product | Year | Ref. |
|---|---|---|---|---|---|---|---|---|
| Catalysis | Ligand assisted | D-Penicillamine capped Cu NCs | Cu6L4(L = D-penicillamine) | Reduction | Methylene blue | Lucomethylene blue | 2014 | 9 |
| 1-Dodecanethiol capped Cu NCs | Cu8DT8 | Reduction | O2 | OH* | 2014 | 10 | ||
| Tetraoctylammonium formate capped Cu NCs | — | 1,3-Dipolar cycloaddition | Azides and terminal alkynes | 1,2,3-Triazoles | 2005 | 11 | ||
| Double ligand capped Cu NCs | [(Cu20(CCPh)12(OAc)6)] supported on silica | 1,3-Dipolar cyclo addition | Azides and terminal alkynes | 1,2,3-Triazoles | 2018 | 12 | ||
| Protein templated | BSA–Cu NCs | — | Oxidation | Styrene | Benzaldehyde | 2014 | 13 | |
| Photo-reduction | Cu/CuO NCs on TiO2 nanotube arrays | — | Reduction | Methylene blue | Lucomethylene blue | 2017 | 14 | |
| 4-Nitrophenol | 4-Aminophenol | |||||||
| Physical | Cu NCs deposited on MgO | — | Reduction | 4-Nitrophenol | 4-Aminophenol | 2018 | 15 | |
| Cu NCs based on the decomposition of surface | — | HER | CO and H2O | H2 and CO2 | 2016 | 16 | ||
| Hydro-thermal | Cu NCs loaded on nanocrystalline Cr2O3 | — | Oxidation | Cyclohexane | Cyclohexanone | 2012 | 17 |
| Application | Synthesis method | Cu NCs | Cluster formula | Target | Limit of detection (LOD) | Linear range | Year | Ref. |
|---|---|---|---|---|---|---|---|---|
| Sensors | Organic polymer-templated | PEI–Cu NCs | — | TNT | 14 pM, 0.05 nM | 0–8 nM | 2018 | 18 |
| PVP–Cu NCS | Cu11L10 [Cu12L10 + 3H] + [Cu13L11 + H] + L = N-vinyl-pyrrolidone | Trinitrophenol | 0.391 μM | 0–27 μM | 2019 | 19 | ||
| PVP-protected Cu NCs | — | Temperature | — | 20–75 °C | 2017 | 20 | ||
| PVP-protected Cu NCs | — | Glutathione (GSH) | 17 μM | 0–0.14 mM | 2019 | 21 | ||
| PEI–Cu NCs | — | pH | — | 2.0–13.2 | 2016 | 22 | ||
| PEI–Cu NCs | — | Cysteine | 0.34 μM | 1–25 μM | 2018 | 23 | ||
| PEI–Cu NCs | — | Protein kinase | 0.038 U mL−1 | 0.1–6.0 U mL−1 | 2017 | 24 | ||
| Protein/peptide-templated | Ovalbumin-directed Cu NCs | — | Doxycycline | 270 pM | 1–1000 μM | 2018 | 25 | |
| BSA–Cu NCs/MOF | — | Tramadol | 0.8 nM | 0.003–2.5 μM | 2019 | 26 | ||
| BSA–Cu NCs | — | RDX | 1.62 nM | 0–0.238 μM | 2019 | 27 | ||
| Silk fibroin–Cu NCs | — | S2− | 0.3 μM | 5–110 μM | 2019 | 28 | ||
| BSA–Cu NCs | — | Gossypol | 25 nM | 0.1–100 μM | 2019 | 29 | ||
| Metallothionein–Cu NCs | — | Hg2+ | 43.8 nM | 97 nM–2.3 μM | 2019 | 30 | ||
| Pb2+ | 142.0 nM | 0.7–96 μM | ||||||
| Natural silk fibroin-stabilized Cu NCs (SF@Cu NCs) | — | pH | — | 6.08–10.05 | 2016 | 31 | ||
| Human serum albumin (HSA) templated Cu NCs | — | Bilirubin | 3.5 × 10−8 M | 1.25 × 10−6 to 7.50 × 10−6 M | 2019 | 32 | ||
| 1.45 × 10−7 M | 5.00 × 10−6 to 2.875 × 10−5 M | |||||||
| Carbon dots/BSA–Cu NCs | — | Dopamine | 32 nM | 0.1 to 100 μM | 2018 | 33 | ||
| BSA–Cu NCs | — | Rutin | 0.02 μM | 0.1–100 μM | 2018 | 34 | ||
| BSA–Cu NCs | — | Protamine | 0.12 ng mL−1 | 3–12 ng mL−1 | 2019 | 35 | ||
| Heparine | 0.0406 ng mL−1 | 6–9 ng mL−1 | ||||||
| Ovalbumin stabilized Cu NCs | — | Folic acid | 0.18 μM | 0.5–200 μM | 2019 | 36 | ||
| Cu NC-encapsulated liposomes | — | Cardiac Troponin T (cTnT) antigen | 0.03 pg mL−1 | 0.1 to 2 pg mL−1 | 2018 | 37 | ||
| DNA-templated | Reticular DNA template Cu NCs | — | Hg2+ | 16 pM | 50 pM–500 μM | 2018 | 38 | |
| TCSDA and dsDNA-templated Cu NCs | — | ATP | 5 pM | 0.01–100 nM | 2017 | 39 | ||
| dsDNA–Cu NCs/GO | — | Protein kinase | 0.039 U L−1 | 0.1–5 U L−1 | 2018 | 40 | ||
| poly-(T) ssDNA templated Cu NCs | — | Uracil-DNA glycosylase (UDG) | 5.0 × 10−5 U mL−1 | 1.0 × 10−4 × 0.01 U mL−1 | 2019 | 41 | ||
| dsDNA templated Cu NCs | — | Human immunoglobulin G (IgG) | 7 pg mL−1 | 0.05–12 ng mL−1 | 2019 | 42 | ||
| dsDNA templated Cu NCs | — | T4 polynucleotide kinase phosphatase | 0.06 U mL−1 | 0.07 U mL−1 to 15 U mL−1 | 2019 | 43 | ||
| ssDNA bivalent aptamer–Cu NCs | — | Vascular endothelial growth factor (VEGF) | 12 pM | 10–800 pM | 2019 | 44 | ||
| ssDNA (elongated poly(T))-templated Cu NCs | — | Biotin | 3.1 nM | 10–1000 nM | 2017 | 45 | ||
| ssDNA (elongated poly(T))-templated Cu NCs | — | Streptavidin | 0.47 nM | 1–200 nM | 2017 | 45 | ||
| ssDNA (elongated poly(T))-templated Cu NCs | — | Uracil-DNA Glycosylase (UDG) Activity | 0.00005 U mL−1 | 0.00005–0.002 U mL−1 | 2019 | 46 | ||
| 0.000002 U mL−1 (hyperbranched DNA templates) | ||||||||
| DNA-templated Cu NCs | — | MicroRNA-155 | 1.1 × 10−11 M | 5.0 × 10−11 M to 1.0 × 10−8 M | 2017 | 47 | ||
| Hairpin DNA-templated Cu NCs | — | MicroRNA-155 | 2.2 × 10−12 M | 5.0 × 10−12 to 1.0 × 10−8 M | 2017 | 48 | ||
| DNA-templated Cu NCs | — | Abasic sites in dsDNA | 1 lesion in 74 nucleotides | — | 2018 | 49 | ||
| AT-rich dsDNA-template Cu NCs | — | microRNA | 36 aM | — | 2018 | 50 | ||
| Ligand-assisted | Cysteine–Cu/Mo NCs | — | Methotrexate | 13.7 nM | 50 nM–100 μM | 2019 | 51 | |
| CTAB–Cu NCs | — | Carbamazepine | 0.08 μg mL−1 | 0.2–20 μg mL−1 | 2019 | 52 | ||
| Dithiothreito–Cu NCs/carbon nitride nanosheets | — | Hg2+ | 0.01 nM | 0.5–10 nM | 2019 | 53 | ||
| GSH–Cu NCs | — | o-Phenylenediamine (OPD) | 93 ng L−1 | 0.15–110 μg L−1 | 2019 | 54 | ||
| L-Cys–Cu NCs | Cu4(Cys)3–5 | m-Dinitrobenzene | 0.13 μM | 99 nM–1.3 μM | 2019 | 55 | ||
| Cys–Cu NCs–DPA | — | Folic acid | 68.9 nM | 0.1–10 μM | 2019 | 56 | ||
| Nitrite | 95.4 nM | 0.1–80 μM | ||||||
| GSH–Cu NCs | — | Cu2+ | 0.17 μM | — | 2019 | 57 | ||
| Bi-ligand (thiosalicylic/cyste-amine)–Cu NCs | — | Cr6+ | 0.03 μM | 0.1–1000 μM | 2019 | 58 | ||
| GSH–Cu NCs/carbon dots | — | Humidity | — | 40–80% | 2019 | 59 | ||
| AMTD–Ac–Cu NCs | Cu3L, Cu3LX, Cu3LX2 | H2O | 0–25.92 v/v% | 0.036% (EtOH) 0.018% (THF) 0.024% (ACT) | 2018 | 60 | ||
| AMTD: (2-amino-5-mercapto-1,3,4-thiadiazole) | L = AMTD | 0.026% (MeCN) | ||||||
| X = Ac | ||||||||
| Adenosine-stabilized Cu NCs | — | Nitrofurantoin | 30 nM | 0.05–4 μM | 2018 | 34 | ||
| Cysteamine-capped Cu NCs | — | Al3+ | 26.7 nM | 1–7 μM | 2018 | 61 | ||
| Curcuma root extract (CRE)–Cu NCs | — | Hg2+ | 0.12 nM | 0.0005–25 μM | 2018 | 62 | ||
| D-Penicillamine-coated Cu/Ag alloy NCs | [Cu3Ag2L4 + H]+ | Ag+ | 5.9 μmol L−1 | 0–0.79 mmol L−1 | 2018 | 63 | ||
| [Cu2Ag2L3 + H]+ | ||||||||
| [Cu3AgL3 + H]+ | ||||||||
| [Cu2Ag2L2 + Na]+ L = D-penicillamine | Halides | — | 095–1.72 mM | |||||
| Cytidine-protected Cu NCs | — | Cr2O72− | 24 nM | 0.05–7.0 μM L−1 | 2017 | 64 | ||
| Thiosalicylic acid (TA) capped Cu NCs | — | CN− | 5 nM | 0.01–1 μM | 2017 | 65 | ||
| NO2− | 5 μM | 15–50 μM | ||||||
| Bimetallic Cu/Ag NCs | — | Temperature | — | 4–55 °C | 2017 | 66 | ||
| D-Penicillamine–Cu NCs | — | Acid phosphatase | 0.8 U L−1 | 2.2–45.5 (100) U L−1 | 2017 | 67 | ||
| 4-Methylthiophenol-capped Cu NCs | — | β-Galactosidase | 0.9 U L−1 | 2.5–212.0 U L−1 | 2017 | 68 | ||
| L-Histidine protected Cu NCs | — | Alkaline phosphatase | 45 μU mL−1 | 0.5 mU mL−1 to 40 mU mL−1 | 2019 | 69 | ||
| GSH–Cu NC | Cu2L4 | Glucose | 3.2 × 10−5 M | 0.1–2.0 mM | 2019 | 70 | ||
| Cu4L3,(L = C10H16O6N3S) | ||||||||
| Etching | L-Cysteine Cu NCs | — | TNT | 9.1 nM | 0–4.8 μM | 2017 | 71 |
| Application | Synthesis method | Cu NCs | Cluster formula | Target | λ ex/λem | PL QY | Year | Ref. |
|---|---|---|---|---|---|---|---|---|
| Imaging and theranostics | Organic polymer-templated | α-Methoxy-ω-thioacetate poly(ethylene glycol) capped Cu NCs | — | Nuclear staining of DNA in HeLa cell line | 390 nm, 500 nm, and 530 nm/583 nm | 30% in water | 2015 | 5 |
| 67% in THF | ||||||||
| Protein/peptide-templated | Egg white capped Cu NCs | — | Bacillus subtilis cells | 344 nm/600 nm | 6.7% | 2016 | 72 | |
| Ligand-assisted | GSH capped Cu NCs | — | Thermometry inside MDA-MB-231 cell line | 400 nm/610 nm | 4.5% | 2017 | 73 | |
| Ligand-assisted | Folic acid modified Cu NCs | — | HeLa cell line | 350 nm/440 nm | 11.6% | 2018 | 74 | |
| Ligand-assisted/organic polymer-templated | Cysteine-stabilized Cu NCs in chitosan matrix | C5L | MCF-7 and HEK 293 cell line | 405 nm/488 nm | — | 2018 | 75 | |
| C6L | ||||||||
| L = L-cysteine | ||||||||
| Protein/peptide-templated | 64Cu NC@BSA-LHRH. | — | Orthotopic lung tumors | 680 nm/740 nm | — | 2015 | 76 | |
| Protein/peptide-templated | Tf-Stabilized Cu NCs | — | Tf receptor positive Daltons lymphoma ascites bearing mice | 375 nm/460 nm | — | 2018 | 77 | |
| Ligand-assisted | 64Cu NCs functionalized with temozolomide | — | Glioblastoma | — | — | 2018 | 78 | |
| Organic polymer-templated | PVP-templated Cu NCs | Cu11L10 | THP-1 macrophages | 392 nm/618 nm | 44.67% | 2019 | 19 | |
| Cu12L11 | ||||||||
| Cu13L11 | ||||||||
| L = PVP |
| Application | Synthesis method | Cu NCs | Cluster formulation | Morphology | λ ex/λem | PL QY | Year | Ref. |
|---|---|---|---|---|---|---|---|---|
| Light harvesting | Ligand-assisted | Cysteine capped Cu NCs | Cu7L3 | Spherical | 365 nm/489 nm | 6.2% | 2018 | 79 |
| L = L-cysteine | ||||||||
| Solar cells | Double ligand capped Cu NCs | [Cu53(RCOO)10(CCtBu)20Cl2H18]+ | Spherical | — | — | 2019 | 80 | |
| LEDs | 1-Dodecanethiol capped Cu NCs | Cu14DT10 | Nano sheets | 365 nm/400–700 nm | 6.5% | 2015 | 81 | |
| Thiophenol based compounds stabilized Cu NCs | Cu9L7 | Nano ribbons | 365 nm/400–700 nm | 15.6% | 2017 | 82 | ||
| Cu8L8 | ||||||||
| Cu9L6 | ||||||||
| Cu8L6 | ||||||||
| Cu10L10 | ||||||||
| L = 4-X thiophenol | ||||||||
| GSH capped Cu NCs in a biopolymeric matrix | — | Nano aggregates | 365 nm/600 nm | 42% | 2018 | 83 | ||
| Ce3+ crosslinked L-cysteine capped Cu NCs | — | Mesoporous Spheres | 395 nm/617 nm | 8.3% | 2018 | 84 | ||
| Organic polymer-templated | PVP–Green Cu NCs with DHLA | — | Spherical | 398 nm/491 nm | 2.46% | 2019 | 85 | |
| PVP–Green Cu NCs with DHLA | — | Nano aggregates | 424 nm/610 nm | 4.83% | ||||
| Red Cu NCs |
2. Optical properties of metal NCs and their aggregation/assembly induced emission
The ultrafine size (usually less than 2 nm) of metal NCs induce electronic transitions between separated energy levels giving rise to light absorption at a given wavelength and emission with a longer wavelength.2,86,87 Bornacelli and coworkers88 synthesized bare Pt, Ag and Au NCs by ion-implantation in sapphire plates, and employed the Jellium model, EFermi/N1/3, to simulate their emission spectra. They concluded that the optical emission of these clusters can be explained based on the quantum confinement effects. Hovewer, the Jellium model would not work for ligand-functionalized NCs such as the thiolate-protected ones, because it does not take into account the contribution of ligands towards the electronic structure and the optical transitions of such structures. The PL origin of ligand-protected metal NCs and relevant parameters that contribute to their emission have been summarized by Xie and co-workers in a recent book,89 pointing out the effect of varying core sizes and the type of ligands.90 Ligand exchange further confirmed their effects on the PL of metal NCs.91 Besides the effects of the ligands and the metal core, there are some other variables such as the thiol-to-metal ratio and the oxidation state of the metal core that affect the PL response of metal NCs.89It has been demonstrated in a plenty of studies that the emission of the metal NCs can be significantly improved as a result of their aggregation. The aggregation-induced emission (AIE) phenomenon which was discovered for molecular dyes in 2001 by Tang and coauthors is also valid for the metal NCs, so far.89,92 AIE characteristics of the metal NCs include sufficiently high PL QYs (typically in the range of 10–50%), large Stokes shift, and long excited state lifetimes.89,93 Among the different ligand stabilized metal NCs, those capped with glutathione, 1-dodecanethiol, penicillamine, and cysteine are the most often reported ones with AIE. The AIE effect in metal NCs can be conveniently triggered by post-synthetic treatment, when changes of the solvent polarity, pH, and/or addition of some ions induce their self-assembly and aggregation.89,92 Although the exact mechanism of AIE in metal NCs still requires further studies, it has been generally stated that restriction in molecular rotational and vibrational motions of the capping ligands after aggregation blocks non-radiative pathways and opens radiative ones, and thus subsequently leads to PL enhancement. As illustrated in Fig. 1, the emission of isolated metal NCs relies on the S1 to S0 transition (fluorescence), while aggregated ones emit based on the T1 to S0 transition (phosphorescence).89,92,94 An additional advantage of the AIE of aggregated metal NCs is that their emission color often appears in the red and near-infrared spectral regions, which means they can be employed for biosensing, with little interference with the autofluorescence of biological materials.94
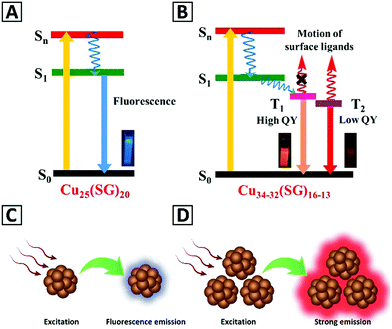 | ||
| Fig. 1 Schematics of the energy levels and excitation/emission processes in (A) isolated Cu25(SG)20 NCs and (B) aggregated Cu34–32(SG)16–13 NCs. Adapted with permission from ref. 92, Copyright 2019, ACS. (C and D) Give a schematic representation of the emission from the isolated metal NCs and the aggregated clusters, respectively. | ||
3. Synthesis of copper NCs
Fig. 2A and Table 1 summarize the main chemical and physical preparation methods that have been utilized for the synthesis of Cu NCs so far. The synthetic procedures can be categorized into 6 groups, including ligand assisted, template-assisted, electrochemical, etching, and physical methods, and assembly and aggregation.64 Chemical methods rely on the reduction of Cu2+ ions into Cu atoms in a solution phase.95,96 Ligands and templates are often employed to stabilize the clusters and to protect them from aggregation and oxidation;2,5,97,98 therefore, assembly and aggregation are considered as a subset of the ligand assisted method, which results in two- and three-dimensional cluster morphologies. Representative electron microscopy images of Cu NCs with different morphologies are presented in Fig. 2B.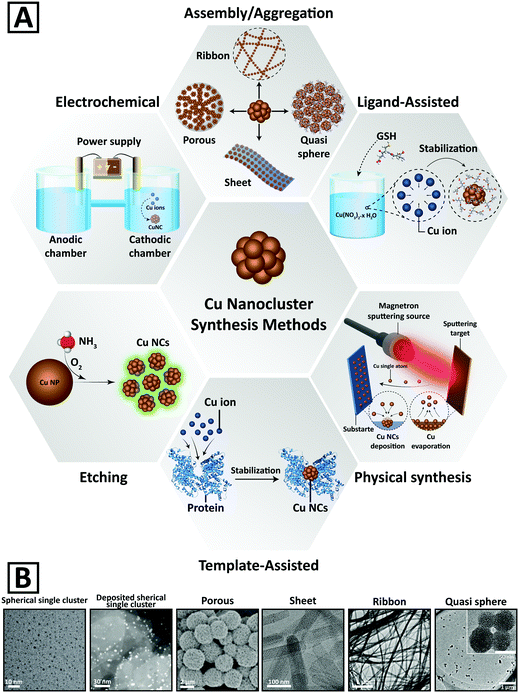 | ||
| Fig. 2 (A) Schematic representation of different chemical and physical methods developed for the synthesis of Cu NCs. (B) Representative electron microscopy images of Cu NCs with different morphologies. Adapted with permission from ref. 15, 82, 84 and 100–102, Copyrights 2016–2018, Wiley-VCH, RSC, and ACS. | ||
Several characterization protocols of Cu NCs are in place to confirm different aspects of their structure. NMR spectroscopy and X-ray absorption spectroscopy are often used, as are chromatographic methods to assess their purity.99 X-ray photoelectron spectroscopy (XPS) is applied to study the oxidation states of the metallic core of Cu NCs, while it has to be noted that the unambiguous recognition between Cu(0) and Cu(I) states is challenging.79 Absorption and steady-state/time-resolved PL spectroscopy are routine while essential measurements to provide basic optical characterization of NCs, and for the determination of their PLQY.92
3.1. Ligand assisted synthesis of Cu NCs
During the synthesis and formation of Cu NCs, there is a high tendency of aggregation in order to decrease the surface energy, and ligands can prevent this by steric effects which rely on non-bonding interactions. Ligands can also influence the reactivity of precursor ions and molecules, which determines the final size and shape of NCs.103 Controlling the kinetics of Cu2+ reduction in the presence of ligands is crucial to attain monodisperse Cu NCs. Different parameters such as ligand concentration, reducing agent concentration, the pH of solution, the temperature, and the reaction time can affect the formation of desirable products. Choosing different ligands not only affects the PL intensity and the catalytic activity of ligand functionalized Cu NCs, but also plays a role in sensitivity to various environments.104 Ligands with thiol and carboxyl groups are the two most important organic molecules which have widely been utilized for the synthesis of Cu NCs,64 as will be presented below.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 which resulted in Cu NCs with a red emission at 600 nm. Mukherjee and co-workers107 mixed Cu2+ and GSH with a molar ratio of 1
1 which resulted in Cu NCs with a red emission at 600 nm. Mukherjee and co-workers107 mixed Cu2+ and GSH with a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and prepared Cu NCs (Cu15(GSH)4) having a blue emission under UV irradiation. Through a similar approach, Huang et al.108 synthesized Cu NCs with an intense blue fluorescence under 365 nm UV irradiation. They added ascorbic acid (AA) to a solution of Cu2+ and GSH and stirred the mixture for 4 h at 65 °C/pH = 6. Cu NCs with a red emission at 620 nm under UV excitation (365 nm) were also prepared by mixing GSH and Cu2+ at a molar ratio of 5
1 and prepared Cu NCs (Cu15(GSH)4) having a blue emission under UV irradiation. Through a similar approach, Huang et al.108 synthesized Cu NCs with an intense blue fluorescence under 365 nm UV irradiation. They added ascorbic acid (AA) to a solution of Cu2+ and GSH and stirred the mixture for 4 h at 65 °C/pH = 6. Cu NCs with a red emission at 620 nm under UV excitation (365 nm) were also prepared by mixing GSH and Cu2+ at a molar ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The maximum PLQY (24%) was attained at pH = 5.101,109
1. The maximum PLQY (24%) was attained at pH = 5.101,109
More recent advances are the synthesis of GSH-capped Cu NCs with different sizes by heating the solution (up to 80 °C) under N2 flow, followed by separation of clusters through gel electrophoresis, capillary electrophoresis, and liquid chromatography. The resulting Cu NCs had a composition of Cu5 (GSH)6, Cu6(GSH)6, Cu7(GSH)6, Cu8(GSH)6, and Cu9(GSH)6. We note that some of these Cu NCs have larger sizes than Au25 (GSH)18 prepared by gel electrophoresis.110 Han et al.111 developed a one-pot synthesis protocol to prepare GSH-protected Cu NCs encapsulated in metal–organic frameworks (MOFs). To obtain Cu NCs@GSH/MOF-5, 1,4-benzene dicarboxylic acid and Zn (NO3)2.6H2O were dissolved in DMF, and under stirring Cu2+ salt and GSH solutions were introduced. Through adjusting pH to 5, a white turbid solution was attained after 24 h at room temperature (RT). The fluorescence intensity of Cu NCs in the composite nanostructure was enhanced 35-fold, while the stability improved from 3 days to 3 months.
Another approach utilizes thiolates as ligands for an organic-phase synthesis of GSH stabilized Cu NCs with a red PL emission under UV irradiation. Huang et al.112 prepared Cu NCs in DMF by a simple mixing method and used them for sensitive detection of water in organic solvents. GSH capped Cu NCs were synthesized by injection of a saturated GSH aqueous solution into a solution of Cu2+ in EtOH, and the aggregation was triggered by supersaturation of GSH in EtOH as a poor solvent. The PL QY was 48% in the solid state, which exceeded the value reported for Zn-coordinated GSH-capped Au NCs.93,113
Zhao and co-workers67 used D-penicillamine as a suitable ligand for the synthesis of Cu NCs. They obtained Cu NCs with a bright red luminescence at 37 °C and at an incubation time of 4 min. Other groups also used penicillamine as a protecting ligand for the synthesis of Cu NCs.9,114–116 Chen and co-workers96 used 2-mercapto-5-n-polypyrimidine as a ligand and NaBH4 as a reducing agent. They obtained clusters with the composition of [Cu8(C7H9N2S)4] which had PL with dual emissions at 423 nm and 593 nm and PLQY of 3.5% and 0.9%, respectively. Chang's group65 has shown that thiosalicylic acid can also be used as a ligand; by mixing it with THF in DMF and then adding Cu2+, Cu NCs were obtained. Mercaptosuccinic acid (MSA) was also utilized as a ligand in the presence of NaBH4 as a reducing agent.117 Another molecule that has often been used as a ligand is L-cysteine. Borghei et al.118–120 have shown that Cu NCs can rapidly be prepared in the presence of cysteine at pH = 12 at RT, which exhibited a PL emission in the range of 410 to 580 nm depending on the cysteine concentration. Su and Liu121 employed L-cysteine with an equal molar ratio with Cu2+ to prepare Cu NCs with a pale red emission color under UV irradiation (365 nm). Blue emitting Cu NCs (at 428 nm) were prepared with 4,6-diamino-2-mercaptopyrimidine (DAMP) as a ligand by Khonkayan et al.122 Stable red emissive Cu NCs were fabricated from CuCl2 using L-cysteine as a capping ligand and a mild reducing agent at RT.121 These pH responsive Cu NCs were used for producing hybrid nanostructures with bovine serum albumin (BSA), which were soluble in water at pH = 3.0 and had PLQY of 6.3% and 2.1% in the solid state and aqueous solution, respectively.121
The cation cross-linking procedure has been used to fabricate aggregated L-cysteine capped Cu NCs with a mesoporous morphology and improved PLQY.84 For this purpose, 100 μL of 0.1 M CuSO4·5H2O was added to 2 mL of 0.1 M L-cysteine in water; the mixture was subjected to vortexing for 10 min, and a suitable amount of 0.1 M Na2CO3 was added to induce aggregation. Thereafter, different amounts of Ce3+ were titrated to produce cross-linked aggregates; PLQY before and after Ce3+ addition was 3.4% and 8.3%, respectively.84 Yet another strategy for improving the PLQY of Cu NCs is direct metal doping. For example, central doping of Au as a single atom into dichalcogenolate-protected Cu NCs led to strong enhancement of their PLQY, which reached 59% when measured at 77 K in 2-MeTHF for [Au@Cu12(S2CNnBu2)6(CCPh)4]+ clusters.123
Self-assembly of Cu NCs into wires, ribbons, and sheets has been realized by employing 1-dodecanethiol (DT) as a ligand. For the synthesis of ribbons, for example, 30 mg of copper(II) acetylacetonate was dissolved in a mixture of dibenzyl ether (2 mL) and liquid paraffin (5 mL). Then, 1 mL DT was introduced, and the mixture was stirred at 120 °C for 30 min.10
Several other thiolate ligands were also employed for the synthesis of Cu NCs, namely, cysteamine,124,125 phenylethanethiol,114 dihydrolipoic acid,126 mercaptobenzoic acid,127 3-mercapto-trimethoxy silane,128 and 2-mercapto-1-methylimidazole.129
3.2. Template-assisted synthesis of Cu NCs
To prepare Cu NCs and at the same time avoid their aggregation, templates such as DNA, protein, peptides, and polymers have been widely utilized. Cu2+ ions are able to bind to those templates and are then reduced to form clusters, whose aggregation is prevented by steric hindrance. Different templates can be chosen based on the types of potential application. Parameters such as pH, temperature, ratio of Cu2+ and templates can control the ratio of functional ligand groups and the metal core. For example, in Cu NC synthesis utilizing the protein template, the increase of pH facilitates breaking of disulfide bonds and stabilizes Cu NCs within the protein scaffold by thiol groups. A recent study has shown that by modifying the synthetic conditions in the template-assisted method, high PLQY exceeding 44% in water can be achieved.19 A brief overview of the resulting Cu NCs is given below.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Examination of different ssDNA templates like random ssDNA, poly A, poly T, poly C and poly G at pH 7.6 by 3-morpholinopropane-1-sulfonic acid has indicated that only poly T template yielded luminescent Cu NCs with a PL peak at ∼600 nm when excited at 340 nm.97,136 The reason for this is that Cu+ is an intermediate product of Cu2+ reduction to Cu, and T has the lowest affinity for binding to Cu+ (while G has the highest affinity for binding to Cu+).
1. Examination of different ssDNA templates like random ssDNA, poly A, poly T, poly C and poly G at pH 7.6 by 3-morpholinopropane-1-sulfonic acid has indicated that only poly T template yielded luminescent Cu NCs with a PL peak at ∼600 nm when excited at 340 nm.97,136 The reason for this is that Cu+ is an intermediate product of Cu2+ reduction to Cu, and T has the lowest affinity for binding to Cu+ (while G has the highest affinity for binding to Cu+).
3.3. Electrochemical synthesis
Electrochemical synthesis is a method that has widely been used to prepare various metal NPs and NCs. During the process, electrolysis of the copper anode releases Cu2+ ions, which are then reduced and capped at the cathode in the form of Cu NCs; by controlling the current density, the final size of NCs can easily be adjusted.160 The advantages of this procedure are that Cu NCs can be synthesized at low current densities and without any additional ligands or surfactants. Under optimum conditions, PLQY of such “naked” Cu NCs could be as high as 13%, which favorably compares with many of Cu NCs prepared by wet chemical methods.161 Huseyinova et al.162 synthesized stable surfactant-free Cu5 clusters with a blue PL emission (305 nm) under excitation at 224 nm. Vilar-Vidal et al.161 produced highly stable ligand-free Cu13 NCs with a blue PL emission (410 nm) by using tetrabutylammonium nitrate as an electrolyte and showed that the number of atoms in the copper core (up to Cu20) can be regulated by heating these clusters at 80 °C, followed by re-dissolution in acetonitrile. Green fluorescent Cu NCs with smaller dimensions (down to Cu5 core atoms) have been produced by centrifugation in ethanol through a purification process.1633.4. Etching methods
Chemical etching can be considered as a top-down method for the synthesis of MNCs from larger, non-luminescent NPs, often in the presence of suitable excess ligands or excess metal salts.64 NCs can be synthesized from poly-disperse NPs through a digestive ripening or core etching mechanism. Using an etching approach for formation of Au and Ag NCs has been rather popular, while there are only a few examples in the case of Cu NC synthesis.92 Wang and co-workers22 added Cu2+ to oleyl amine (OA) and 1-octadecene (ODE) solution to prepare OA–Cu NPs. Etching of Cu NPs was performed by adding an aqueous solution of polyethyleneimine (PEI) to OA–Cu NPs in chloroform at 50 °C. In another experiment,164 Cu2+ and AA (serving as both a reducing agent and a capping agent) were used to prepare Cu NPs, which were added to GSH aqueous solution to provide the etching process. The resulting Cu NCs exhibited a strong emission peak at 600 nm. Instead of using GSH for etching, other thiol ligands like cysteine164,165 and penicillamine164 can also be utilized. Xie et al.166 reduced Cu2+ by NaBH4 to attain Cu NPs protected by GSH, which were transferred to an organic phase through the addition of cetyltrimethylammonium bromide (CTAB), which were incubated for 24 h to obtain Cu NCs with a blue PL emission at 438 nm. Patra's group92 synthesized red and blue emitting Cu NCs through the etching of Cu NPs by addition of a highly concentrated GSH. The as-synthesized red emitting Cu34–32(SG)16–13 NCs showed a PL peak at 625 nm with a low PLQY of 0.03%, which could be increased 36-fold via addition of EtOH with fv = 90%. The as-prepared blue-emitting Cu25(SG)20 NCs showed a PLQY of 9.7%, which could be further enhanced two-fold by addition of EtOH with fv = 75%. Deng et al.100 developed a method to etch Cu NPs with ammonia (NH3); green fluorescent Cu NCs with a PLQY of 6.6% were attained after ∼15 min.3.5. Other methods
There are few other techniques used for fabrication of Cu NCs which are somewhat hard to be classified into the above-mentioned groups. Lopez-Quintela and coworkers167 employed the water-in-oil micro-emulsion technique to prepare blue emitting Cu NCs under UV irradiation, using Cu2+ as an aqueous solution, cyclohexane as an oily phase, NaBH4 as a reducing agent and sodium dodecyl sulfate with isopentanol as a surfactant. Recently, Koninti et al.168 synthesized Cu NCs inside the water pool of reverse micelles, which were made of sodium bis(2-ethyl-1-hexyl)-sulfosuccinate or Triton X-100 or CTAB. Quite remarkably, the stability of L-cysteine protected Cu NCs increased from 40 days in bulk water to 180 days in reverse micelles due to the protection of the as-synthesized clusters from aerial oxidation. Kawazaki et al.169 employed a microwave-assisted polyol synthesis to produce oxidation resistant Cu NCs through in situ ethoxylation reaction of ethylene glycol, without the need for any additional ligands or surfactants. Toh et al.170 embedded Cu NCs in ZnO thin films prepared by the radio frequency magnetron sputtering method to study their magnetic properties. The film deposition was carried out at 170 K171,172 under the gas flow of Ar and He (100–200 cm3 min−1). Then, Cu NCs were formed on the ZnO film by using nanocluster beam deposition. The clusters had an average size of 8 to 10 nm while secondary phase nanoclusters (CuO and Cu2O) were also formed. After depositing another ZnO layer on top of Cu NCs, improved ferromagnetism at RT was attained through the overlapping of the p-orbital from O contributed by ZnO and the d-orbital contributed by Cu. Stable and ligand-free Cu NCs were synthesized via an anti-replacement method.173 A mixture of 3 mL of CuCl2 solution (20 mg mL−1) and 2 mL of Au NP solution (0.01 mg mL−1) was irradiated by a pulse laser (532 nm) for 5 min; Cu NCs with 2.0 ± 0.4 nm diameter were formed as a result and were separated by centrifugation, and they emitted at 440 nm under 350 nm excitation wavelength. The formation mechanism of Cu NCs under laser irradiation was ascribed to the generation of “hot electrons” in Au NPs, which consequently reduced adjacent Cu2+ ions.4. Applications of Cu NCs
4.1. Catalysis
Metal NCs have become quite important from the point of view of their catalytic activity, due to their ultrasmall size and high surface energy,174,175 which capitalized on the previously conducted catalytic related studies of larger MNCs.140 Although NCs based on noble metals display good catalytic performance, it is highly desirable to offer alternative, less expensive while catalytically active Cu NC analogues. Computational aided methods can suggest suitable strategies for developing active catalysts based on Cu NCs.104 In that respect, theoretical studies on the surface chemistry of Cu NCs aimed at the exploration of adsorption steps of different chemical compounds are helpful in the understanding of various types of possible catalytic reactions. For instance, theoretical comparisons between adsorption of CO and H2 molecules on Cu NCs by molecular dynamics simulations have shown that CO is adsorbed more easily than H2, and this has a greater impact on the cluster structure.176 As depicted in Scheme 2, catalytic reactions which have been performed so far on Cu NCs can be classified into four groups, including reduction, oxidation, hydrogen evolution reaction (HER), and 1,3-dipolar cycloaddition. Examples of Cu NCs used in these kinds of catalytic reactions are outlined below and listed in Table 1.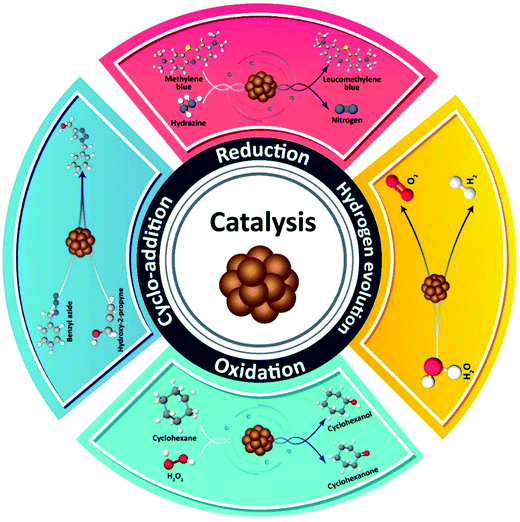 | ||
| Scheme 2 Schematic representation of four different types of chemical reactions which can be catalyzed by Cu NCs. | ||
Titanium dioxide is a very well-known photocatalyst that has attracted researchers’ attention in the last decades. To improve visible light sensitivity of TiO2, Liu et al.177 prepared Cu(II) NC-grafted Nb-doped TiO2. Nb ions reduced the energy level below the conduction band of TiO2 to be matched with the ECu2+/Cu+ (redox potential) of the Cu(II) NC complex on the surface of the catalyst. The resulting Cu(II)–NbxTi1−xO2 nanocomposite could decompose 2-propanol as an organic compound into CO2 under visible light. A theoretical and experimental study demonstrated that deposition of Cu5 NCs on the surface of TiO2 NPs could extend visible light absorption and lead to the formation of an efficient photocatalyst.178
Cai et al.15 deposited bimetallic Cu/Au NC catalysts onto a MgO powder support for the reduction of 4-NP to 4-AP by NaBH4. Cu-rich NCs, Au-rich NCs or Cu/Au-equal NCs formed an alloy structure randomly. The reaction rate constant of 3.49 × 104 min−1 mole of metal−1 was attained from the Cu/Au-equal NC catalyst, which was 6.6 (8.9) fold higher than that for the Cu-rich (Au-rich) NCs. Theoretical modeling determined the balancing of the adsorption of 4-NP and desorption of 4-AP, which enhanced the activity of Cu/Au-NC catalysts. It was also found that the activity of Cu/Au-equal NCs prepared by beam deposition was 25 fold higher than that of the catalyst synthesized by wet chemistry.
The first report on the catalytic activity of copper-based structures for reduction of CO2 appeared in 1981.179 Tang and co-workers180 utilized Cu32H20L12 NCs (L is dithiophosphate ligand) to reduce CO2 to CO and HCOOH. After adsorption of CO2 on the surface of the catalyst, the main step was where the hydrogen was added: combination of hydrogen with C would facilitate the reaction to form HCOOH, while addition of hydrogen to O would produce CO. The turnover number of Cu13H20L12 NCs has been determined as ∼1740 mole HCOOH per mole of Cu32H20L12.
Although use of the thiolate ligands may become detrimental for achieving high catalytic activity, there have been reports on thiolated Cu NCs showing decent catalytic performance.181 Based on the density functional theory (DFT) simulations, thiolated [Cu25H10(SPhCl2)18]3− NCs were introduced as a catalyst for reduction of ketones to alcohols in the presence of hydrogen at RT.182 Other studies suggested that the migration of ligands and their partial removal activate catalytic performance of thiolated metal NCs.183,184
Even though Au NCs are most popular for electrochemical reduction,185 several studies96,186 have shown that Ag and Cu NCs are also electro-catalytically active. It was reported that Cun (n < 9) NCs have high catalytic activity for the oxygen reduction reaction (ORR).187 Results of electrochemical cyclic voltammetry (CV) also indicated that Cu NCs with smaller core sizes exhibited higher electro-catalytic activity for oxygen reduction.96,188 Ligand-free Cu NCs were introduced as electrocatalysts for the ORR.173 Reduction currents were observed when Cu NCs were immersed in 0.1 M KOH solution saturated with either O2 or N2, with respective current density 22 and 1500 times higher than that of Au NCs and protected Cu NCs of similar size (Fig. 3A). The mass activity of these Cu NCs was 1.6 and 4.2 fold higher than that of the target set by the U.S. Department of Energy and the commercial Pt catalyst. Such a high electrocatalytic activity was ascribed to the clean surface in the absence of ligands, which greatly facilitated the electron transfer. Interestingly, ligand-free Cu NCs were also quite durable: The current–time response of Cu NCs retained 88.1% of its initial value after 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 s (Fig. 3A), which was better than that for the ligand-functionalized Cu NCs used for comparison.
000 s (Fig. 3A), which was better than that for the ligand-functionalized Cu NCs used for comparison.
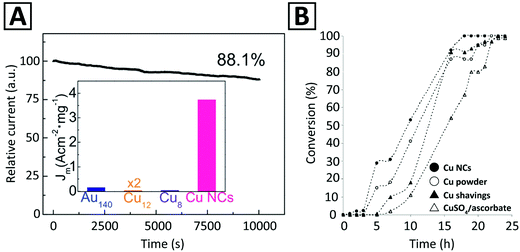 | ||
| Fig. 3 (A) Time-dependent reduction current (in the O2-saturated solution, at 0.9 V) of ligand-free Cu NCs used as an electrocatalyst for the ORR. The inset compares the current density (Jm) per mass unit of the ligand-free Cu NCs with that of several ligand-functionalized MNCs, namely, Au140(S(CH2)5CH3)53, Cu12DT8Ac4, and Cu8(C7H9N2S)4. Adapted with permission from ref. 173, Copyright 2018, RSC. (B) Observation of time-dependent conversion profiles for the 1,3-cycloaddition of prop-2-yn-1-ol to benzyl azide employing different Cu catalysts. Adapted with permission from ref. 11, Copyright 2005, Wiley-VCH. | ||
By employing DFT, the effect of chalcogen (oxygen family elements) doping on the catalytic activity of Cu4 NCs was systemically investigated.189 Doped Cu NCs could electrochemically reduce CO2 to CH4 and CH3OH. It was also shown that the activation energy barrier for CO2 hydrogenation to CH4 was reduced by Cu4S and Cu4O NCs by 0.15 eV and 0.37 eV, respectively, and over-potentials for the reaction changed in the order of Cu4S < Cu4O < Cu4Se.
Sarkar and co-workers17 synthesized Cu NCs supported on Cr2O3 for the oxidation of cyclohexane. Cr2O3–4.4wt% Cu showed the highest selectivity and conversion in comparison to CuO, Cu2O, and other proportions of Cu–Cr2O3 catalysts. This catalyst exhibited 86% conversion to cyclohexanone and cyclohexanol, with 85% selectivity of formation of cyclohexanone, and its TOF was about 52.5 and did not change after 4 successive runs. In another study, a 0.9%Cu/SiO2–MnO2 catalyst for oxydehydration of glycerol was used,192 with the main products being acrylic acid and acrolein. The conversion efficiency of glycerol was 77.1%, while the attained acrylic acid selectivity was 74.7%, and the TOF for this catalyst was 32.9 h−1. Commercial MnO2 showed only 5.5% conversion of glycerol under the same conditions. Athawale and Bhagwat193 used a Cu NCs/polyaniline composite as a catalyst for a Wacker oxidation reaction, where 1-decene was converted into 2-decanone in the presence of molecular oxygen. Hu et al.194 showed that 3,3′,5,5′-tetramethylbenzidine could be oxidized by H2O2 in the presence of Cu NCs; the reaction would not proceed without a catalyst. In a theoretical study, Tang et al.195 investigated the catalytic action of Cu55 NCs on the oxidation of CO to CO2. It was found that the oxidation process is catalyzed by Cu NCs through two mechanisms: Eley–Rideal (adsorption of oxygen molecules on the cluster surface and then interaction with CO) and Langmuir–Hinshelwood (co-adsorption of O2 and CO).
The densest crystallographic plane of copper with the lowest surface energy is the (111) plane.197 This surface plane can become unstable under CO exposure (0.1 to 100 Torr) at RT to form Cu NCs, with the edge of Cu atoms decorated by CO molecules.16 DFT simulations have shown that the energy of CO binding to low-coordinated Cu atoms and the weakening of binding of Cu to neighboring atoms drive this process. Cu NC formation could activate the surface for the water dissociation reaction (e.g. CO + H2O ↔ CO2 + H2) leading to the production of H2. It is noteworthy that no clustering was detected on the surface of Pt(111) under CO exposure.16 Even though the high surface area of Cu NCs provides high catalytic selectivity, its kinetic rate for H2 dissociation is very low.198 Very recently, Hoyt et al.104 employed DFT to explore the mechanism of how Cu NCs improve the catalytic activity for the HER. They showed that an icosahedral Cu13 NC has a large magnetic moment, which influences the catalytic behavior. The most capable transition state for H2 dissociation has lower energy of activation than that of single-crystal Cu surfaces, but needs a magnetization change from 5 to 3 μB. Fragile spin–orbit coupling delays this change, reducing the kinetic rate of H2 dissociation by a factor of 16. Strategies to aid magnetization change through environmental magnetic stimulus can improve the catalytic efficiency of Cu NCs.
Other studies suggested that organometallic Cu NCs are more active than thiolated ones.12 The catalytic activity of [Cu20(CCPh)12(OAc)6)], both in a bare form and immobilized on silica, in the Huisgen cycloaddition was examined. Unsupported Cu NCs used as a homogeneous catalyst were unstable and dissociated into smaller NCs and/or discrete Cu(I) cations; on the other hand, silica-supported Cu NCs displayed comparable yields even after recycling.
4.2. Sensing
Cu NCs have been widely applied in sensing using several strategies, including electrochemical signaling, enzyme mimetic activity, and optical responses. Among these sensing schemes, the detection based on absorption and emission properties has been employed more frequently, and included in situ synthesis of fluorescent Cu NCs (through the turn on/turn-off mechanism), fluorescent quenching through the inner filter effect (IFE), FRET, and electron transfer, and enhanced fluorescent intensity (mostly due to AIE mechanisms). Accordingly, a lot of studies have recently been conducted on potential applications of Cu NCs as a fluorescent sensor for detection of various compounds such as anions, namely, halides (Cl−, Br− and I−),63,201 S2−,28,153,165,202,203 NO2−,65,131 CN−,65 Cr2O72−,64 and phosphate;204 cations, such as Ag+,63,205 Hg2+,38,62,147,206,207 Al3+,61,158,208 Zn2+,208,209 Fe3+,4,30,108,138,210–213 Au3+,214 Mn2+,133 Pb+,215 chromium(IV),216 Cr6+,217 Cu2+;218–220 various organic compounds; water in organic solvents;60,221 and H2O29,117,126,137 and H2S156,222,223 in aqueous systems. Regarding organic compounds, Cu NC based sensors have been applied for detection of various medications including tramadol,26 methotrexate,51 carbamazepine,52 nitrofurantoin,42o-phenylenediamine,224 doxycycline,25 and tetracycline;225 biocidal and herbicidal compounds such as thiram and paraquat;72 explosive compounds including trinitrotoluene (TNT),71,226,227m-dinitrobenzene,55 nitrofurantoin,34 RDX,27 and trinitrophenol (TNP);19,228 food additives such as melamine,229 folic acid,56 quinoline yellow,224 gossypol;29 humidity and ethanol.59 Representative examples are presented in Table 1 and are briefly outlined below.Orange-fluorescent Cu NCs (560 nm) with a PLQY of 5.8% were synthesized using ovalbumin and applied for detecting doxycycline with a linear range from 1 to 1000 μM, and LOD of 270 nM.25 This sensing probe was also applicable for detecting doxycycline in urine specimens as a real sample with recoveries over 90%. In the presence of doxycycline, Cu NCs@OVA exhibited a high PL enhancement due to the interaction between doxycycline and ovalbumin that results in the formation of a more compact structure of the nanoclusters. Under UV excitation, a change in the solution color containing a different concentration of doxycycline (from orange to yellow) occurred. Additionally, Cu NCs@OVA showed high selectivity for doxycycline detection compared with other antibiotics such as penicillin, ampicillin, streptomycin, lincomycin, and norfloxacin. It was emphasized that the aqueous solubility of these clusters provided by multiple hydroxyl groups on the surface as well as their low toxicity and biocompatibility features could make them suitable for in vivo applications.25,72 They also exhibited good stability in hyper-saline environments (at 40 °C and under photobleaching conditions) and in the presence of several organic solvents and metal ions.25
A label-free, “turn off” analytical strategy using water-soluble adenosine-stabilized Cu NCs has been developed by Wang et al.34 This fluorescent sensing probe showed fast, sensitive and selective response to nitrofurantoin (NFT) in a wide linear range of 0.05–4.0 μM with a detection limit of 30 Nm, and has been used for the analysis of lake water samples with a recovery of 96–105% and relative standard deviation lower than 2%. The emission intensity of the adenosine-stabilized Cu NCs reduced with a gradual shift towards longer wavelengths at higher NFT concentrations. A small variation in the PL spectra of Cu NCs in the presence of many other substances such as phenylalanine, proline, isoleucine, valine, alanine, glutamic acid, serine, glycine, lysine, leucine, glutamine, methionine, tyrosine, aspartic acid, asparagine, threonine, arginine, cysteine, glucose, and uric acid (at 1.0 μM) proved the high selectivity of this probe to NFT. The quenching mechanism has been proposed to be based on the IFE between adenosine-stabilized Cu NCs and NFT, because of the overlap between the absorption band of NFT at 250–430 nm and the excitation and emission of Cu NCs. A ratiometric sensor based on GSH-stabilized Cu NCs for the detection of o-phenylenediamine (OPD), as an intermediate in medicine production, has been developed, and exhibited a linear detection range of 0.15 to 110 μg L−1 and LOD of 93 ng L−1. In the samples of river water and textile dyeing wastewater, its recovery rate was 96.8% and 100.3%, respectively.224
Quantitative detection of explosive compounds such as nitroaromatics (e.g. trinitrophenol, dinitrobenzene, TNT) and heterocyclic nitramines (e.g. trinitrotriazine, RDX) using Cu NCs as a turn-on/turn-off luminescent probe has been reported. Cysteine-stabilized Cu NCs were employed for selective sensing of dinitrobenzene and picric acid based on a turn-on fluorescence mechanism.55 The sensor exhibited a linear range of 99 nM to 1.3 and LOD of 0.13 μM. Quantitative detection of RDX with a LOD of 1.62 nM in a linear concentration range of 0 μM to 0.238 μM was performed using BSA-stabilized Cu NCs;27 it was based on the turn off–on fluorescence mechanism using Zn2+ ions as a modulator. Upon adding RDX, due to the Lewis acid–base interactions between Zn2+ and trinitrotriazine, fast recovery of Zn2+ induced fluorescence quenching of Cu NCs was realized. A TNT probe based on L-cysteine modified Cu NCs with a LOD of 9.1 nM,71 and a luminescence sensor using a Cu NCs-ZIF-8 nanocomposite with a LOD of 8.5 μM227 have been demonstrated. Recently, a fluorescent and colorimetric sensor for selective and sensitive detection of trace amounts of TNT both in solution (LOD = 14 pM) and in the gas phase (LOD = 0.05 nM) based on water-soluble PEI-capped Cu NCs has been developed.18 Their high sensitivity arises from selective binding between PEI and an electron deficient molecule such as TNT; it can result in a charge transfer complexing interaction between the aromatic ring of TNT and amino groups of PEI-capped Cu NCs through a photo-induced electron transfer (PET) reaction. In the presence of TNT, the emission of NCs was quenched, which could be related to the formation of a Meisenheimer complex through acid base pairing, hydrogen bonding, and electrostatic interactions. The selective sensing performance of the PEI-capped Cu NCs compared with some organic molecules such as toluene, nitrobenzene (NB), 4-nitrotoluene (4-NT), trinitrophenol (TNP), dinitrophenol (DNP), RDX and 4-nitrophenol (4-NP) could arise from the absence of the Meisenheimer complex. Preparing paper strips of PEI-capped Cu NCs with storage stability over one month has also been reported, which could be useful for rapid onsite and visual detection of TNT.18 These paper strips showed a detection limit of 10 nM within 1 min for sensing of TNT in vapor form. Green fluorescent PVP-stabilized Cu NCs with a large Stokes shift and a high PLQY (>44%) were used for detection of trinitrophenol (TNP) with a LOD of 0.391 μM.19
A chemiluminescent sensor based on L-Cys stabilized Cu NCs-DPA was developed for the detection of folic acid with a linear concentration range of 0.1–10 μM and LOD of 69.8 nM. A similar sensor based on Cu NCs–DPA–FA was used for detection of nitrites, and exhibited a linear range of detection within 0.1–80.0 μM and LOD of 0.0954 μM.56 Blue emitting L-Cys stabilized Cu NCs were used for detection of quinoline yellow as a food colorant, with a linear concentration range from 0.2 to 5.5 μM (LOD of 0.11 μM) and high selectivity compared with other yellow colorant additives such as sunset yellow and tartrazine.224 Formation of a stable complex between gossypol and BSA in a BSA–Cu NCs has been used in a fluorescence-based probe for detection of gossypol, with a linear range of 0.1–100 μM and LOD of 25 nM. This sensor was used in real samples (cottonseed meal and oil), and exhibited a selective fluorescent quenching behavior in the presence of interfering compounds such as Na+, K+, Ca2+, Mg2+, Zn2+, glucose, glycine, and palmitic acid.29
Detection of H2O2 in aqueous solutions in a concentration range from 1 μM to 1 M by a direct and fast colorimetric sensing probe and without using any chromogenic reagent or expensive instrument has been reported by Du et al.117 The sensing probe has been prepared using water-soluble mercaptosuccinic acid stabilized Cu NCs (MSA-capped Cu NCs). The color of a diluted Cu NC solution exhibited a change from claret-red at low concentrations of H2O2 (about 0.001 mM) to saffron yellow at high concentrations (about 1000 mM) (Fig. 4A-I). With increasing concentration of H2O2 from 0 to 1 mM, the absorption peak at 520 nm was gradually quenched due to the aggregation and growth of Cu NPs via the cleavage of Cu–S bonds and detachment of MSA from the Cu NP surface (Fig. 4A-II). By a further increase in the H2O2 concentration from 1 mM to 1000 mM, the absorption peak at 375 nm was progressively increased because of Cu2O formation (Fig. 4A-III). Therefore, the MSA-capped Cu NCs could be applied as a colorimetric H2O2 sensor in a wide range of 0.001 mM to 1000 mM. These Cu NCs also revealed a desirable selectivity and salt tolerance capability for H2O2 sensing in real water samples with recoveries in the range of 96.7% to 104.1% and a relative standard deviation lower than 4%. They also demonstrated a fast kinetics reaction of about 60 s, a slight influence of temperature (20–40 °C) on the absorption response, and storage stability of the purified and freeze-dried Cu NCs for 10 months (under dark conditions).
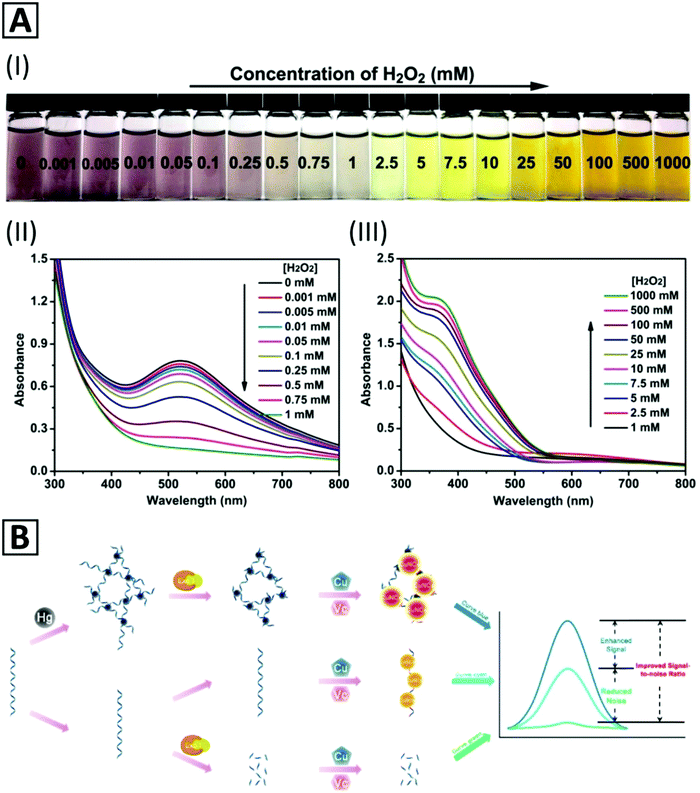 | ||
| Fig. 4 (A) Color change of PBS solution (pH = 7.4, 10 mM, 37 °C) containing MSA-capped Cu NCs with the addition of H2O2 of different concentrations (I); UV-Vis absorption spectra of the MSA-capped Cu NCs in the presence of H2O2 with concentrations of (II) 0.001 mM to 5 mM and (III) 5 mM to 1000 mM. Adapted with permission from ref. 117, Copyright 2017, ACS. (B) Schematic representation of a Hg2+ biosensor based on reticular DNA templated–Cu NC aggregates. Adapted with permission from ref. 38, Copyright 2018, ACS. | ||
A fluorescent chemodosimeter for sensitive and selective detection of Al3+ in real water samples with a recovery of 92–101% and a relative standard deviation of less than 4% has been reported by Boonmee et al.61 using cysteamine–Cu NCs as a sensing probe. This sensor worked in a concentration range of 1–7 μM with a low detection limit of 26.7 nM. The Cu NCs were synthesized by a ligand-assisted method in which cysteamine (Cys), a weak base containing –NH2 and –SH moieties, was applied as both a capping agent and a reducing agent. The –SH moiety attached to the surface of Cu NCs through Cu–S bonding, and the –NH2 moiety acted as a receptor interacting with metal ions through different interactions such as electrostatic interactions under acidic conditions. Al3+ ions could interact with –NH2 moieties on the surface of Cu NCs resulting in the aggregation of the NCs and subsequently an enhancement of the fluorescence intensity depending on the concentration of Al3+. According to the aggregation-induced emission (AIE) mechanism, the fluorescent intensity of Cys–Cu NCs at 380 nm linearly increased with Al3+ concentration.
D-Penicillamine capped-bimetallic AuCu NCs have been used as a fluorescent probe for Fe3+ detection in rain and river water, as well as in human blood serum.213 A high selectivity in the presence of various anions and cations especially Fe2+ has been demonstrated. Quenching of the PL intensity in the presence of Fe ions occurred through the IFE mechanism because of the overlap between the Fe3+ absorption peak and excitation peak of bimetallic AuCu NCs located at around 275 nm. The sensor exhibited a linear detection range of 0.5–7.0 μM and 7.0 μM–0.1 mM, and a LOD of 0.1 μM. Synthesis of water-soluble L-histidine-capped Cu NCs, stable against photobleaching and with long-term storage stability for sensitive and selective detection of Fe3+ has been performed by Lin et al.210 The LOD of this sensor was 82 nM in a concentration range of 0.10 to 20 μM. High quenching efficiency and stable sensing conditions were achieved at pH = 4.1. The sensing mechanism was based on a fluorescence “turn off–on” mechanism in which the fluorescence intensity of the L-His–Cu NCs was quenched due to the aggregation of the particles through bonding between Fe3+ ions with amine groups of L-histidine. The fluorescence intensity increased after adding ethylenediaminetetraacetate (EDTA) into the mixture of L-His–Cu NPs with Fe3+. This chemosensor was employed to detect Fe3+ in real water samples, including tap water and river water with recoveries in the range of 82.8–107.4%. L-Histidine-capped Cu CNs exhibited a selective sensing performance towards Fe3+ compared with various interfering ions such as Na+, K+, Li+, Ca2+, Mg2+, Co2+, Ni2+, Pb2+, Zn2+, Mn2+, Cu2+, Hg2+, Cd2+, Al3+, and Fe2+, as well as some common anions such as F−, Cl−, Br−, I−, Ac−, NO32−, PO43−, CO32−, SO42−, and SO32−.
A turn-on PL sensor based on silk fibroin (SF) protected–Cu NCs (SF@Cu NCs) has been developed by Zhang et al.203 It was used to detect S2− in aqueous solutions with a LOD of ∼0.3 μM and a linear range of 5–110 μM. The working mechanism of the sensor was ascribed to the assembly induced emission enhancement (AIEE). In the presence of S2− ions, SF@Cu NCs aggregated to larger, rod-shaped nanoparticles, which led to an increase of the PLQY from 1.6% to 4.9%. High selectivity was observed towards S2− in the presence of different interfering ions and macromolecules, and in particular sulfur-containing ions such as SO32−, SO42−, S2O42− and SCN−. A DNA templated–Cu/Ag NC fluorescent probe was developed by Ding et al.156 to detect S2− with a LOD of 3.75 pM and a wide linear concentration range from 10 pM to1 mM. This sensor worked due to the PL quenching of DNA–Cu/Ag NCs in the presence of sulfide; application for measuring sulfide amount in mouse blood (H2S poisoned blood sample) was demonstrated. Sulfide ion detection using water-soluble Zn-modified Cu NCs based on a protein/peptide templated method has been reported by Li et al.202 As compared with bare Cu NCs, Zn-modified Cu NCs exhibited an enhanced fluorescence intensity (by about 3.5-fold), and their PLQY reached 6.2%. Due to the degradation of the copper shell and change in conformation of proteins, as well as the formation of Zn(OH)x at high pH values (>8), the optimum pH ranged from 6 to 8. CuSx is an insoluble salt of copper and due to high oxidation/decomposition stability of Zn-modified Cu NCs in water, this sensor could be used for sensitive detection of sulfide ions by fluorescence quenching at 663 nm. The recovery of this ratiometric sensor in real water samples such as lake and tap water containing 20–80 μM S2− was found to be 101–109.9%. This sensing system also showed a selective sensing performance for S2− detection in water samples containing other common species.
Shen et al.220 employed in situ synthesized Cu NCs for the detection of Cu2+ through an AIE phenomenon. Rapid reduction of Cu2+ ions in solution, and cluster formation using thiol-containing glutathione as both reducing and stabilizing agents occurred when THF, acetonitrile, and DMF were added. Aggregation of NCs induced a bright emission which increased through addition of more copper ions.
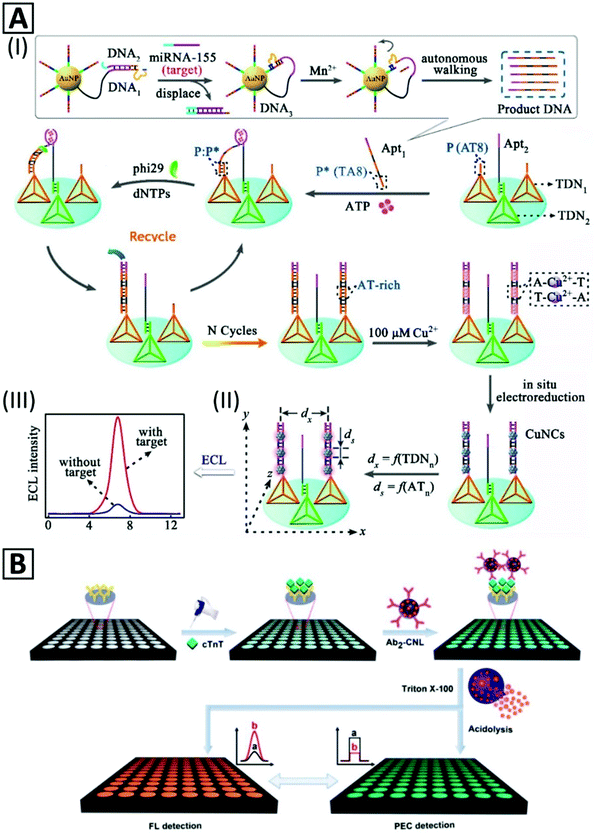 | ||
| Fig. 5 (A) Schematic representation of miRNA155 detection based on nanocrane-like DNA structures. (I) Strand displacement by the target molecule leads to amplified production of DNA oligonucleotide containing an ATP binding sequence (Apt1). (II) Tetrahedral DNA nanostructures (TDN) modulate the efficiency of the ECL signal of Cu NCs which are formed by in situ electroreduction via programming the lateral spacing (dx) and size controlling (ds) through an AT periodic sequence. (III) ECL signal in the presence and absence of the target. Adapted with permission from ref. 50, Copyright 2018, ACS. (B) Representation of two complementary detection strategies (photoelectrochemical and fluorescent) for an immunoassay with Cu NC-encapsulated liposomes as signal generators. Adapted with permission from ref. 37, Copyright 2018, ACS. | ||
Cu NCs were also used for protein detection. An immunoprobe was prepared by blocking unreacted sites of antibody-immobilized Pt NPs by BSA–Cu NCs.235 The immunosensor could detect prostate specific antigen through a sandwich assay with a LOD of 145.69 fg mL−1 with a linear range from 0.5 pg mL−1 to 100 ng mL−1. In another study,37 an immunosensor was developed for detection of cardiac Troponin T (cTnT) antigen based on photoelectrochemical (PEC) signals generated after lysis treatment of secondary antibody-bonded liposomes. As illustrated in Fig. 5B, the secondary antibody was bonded to the external surface of Cu NC-encapsulated liposomes. This immunosensor could sensitively detect cTnT by a signal-off method through the PEC mechanism, with a LOD of 0.03 pg mL−1.
Cao et al.45 presented a strategy for detecting a small molecule and its interacting protein, using streptavidin and biotin as a model (Fig. 6A). Protein molecules immobilized on magnetic nanoparticles (MNPs) were one part, and ssDNA oligonucleotides with a small molecule at their 5′-end were another part of the detection system. The signal was generated via the formation of Cu NCs which were synthesized by a polythymin templating method. Supernatant or precipitant location of the generated signals determined the interacting proteins. This biosensor enabled detection of streptavidin and biotin in the linear range of 1–200 nM and 10–1000 nM, with LOD of 0.47 nM and 3.1 nM, respectively.
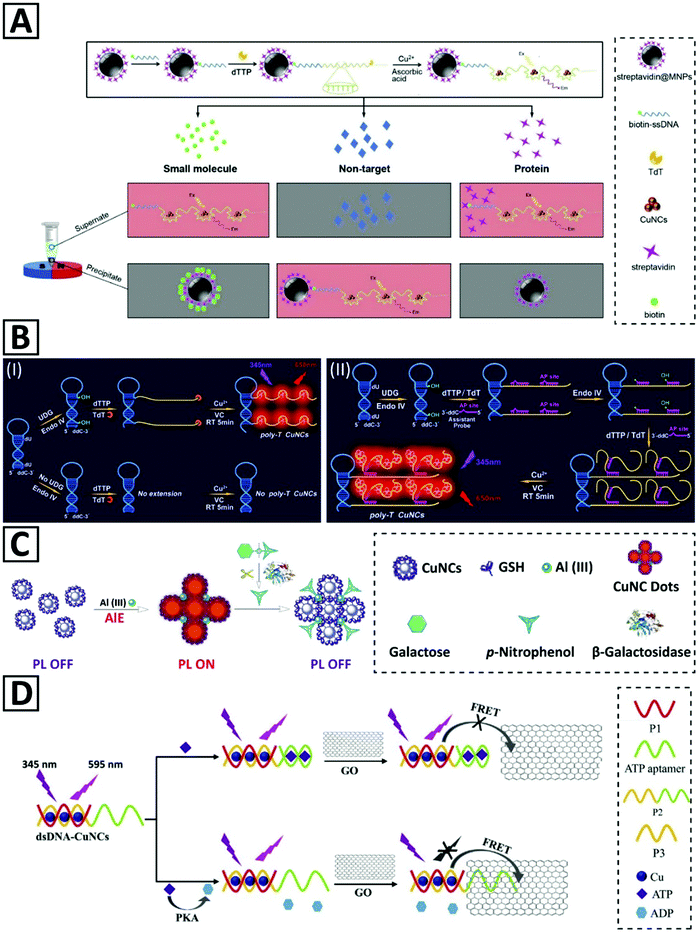 | ||
| Fig. 6 (A) Representation of the detection strategy for small molecules and proteins relying on magnetic separation and opposite fluorescent signaling in the presence of Cu NCs. Adapted with permission from ref. 45, Copyright 2017, Elsevier. (B) Schematic illustration of (I) the proposed UDG assay principles and the roles of TdT and endonuclease IV in Cu NC template formation. (II) Hyperbranched extension of the Cu NC template helped by the assistant probe. Adapted with permission from ref. 46, Copyright 2019, Elsevier. (C) Schematic illustration of β-galactosidase activity measurement via emission quenching of aluminum-assisted self-assembled Cu NCs. Adapted with permission from ref. 105, Copyright 2017, RSC. (D) Schematic illustration of the PKA activity detection via a FRET-based fluorescent biosensor. Adapted with permission from ref. 40, Copyright 2018, Elsevier. | ||
Hybrid nanostructures of Cu NCs and glucose oxidase (GOx) enzyme were employed for glucose sensing.121 Enzymatic oxidation of glucose led to H2O2 production that would quench the emission of Cu NCs. The GOx/Cu NC assembly selectively measured glucose with a LOD of 1.5 μM in a linear range of 5–100 μM. Another kind of biosensor for glucose via the turn-off fluorescence strategy has been designed based on lysozyme-stabilized Cu NCs (Lys-CCs).236 A glucose biosensor using a pH-responsive fluorescent solution, a mixture of luminescent Cu NCs and CaCO3 nanoparticles, and alginate, has been reported.70 In the presence of glucose, the produced H+ could release Ca2+ from CaCO3 nanoparticles, which turned the solution into a gelated phase with an enhanced emission. This AIE-based sensor could detect glucose with a linear range of 0.1 to 2.0 mM and LOD of 3.2 × 10−5 M. Zhang et al.237 employed OVA@Cu NCs for L-lysine (L-Lys) detection with a LOD of 5.5 μM and a linear range of 10 μM–1 mM. They showed that OVA@Cu NCs could selectively detect L-Lys among many other amino acids and cations based on the PL enhancement via coordination of copper by L-Lys functional groups and surface electron density increment on Cu NCs. Yang and co-workers25 employed OVA@Cu NCs to detect Vitamin B1 (VB1) and doxycycline based on turn-off and turn-on PL strategies. VB1 quenched PL of OVA@Cu NCs through aggregation, while doxycycline enhanced the emission due to its strong interaction with OVA, and making a more compact structure. OVA–Cu NCs with a red emission at 625 nm and a PLQY of 3.95% were used for folic acid detection with a LOD of 0.18 μM. Evaluation of FA absorption (360 nm) and OVA–Cu NC excitation spectra (348 nm) as well as fluorescence lifetimes of Cu NCs in the presence and absence of FA indicated that the detection mechanisms should be the static quenching and the IFE.36 Quenching of BSA–Cu NCs by rutin (a kind of flavonoid) through hydrogen bonding and electrostatic interactions between BSA and rutin was used for naked eye fluorescent detection in aqueous solution as well as on a paper filter.34Via the same mechanism, detection of mangiferin, a kind of flavonoid, was performed in real samples by BSA–Cu NCs.238 In another biosensing method, Kojic acid, a fungal metabolite, was measured through Cu NCs FL quenching.239 The fluorescence of BSA–Cu NCs was diminished due to its binding to copper ions and the formation of copper Kojate on the surface of Cu NCs.
Quenched fluorescence of NCs triggered by electron transfer of metal ions can be recovered by strong interactions between analytes and those ions.240 The turn-on fluorescence of PEI–Cu NCs was employed for detecting biothiols (e.g. cysteine and GSH).23 Herein, –SH functional groups interacted strongly with Cu2+ (introduced into the reaction mixture) as a quencher. Acetylcholinesterase (AChE), which is capable of hydrolyzing acetylthiocholine into thiocholine, has a strong tendency in forming a complex with Cu2+; hence, it could be assayed by PEI–Cu NCs with a linear range of 3–200 mU mL−1 and LOD of 1.38 mU mL−1.23 This approach was also used for detecting an AChE typical inhibitor, tacrine.23 In another study, a dual-emitting nanohybrid was prepared based on blue emitting carbon dots modified with 3-aminophenyl boronic acid (APBA-CDs) and BSA–Cu NCs for the detection of dopamine.33 Red-emitting BSA–Cu NCs acted as an internal reference for measuring the PL quenching of APBA-CDs in the presence of dopamine, and a decrease in fluorescence intensity of APBA-CDs could be detected by the naked eye. Wang et al.39 presented a molecular biology technique termed target-cycling strand displacement amplification (TCSDA) to detect adenosine 5′-triphosphate (ATP) in a broad dynamic range from 0.01 nM to 100 nM with a LOD of 5 pM. In the presence of ATP, an oligonucleotide hairpin probe containing an ATP aptamer sequence was structurally switched so that the TCSDA reaction was started by a DNA polymerase Klenow fragment (KF polymerase). This approach could detect ATP in a broad dynamic range from 0.01 nM to 100 nM, with a LOD of 5 pM. The PL intensity of PVP-stabilized Cu NCs was quenched after formation of a MnO2–Cu NC complex through electrostatic interactions with MnO2 nanospheres. GSH was selectively detected by this complex due to its capability of recovering the PL signal of Cu NCs (acting as donors) via digesting the MnO2 nanospheres (energy acceptors). This FRET-based biosensor could detect GSH with a LOD of 17 μM.21
In addition to PL properties, the catalytic activity of Cu NCs was employed for biosensing, as well. Xu and co-workers241 used the advantage of the peroxidase-mimetic activity of Cu NCs to detect cholesterol with a broad linear range of 0.05–10 mM and LOD of 1.5 μM. The assay was based on chemiluminescent signals produced by coupling two reactions. The first reaction was the oxidation process of cholesterol catalyzed by cholesterol oxidase enzyme, which produced H2O2 as a by-product. The second reaction was the luminol oxidation by H2O2, catalyzed by Cu NCs.
The AIE effect is yet another common strategy that has been used for the activity detection of pyrophosphatase (PPi) by GSH-capped Cu NCs.249,250 In the presence of Al3+, the aggregation of Cu NCs occurred, leading to the PL enhancement due to AIE, providing a sensitive probe with a LOD of 1.3 mU mL−1. Zhao et al.68 prepared stable Cu NCs with improved AIE properties by employing a hydrophobic capping agent (4-methylthiophenol) during synthesis. The as-synthesized Cu NCs exhibited a weak emission due to their hydrophobic protecting ligands, but through subsequent processing, they were self-assembled into highly red emissive particles. Addition of a hydrophobic electron acceptor molecule (4-nitrophenol) could quench the emission of these Cu NCs by about 80%. Through this quenching strategy and using 4-nitrophenyl-β-D-galactopyranoside (NPGal) as the synthetic substrate for β-galactosidase enzyme, the enzymatic activity of β-galactosidase in serum could be measured with a LOD of 0.9 U L−1 in the linear range of 2.5–212.0 U L−1. Huang et al.105 used a similar strategy to monitor the β-galactosidase activity. They employed aluminum cations to self-assemble GSH-capped Cu NCs into so-called Cu NC dots with strong PL intensity, which was quenched upon enzymatic release of 4-nitrophenol from 4-nitrophenyl-β-D-galactopyranoside (Fig. 6C). The same PL-off or PL-on switching mechanism was used for the real-time monitoring of acid phosphatase activity, in which after hydrolyzing the bond between 4-nitrophenol and the phosphate group in p-nitrophenyl phosphate disodium, the PL quenching occurred.251 In another study,67 the acid phosphatase (ACP) enzyme activity was measured by redox-responsive emission of Cu NC aggregates. It was shown that D-penicillamine-capped Cu NCs with pH and temperature responsiveness could aggregate to form particles with stronger red luminescence. The existence of oxidant species such as free ferric ions (Fe3+) could quench the PL intensity of CNC aggregates by 80% due to oxidation of copper atoms, providing an assay with 0.8 U L−1 LOD and a broad linear scope of up to 100 U L−1. This strategy also was applied for ACP activity measurement in diluted serum samples as complex medium.67
Wang et al.24 have introduced a novel assay for protein kinase A (PKA) activity based on the overlap of the emission of PEI-capped Cu NCs (with an emission peak around 515 nm) and the UV-Vis absorption spectrum of Au NPs. The already mentioned inner-filter effect (IFE) could quench PL of PEI–Cu NCs up to 59%. Peptide-functionalized Au NPs after phosphorylation by PKA enzyme with the assistance of adenosine 5′-triphosphate (ATP) molecules tend to aggregate in the presence of multivalent Zr4+ cations. This fluorescent biosensor for PKA activity measurement with a linear range of 0.1–6.0 U mL−1 and LOD of 0.038 U mL−1 also was employed for measuring the cellular kinase activity in HepG-2 cell lysates. In another study,40 a biosensor for the PKA activity was developed based on the quenching effect of graphene oxide (GO) plates on dsDNA–Cu NCs through fluorescence resonant energy transfer (FRET) (Fig. 6D). Two-domain oligonucleotides including an ATP aptamer sequence were hybridized with a shorter complementary sequence as a template for Cu NC formation. Binding ATP molecules to aptamer sequences and folding ssDNA inhibited proximity between GO and dsDNA–Cu NCs and kept Cu NCs fluorescent. ATP hydrolysis to ADP by the PKA enzyme made the ATP domain of the oligonucleotide to be single-stranded, which could adsorb on GO via electrostatic and π–π stacking leading to PL quenching of dsDNA–Cu NCs. The linear range and LOD of this assay were 0.1–5.0 U mL−1 and 0.039 U mL−1, respectively. A poly(AT–TA) dsDNA with a restriction site for EcoRI endonuclease was also used for endonuclease activity detection.245 In the presence of the enzyme, dsDNA was cut even in the presence of reducing agent AA to affect the fluorescence intensity. This assay could detect EcoRI endonuclease concentration from 0.002 U μL−1 to 0.1 U μL−1, with a LOD of 0.00087 U μL−1.
Nano-thermometers have plenty of applications, especially in monitoring the temperature within biological cells and microfluidic devices.255,256 Shi et al.20 synthesized stable, blue-emitting Cu NCs with a PLQY of 12% and have shown that their fluorescence signal linearly decreased by 73% with increasing temperature from 20 to 75 °C, without any shifts of the emission peak (425 nm). Only 5% deviation was recorded after 10 cycles of heating and cooling between 20 and 75 °C. Han et al.66 used bimetallic GSH–Cu/Ag NCs for temperature sensing; by introducing Ag ions during the synthesis of Cu NCs, 9-fold enhancement of fluorescence intensity was attained. These bimetallic NCs with a strong orange-yellow emission and ionic strength stability showed a linear and inverse relationship between fluorescence intensity and temperature changes within 4–55 °C. Reversibility of the thermal response was demonstrated after 7 heating–cooling cycles without a decrease in the luminescence intensity. Bimetallic NCs were employed for confocal fluorescent imaging of HeLa cells after 24 h of incubation at three different temperatures (293 K, 303 K, and 313 K). The mechanism underlying fluorescence intensity quenching was ascribed to the aggregation of NCs.216 Wang et al.98 prepared highly fluorescent GSH-protected Cu NCs with QY 5% and used them for temperature monitoring and confocal imaging of MC3T3-E1 human cancer cells in the range of 15–45 °C.
4.3. Bioimaging
Since UV irradiation necessary to excite blue emitting Cu NCs may damage the cells upon imaging, and induce the auto-fluorescence of the tissue, red emitting Cu NCs emerged as alternative probes. Wang and co-workers13 used BSA–Cu NCs for low energy excitation at 524 nm and imaged 24 h-incubated CAL-27 cells at the peak emission of 625 nm. Kailasa's group72 synthesized egg white supported Cu NCs with two emission wavelengths; confocal laser microscopy revealed a blue signal when excited by a 405 nm laser, and a green signal when excited by a 488 nm laser in Bacillus subtilis. When HeLa cells were treated with PEG-capped Cu NCs, confocal microscopy showed the ability of these Cu NCs to stain the nuclei through cellular uptake by membrane crossing without endocytosis.5 In another study, transferrin (Tf) receptors were targeted by red emitting Trf–Cu NCs.258 High expression of the Tf receptor on HeLa cells caused higher uptake of Tf–Cu NCs in comparison to 3T3 cells.
Temperature changes could be related to some unfavorable biological pathways of alive cells,259,260 making temperature measurements combined with the cellular imaging quite important. Intracellular synthesis of Cu NCs has been performed for determination of temperature in living cells; Cu NCs with sizes of 2.4 ± 0.4 nm and red emission at 610 nm could be synthesized in malignant cell lines such as MDA-MB-231 via a specific biomolecular process.73 Notably, this cannot be done in healthy cells, e.g. L02. Accumulation of these NCs in the target cells was efficient, and sensitivity of the fluorescence signal to the physiological temperature was high; the calibration curve for MDA-MB-231 cells showed 3.18% decrease in PL intensity per 1 °C temperature elevation. Owing to the critical role of Ca2+ in neurodegeneration and the importance of monitoring of Ca2+ pathways in neurons, Tian's group261 developed a Ca2+sensitive probe via Ca2+ ligand modified PEI templated Cu NCs (Fig. 7A-I). Alexa 660 NHS ester was conjugated to Cu NCs as a reference (Fig. 7A-II). The intensity of Cu NC PL in neurons was amplified based on the increase of Ca2+ concentration. The signal ratio of Cu NCs to reference sample could also be calibrated to a high concentration range sensor based on intracellular bioimaging. In another study, non-luminescent cysteine and chitosan protected Cu NCs were employed to specify cell differentiating at pH = 7.4. The kinetics of Cu NC aggregation inside different cell lines was dependent on the cell type so that a novel approach for detection of various cell lines could be developed only by green channel monitoring of confocal microscopy (Fig. 7B).75,262
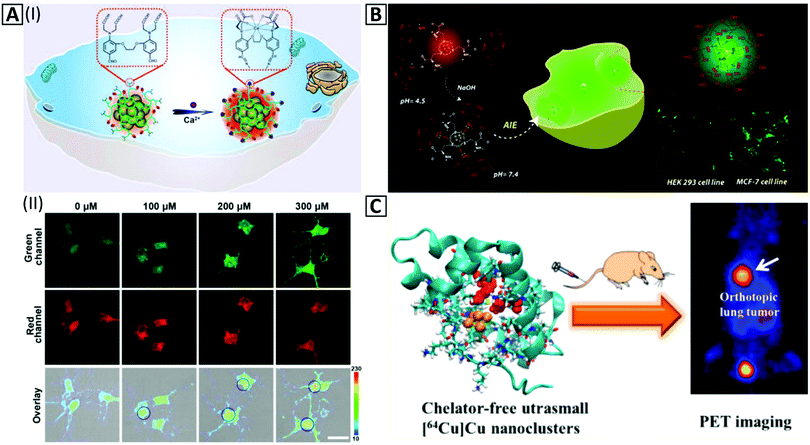 | ||
| Fig. 7 (A-I) Schematic illustration of in vitro imaging based on the amplification of the PL intensity of the Cu NC@Alexa660 probe in the presence of Ca2+. (A-II). Fluorescence confocal imaging exhibits amplification of the green channel intensity ratio to the constant signal intensity of the red channel inside neurons via an increase of Ca2+ concentration from 0–300 μM in the presence of 90 μg mL−1 of the Cu NC@Alexa660 probe. Adapted with permission from ref. 261, Copyright 2019, ACS. (B) Schematic illustration of in vitro imaging of HEK 293 and MCF-7 cells by red-emitting Cu NCs at pH = 4.5, which change their color to green at pH = 7.4 via the AIE effect inside the cell cytoplasm. Adapted with permission from ref. 75, Copyright 2018, ACS. (C) In vivo PET imaging and organ distribution study after 2 h intravenous injection of Cu NCs @BSA-LHRH. Adapted with permission from ref. 76, Copyright 2015, ACS. | ||
4.4. Theranostic applications
Theranostic agents can be used in therapeutics and diagnostics at the same time.8 Ghosh et al.143 developed a hydrogel-based anticancer carrier containing Cu NCs and Cisplatin, using red fluorescent Cu NCs synthesized in an aqueous environment by using poly(vinylpyrrolidone) (PVP) stabilizer and dihydrolipoic acid. Composite fluorescent particles were sensitive to pH variations and exhibited emission changes from red to orange through adjustment of the pH in the range of 4 to 8.5. Fluorescence microscopy showed how this kind of nanocarrier can be used for mammalian cell uptake monitoring; synergetic anticancer activity was attained by coupling the therapeutic effect of Cisplatin alongside with the ability of Cu NCs in killing cancerous cells through generation of reactive oxygen species (ROS). Goswami et al.77 prepared blue emitting Tf-templated Cu NCs and combined them with Dox through electrostatic interactions to formulate sphere-shaped Tf–Cu NC–Dox NPs for active theranostics. Due to FRET occurring in this system, Tf–Cu NC–Dox NPs displayed significant red emission, while the release of Dox inside the cytoplasm of Tf receptor overexpressed cancer cells restored the blue emission of Cu NCs. A synergistic effect of the ROS generation by Tf–Cu NCs and the anticancer activity of Dox for the therapy of mouse models has been demonstrated. Another study revealed that temozolomide loaded positron emitting Cu NCs could upgrade a PET contrast agent to a theranostics agent for glioblastoma.784.5. Other bioapplications
In addition to applications of Cu NCs considered above, there are a few other emerging areas where they can be of interest. For instance, biological staining is frequently used to mark cells in flow cytometry, and to flag proteins or nucleic acids in gel electrophoresis.265 Zhu et al.266 developed a method for in situ staining of DNA in a polyacrylamide gel through the formation of Cu NCs in the presence of a DNA template. Cu NCs have also been utilized for the detection of single nucleotide polymorphisms, which are responsible for various genetic problems of human health.267 Since dsDNA affects the Cu NC environment and thus changes the fluorescence intensity, a “mix-and-measure” strategy has been employed for mismatch detection of dsDNA. Cu NCs can also serve as smart probes opening opportunities for fast and economical bioanalysis.268In recent years, nano-bioelectronics has attracted significant attention as a rapidly expanding interdisciplinary field which utilizes nanomaterials to overcome some current limitations in bioelectronics.269 DNA nanowires are promising materials for this purpose, while Cu NCs with resistance to charge transfer can be employed as the key element for the “ON–OFF” process. To exhibit practical aspects of this phenomena, one end of dsDNA was immobilized on the surface of gold electrodes, and another end was tagged by methylene blue (a redox-active agent). In the presence of AA, Cu2+ was reduced to Cu0 and deposited on the DNA scaffold, which resulted in a switch-off of the charge transfer path. The process was reversible and repetitive by utilizing oxidants that caused the stripping of Cu NCs.270
The antimicrobial effect of Cu NCs has recently been explored.110 Baghdasaryan et al. showed the dose-dependent bactericidal capability of GSH-capped Cu NCs: at low cluster concentrations the growth of bacteria was slowed down, and at high dosages (≥250 μg mL−1) the bacterial replication was fully restricted. Antimicrobial action of metal NCs has been ascribed to intracellular ROS generation.271 To utilize the high antibacterial performance of metal NCs, Ag, Cu, and Au NCs@Bacitracin were synthesized by Wang and coworkers.272 Bacitracin as a peptide antibiotic can damage the bacterial membrane; hence, the synergy of the membrane damage and the ROS generation by different MNCs@Bacitracin clusters could be attained. It was demonstrated that Ag NCs@Bacitracin was the most powerful antibacterial agent against S. aureus. Comparative results of zone inhibitions, growth curve, percentage of PI stained bacteria, and relative ROS levels are provided in Fig. 8. Cu NC-doped kanamycin-loaded hydroxyapatite NPs were introduced as an antibiofilm and antibacterial agent.273
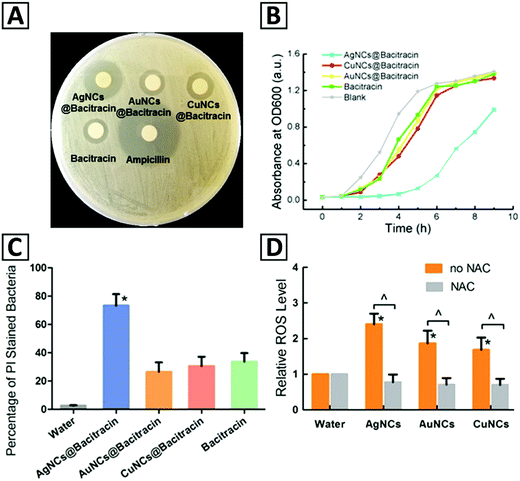 | ||
| Fig. 8 (A) Zone inhibitions against S. aureus by Ag, Au, Cu NCs@Bacitracin, bacitracin, and ampicillin. (B) S. aureus growth curve in the presence of Ag, Au, and Cu NCs@ Bacitracin. (C) The percentage of stained bacteria cells with PI in the presence of water, Ag, Au, Cu NCs@ Bacitracin, and bacitracin. (D) ROS levels in bacteria cells treated with Ag, Au, and Cu NCs@Bacitracin; ROS levels are for two separate groups treated by water with/out NAC. Adapted with permission from ref. 272, Copyright 2019, ACS. | ||
4.6. Applications in photovoltaics and optoelectronics
 | ||
| Fig. 9 (A) Schematic illustration of a Cu NCs/ImRGO nanocomposite, and (B) positions of the HOMO–LUMO levels of Cu NCs and work function (ϕ) of ImRGO. Adapted with permission from ref. 79, Copyright 2018, ACS. (C) Cross-sectional scanning electron microscopy image of a CuI film made from Cu53 NCs deposited on a lead halide perovskite film through iodization, and (D) energy level diagram of the CuI-based perovskite solar cell. Adapted with permission from ref. 80, Copyright 2019, Wiley-VCH. | ||
Another strategy for tuning fluorescence color is metal doping.277 Liu et al.202 showed that Au+ doping upon preparation of Cu nanosheets increased their PL intensity with an emission red-shift. The Au doping induced metallophilic interactions of Au+–Cu+, facilitating excited electrons to experience the radiative relaxation, owing to changes in the ligand-to-metal charge transfer (LMCT) and/or LMMCT. The Au doping also decreased the original Cu-centered triplet energy state inducing an emission red-shift. They revealed that introducing 0.3% Au was enough to give rise to 4-fold PL enhancement with a 100 nm emission red-shift. Such Au+-doped Cu nanosheets have been applied as phosphors for WLED fabrication.
The AIE effects of luminescent metal NCs have also been utilized for fabrication of WLEDs.101,122,278 As already mentioned above, AIE is able to significantly enhance the emission of the Cu NCs by suppression of the rotation and vibration of their capping ligands.279 Wang et al.18 used a solvent-induced aggregation method for synthesis of orange emitting aggregated GSH-capped Cu NCs, which showed PLQY of 24% and 43% in solution and solid state, respectively. In parallel, blue emitting Cu NCs@ PVP were synthesized, with a PLQY of 14%. Both monochrome blue or orange LEDs, and WLEDs were fabricated by depositing respective powders on commercial GaN LED chips providing 370 nm excitation (Fig. 10).101 WLEDs combining blue emitting sulfur QDs with the aggregated orange emitting Cu NCs have been demonstrated, as well.280 The same group used in situ aggregation of Cu NCs in a carboxylated polyurethane matrix to produce remote dual blue/orange emitting composite films,278 which were thermally and mechanically stable, and stretchable. The PLQY of the films was as high as 18% and the CIE chromaticity coordinate was (0.34, 0.29). A dehydration-triggered aggregation of GSH capped Cu NCs occurring in a PVP/poly(vinyl alcohol) matrix resulted in the formation of flexible, large-area polymer composite films with a high PLQY of 30%, which have been used for fabrication of bright orange LEDs.281 A similar dehydration mechanism was used to fabricate protective transparent UV shielding films,282 and biocompatible hypromellose–chitosan copolymers with AIE improved the emission of the Cu NCs reaching 42%.83
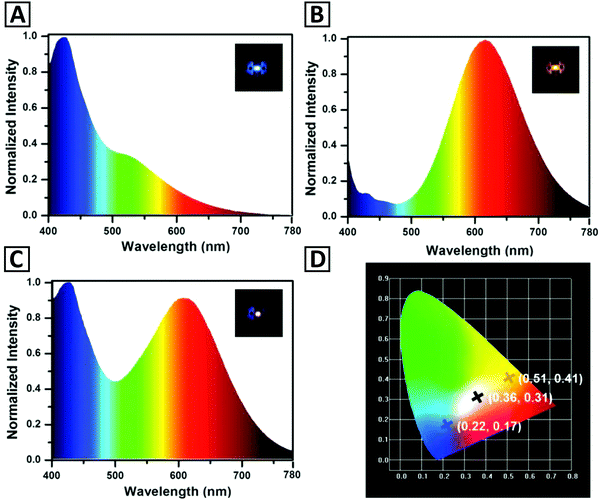 | ||
| Fig. 10 Emission spectra of monochrome down-conversion LEDs fabricated by using (A) blue emitting Cu NCs and (B) orange emitting Cu NCs. Panel (C) shows an emission spectrum of the WLED fabricated by a combination of these two kinds of Cu NCs. Insets in (A–C) provide photographs of operating blue, orange and white LEDs; while panel (D) shows CIE coordinates of three respective LEDs. Reproduced with permission from ref. 101, Copyright 2016, Wiley-VCH. | ||
5. Conclusions and outlook
In this review, rapidly expanding deployment of Cu NCs in a series of important applications such as catalysis, chemical and biological sensing, bioimaging, theranostics, and fabrication of LEDs is summarized. Extensive up to date research has been carried out on developing viable methods for the synthesis of these promising nanomaterials. One important factor that controls the properties of Cu NCs is related to the nature of their functional capping agents. Thus, the main thrust in the field of synthesis of Cu NCs is to identify the most appropriate capping agents (ligands) and related templates. Due to the favorable price considerations and availability of precursor materials, practical catalytic applications of Cu NCs are expected to become widespread as compared to noble metal catalysts, i.e. silver and gold. Also for this application, in order to obtain the desired catalytic activity in appealing reactions e.g. reduction, oxidation, and hydrogen generation, several capping agents/supporting ligands have been investigated. Development of theoretical methods which predict the catalytic performance of Cu NCs or alloys can facilitate the process of synthesis and introduction of catalysts implemented in some industrial applications in the future. Most of the sensors that have been designed using Cu NCs exploit their emission (both quenching and enhancement) as the main analytical signal. Capping ligands or stabilizing scaffolds play an important role in the detection mechanism. As in other fluorescent probes, the emission intensity can decrease, increase, or get quenched as a result of interactions with the analyte; thereby, plenty of detection strategies could be employed for different kinds of target molecules. Additionally, combining two or more signal amplification strategies such as molecular biology-based techniques with methods like in situ synthesis of Cu NCs can enhance the detection capability of the (bio)sensor and significantly lower the detection limits. Non (or low) toxicity, biocompatibility, and low price make Cu NCs attractive for designing label-free fluorescence sensors. Herein, a variety of analyte classes have successfully been analyzed. There are still drawbacks in this field which are currently inherent to Cu NCs, such as moderate PL QY and higher susceptibility to oxidation; improvement of these aspects has been in the focus of researcher's attention. Using various types of scaffolds, which can be conjugated to targeting agents, Cu NCs have also been utilized as fluorophores for both in vitro and in vivo bioimaging. Bioimaging can be used to characterize different types of cancer cells, nanothermometry, and estimation of concentration of analytes inside cells. Cu NCs are able to generate ROS inside eukaryotic and prokaryotic cells and to induce cell death as anticancer and antibacterial agents. 64Cu as a PET imaging probe has been used for the design of new types of PET contrast agents. Recently, Cu NCs have been also used in photovoltaics, which may provide new opportunities for fabrication of solar cells. Light-emitting Cu NCs were also employed as phosphors in down-conversion LEDs, both monochrome ones, in particular in WLEDs; however, both efficiency and long-term stability of such devices still require massive improvements before their actual practical use.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge financial support from the Iran National Science Foundation (INSF No. 95-S-48740), Sharif University of Technology (Grant No. QA970816), the Research Grant Council of Hong Kong S.A.R. (CityU11305617), and the Science Technology and Innovation Committee of Shenzhen Municipality (JCYJ20170818104224667).References
- J. Wilcoxon and B. L. Abrams, Chem. Soc. Rev., 2006, 35, 1162–1194 RSC.
- X. Hu, T. Liu, Y. Zhuang, W. Wang, Y. Li, W. Fan and Y. Huang, TrAC, Trends Anal. Chem., 2016, 77, 66–75 CrossRef CAS.
- L. Zhang and E. Wang, Nano Today, 2014, 9, 132–157 CrossRef CAS.
- H. Cao, Z. Chen, H. Zheng and Y. Huang, Biosens. Bioelectron., 2014, 62, 189–195 CrossRef CAS PubMed.
- M. J. Barthel, I. Angeloni, A. Petrelli, T. Avellini, A. Scarpellini, G. Bertoni, A. Armirotti, I. Moreels and T. Pellegrino, ACS Nano, 2015, 9, 11886–11897 CrossRef CAS PubMed.
- X. Liu and D. Astruc, Coord. Chem. Rev., 2018, 359, 112–126 CrossRef CAS.
- Z. Wang, B. Chen and A. L. Rogach, Nanoscale Horiz., 2017, 2, 135–146 RSC.
- S. Shahsavari and F. Behroozi, J. Nanomed. Res., 2016, 3, 00069 Search PubMed.
- X. Jia, X. Yang, J. Li, D. Li and E. Wang, Chem. Commun., 2014, 50, 237–239 RSC.
- Z. Wu, Y. Li, J. Liu, Z. Lu, H. Zhang and B. Yang, Angew. Chem., Int. Ed., 2014, 53, 12196–12200 CrossRef CAS PubMed.
- L. D. Pachón, J. H. Van Maarseveen and G. Rothenberg, Adv. Synth. Catal., 2005, 347, 811–815 CrossRef.
- A. W. Cook, Z. R. Jones, G. Wu, S. L. Scott and T. W. Hayton, J. Am. Chem. Soc., 2017, 140, 394–400 CrossRef PubMed.
- C. Wang, C. Wang, L. Xu, H. Cheng, Q. Lin and C. Zhang, Nanoscale, 2014, 6, 1775–1781 RSC.
- Z. Jin, C. Liu, K. Qi and X. Cui, Sci. Rep., 2017, 7, 39695 CrossRef CAS PubMed.
- R. Cai, P. R. Ellis, J. Yin, J. Liu, C. M. Brown, R. Griffin, G. Chang, D. Yang, J. Ren and K. Cooke, Small, 2018, 14, 1703734 CrossRef PubMed.
- B. Eren, D. Zherebetskyy, L. L. Patera, C. H. Wu, H. Bluhm, C. Africh, L.-W. Wang, G. A. Somorjai and M. Salmeron, Science, 2016, 351, 475–478 CrossRef CAS PubMed.
- B. Sarkar, P. Prajapati, R. Tiwari, R. Tiwari, S. Ghosh, S. S. Acharyya, C. Pendem, R. K. Singha, L. S. Konathala and J. Kumar, Green Chem., 2012, 14, 2600–2606 RSC.
- R. Aparna, J. A. Devi, P. Sachidanandan and S. George, Sens. Actuators, B, 2018, 254, 811–819 CrossRef CAS.
- Y. Li, L. Feng, W. Yan, I. Hussain, L. Su and B. Tan, Nanoscale, 2019, 11, 1286–1294 RSC.
- Y. E. Shi, S. Luo, X. Ji, F. Liu, X. Chen, Y. Huang, L. Dong and L. Wang, Dalton Trans., 2017, 46, 14251–14255 RSC.
- T. Li, Z. Wang, D. Jiang, H. Wang, W. F. Lai, Y. Lv and Y. Zhai, Sens. Actuators, B, 2019, 290, 535–543 CrossRef CAS.
- C. Wang, Y. Yao and Q. Song, Colloids Surf., B, 2016, 140, 373–381 CrossRef CAS PubMed.
- J. Yang, N. Song, X. Lv and Q. Jia, Sens. Actuators, B, 2018, 259, 226–232 CrossRef CAS.
- S. Song, C. Wang, Y. Zhao, T. Hu, X. Zhou, T. Zhao, M. Yang and Q. Lin, Part. Part. Syst. Charact., 2018, 35, 1700471 CrossRef.
- K. Yang, Y. Wang, C. Lu and X. Yang, J. Lumin., 2018, 196, 181–186 CrossRef CAS.
- A. Yousefzadeh, J. Hassanzadeh, S. M. J. Mousavi and M. Yousefzadeh, Sens. Actuators, B, 2019, 154–162, 154–162 CrossRef.
- R. S. Aparna, J. S. Anjali Devi, R. R. Anjana, J. Nebu and S. George, Sens. Actuators, B, 2019, 291, 298–305 CrossRef CAS.
- G. Zhang, R. Wang, L. Shi, C. Zhang, Y. Zhang, Y. Zhou, C. Dong, G. Li and S. Shuang, Sens. Actuators, B, 2019, 279, 361–368 CrossRef CAS.
- S. Xu, K. Zhou, D. Fang and L. Ma, Molecules, 2019, 24, 95 CrossRef PubMed.
- Z. Shojaeifard, N. Heidari and B. Hemmateenejad, Spectrochim. Acta, Part A, 2019, 209, 202–208 CrossRef CAS PubMed.
- G. Zhang, T. Xu, H. Du, Y. Qiao, X. Guo, L. Shi, Y. Zhang, S. Shuang, C. Dong and H. Ma, J. Mater. Chem. C, 2016, 4, 3540–3545 RSC.
- R. Rajamanikandan and M. Ilanchelian, Mater. Sci. Eng., C, 2019, 98, 1064–1072 CrossRef CAS PubMed.
- W. He, R. Gui, H. Jin, B. Wang, X. Bu and Y. Fu, Talanta, 2018, 178, 109–115 CrossRef CAS PubMed.
- Y. Wang, T. Chen, Q. Zhuang and Y. Ni, Talanta, 2018, 179, 409–413 CrossRef CAS PubMed.
- R. Aparna, J. A. Devi, R. Anjana, J. Nebu and S. George, Analyst, 2019, 144, 1799–1808 RSC.
- X. Li, X. Wu, F. Zhang, B. Zhao and Y. Li, Talanta, 2019, 195, 372–380 CrossRef CAS PubMed.
- L.-P. Mei, X.-Y. Jiang, X.-D. Yu, W.-W. Zhao, J.-J. Xu and H.-Y. Chen, Anal. Chem., 2018, 90, 2749–2755 CrossRef CAS PubMed.
- J. Li, W. Fu, J. Bao, Z. Wang and Z. Dai, ACS Appl. Mater. Interfaces, 2018, 10, 6965–6971 CrossRef CAS PubMed.
- Y. M. Wang, J. W. Liu, L. Y. Duan, S. J. Liu and J. H. Jiang, Microchim. Acta, 2017, 184, 4183–4188 CrossRef CAS.
- M. Wang, Z. Lin, Q. Liu, S. Jiang, H. Liu and X. Su, Anal. Chim. Acta, 2018, 1012, 66–73 CrossRef CAS PubMed.
- Y. Ling, J. Zhou, X. F. Zhang, X. H. Wang, N. B. Li and H. Q. Luo, Sens. Actuators, B, 2019, 286, 46–51 CrossRef CAS.
- R. Li, Q. Liu, Y. Jin and B. Li, Sens. Actuators, B, 2019, 281, 28–33 CrossRef CAS.
- X. Zhang, Q. Liu, Y. Jin and B. Li, Microchim. Acta, 2019, 186, 3 CrossRef PubMed.
- F. M. Moghadam and M. Rahaie, Biosens. Bioelectron., 2019, 132, 186–195 CrossRef CAS PubMed.
- J. Cao, W. Wang, B. Bo, X. Mao, K. Wang and X. Zhu, Biosens. Bioelectron., 2017, 90, 534–541 CrossRef CAS PubMed.
- G. Liu, W. He and C. Liu, Talanta, 2019, 195, 320–326 CrossRef CAS PubMed.
- Y.-S. Borghei, M. Hosseini and M. R. Ganjali, Microchim. Acta, 2017, 184, 2671–2677 CrossRef CAS.
- Y.-S. Borghei, M. Hosseini, M. R. Ganjali and S. Hosseinkhani, Sens. Actuators, B, 2017, 248, 133–139 CrossRef CAS.
- S. Singh, M. K. Singh and P. Das, Sens. Actuators, B, 2018, 255, 763–774 CrossRef CAS.
- Y. Zhou, H. Wang, H. Zhang, Y. Q. Chai and R. Yuan, Anal. Chem., 2018, 90, 3543–3549 CrossRef CAS PubMed.
- Y. Nerthigan, A. K. Sharma, S. Pandey and H.-F. Wu, Microchim. Acta, 2019, 186, 130 CrossRef PubMed.
- A. Hatefi, E. Rahimpour, M. Khoubnasabjafari, M. Edalat, V. Jouyban-Gharamaleki, S. Alvani-Alamdari, A. Nokhodchi, M. H. Pournaghi-Azar and A. Jouyban, Microchim. Acta, 2019, 186, 194 CrossRef PubMed.
- H. Liu, X. Gao, X. Zhuang, C. Tian, Y. Li and A. Rogach, Analyst, 2019, 144, 4425–4431 RSC.
- Y. Ma, Y. Yu, B. Lin, L. Zhang, Y. Cao and M. Guo, Microchim. Acta, 2019, 186, 206 CrossRef PubMed.
- G. Hambarde, S. Bothra, Y. Upadhyay, R. K. Bera and S. K. Sahoo, Microchem. J., 2019, 147, 899–904 CrossRef CAS.
- S. Han and X. Chen, Spectrochim. Acta, Part A, 2019, 210, 315–320 CrossRef CAS PubMed.
- Z. Shen, C. Zhang, X. Yu, J. Li, B. Liu and Z. Zhang, Microchem. J., 2019, 145, 517–522 CrossRef CAS.
- Y.-S. Lin, T.-C. Chiu and C.-C. Hu, RSC Adv., 2019, 9, 9228–9234 RSC.
- Z. Wen, S. Song, C. Wang, F. Qu, T. Thomas, T. Hu, P. Wang and M. Yang, Sens. Actuators, B, 2019, 282, 9–15 CrossRef CAS.
- Y. Cheng, F. Sun and Y. Zhou, J. Lumin., 2018, 197, 376–382 CrossRef CAS.
- C. Boonmee, V. Promarak, T. Tuntulani and W. Ngeontae, Talanta, 2018, 178, 796–804 CrossRef CAS PubMed.
- J. R. Bhamore, B. Deshmukh, V. Haran, S. Jha, R. K. Singhal, N. Lenka, S. K. Kailasa and Z. V. P. Murthy, New J. Chem., 2018, 42, 1510–1520 RSC.
- L. Kong, X. Chu, C. Wang, H. Zhou, Y. Wu and W. Liu, Nanoscale, 2018, 10, 1631–1640 RSC.
- T. Chen, Z. Zhang, Y. Wang and Y. Ni, Gaodeng Xuexiao Huaxue Xuebao/Chemical Journal of Chinese Universities, 2017, 38, 1737–1741 CAS.
- J. Cang, C. W. Wang, P. C. Chen, Y. J. Lin, Y. C. Li and H. T. Chang, Anal. Methods, 2017, 9, 5254–5259 RSC.
- B. Han, X. Hou, R. Xiang and G. He, Anal. Methods, 2017, 9, 4028–4032 RSC.
- M. Zhao, H. Feng, J. Han, H. Ao and Z. Qian, Anal. Chim. Acta, 2017, 984, 202–210 CrossRef CAS PubMed.
- M. Zhao, Z. Qian, M. Zhong, Z. Chen, H. Ao and H. Feng, ACS Appl. Mater. Interfaces, 2017, 9, 32887–32895 CrossRef CAS PubMed.
- Y. Hu, Y. He, Y. Han, Y. Ge, G. Song and J. Zhou, Microchim. Acta, 2019, 186, 5 CrossRef PubMed.
- S. Gou, Y.-e. Shi, P. Li, H. Wang, T. Li, X. Zhuang, W. Li and Z. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 6561–6567 CrossRef CAS PubMed.
- S. Yang, X. Sun and Y. Chen, Mater. Lett., 2017, 194, 5–8 CrossRef CAS.
- J. R. Bhamore, S. Jha, A. K. Mungara, R. K. Singhal, D. Sonkeshariya and S. K. Kailasa, Biosens. Bioelectron., 2016, 80, 243–248 CrossRef CAS PubMed.
- J. Ye, X. Dong, H. Jiang and X. Wang, J. Mater. Chem. B, 2017, 5, 691–696 RSC.
- J.-M. Xia, X. Wei, X.-W. Chen, Y. Shu and J.-H. Wang, Microchim. Acta, 2018, 185, 205 CrossRef PubMed.
- A. Dutta, U. Goswami and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2018, 10, 19459–19472 CrossRef CAS PubMed.
- F. Gao, P. Cai, W. Yang, J. Xue, L. Gao, R. Liu, Y. Wang, Y. Zhao, X. He and L. Zhao, ACS Nano, 2015, 9, 4976–4986 CrossRef CAS PubMed.
- U. Goswami, A. Dutta, A. Raza, R. Kandimalla, S. Kalita, S. S. Ghosh and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2018, 10, 3282–3294 CrossRef CAS PubMed.
- A. Choksi, X. Zhang, G. S. Heo, H. Luehmann and Y. Liu, J. Nucl. Med., 2018, 59, 1122 Search PubMed.
- S. Maity, D. Bain, K. Bhattacharyya, S. Das, R. Bera, B. Jana, B. Paramanik, A. Datta and A. Patra, J. Phys. Chem. C, 2017, 122, 13354–13362 CrossRef.
- P. Yuan, R. Chen, X. Zhang, F. Chen, J. Yan, C. Sun, D. Ou, J. Peng, S. Lin and Z. Tang, Angew. Chem., Int. Ed., 2019, 58, 835–839 CrossRef CAS PubMed.
- Z. Wu, J. Liu, Y. Gao, H. Liu, T. Li, H. Zou, Z. Wang, K. Zhang, Y. Wang and H. Zhang, J. Am. Chem. Soc., 2015, 137, 12906–12913 CrossRef CAS PubMed.
- L. Ai, W. Jiang, Z. Liu, J. Liu, Y. Gao, H. Zou, Z. Wu, Z. Wang, Y. Liu and H. Zhang, Nanoscale, 2017, 9, 12618–12627 RSC.
- Z. Wang, Y. e. Shi, X. Yang, Y. Xiong, Y. Li, B. Chen, W. F. Lai and A. L. Rogach, Adv. Funct. Mater., 2018, 28, 1802848 CrossRef.
- D. Li, G. Wang, L. Cheng, C. Wang and X. Mei, ACS Omega, 2018, 3, 14755–14765 CrossRef CAS PubMed.
- D. Li, G. Wang, Y. Peng, Z. Chen, X. Gao, L. Cheng and X. Mei, Nanoscale Adv., 2019, 1, 1086–1095 RSC.
- J. Li, J.-J. Zhu and K. Xu, TrAC, Trends Anal. Chem., 2014, 58, 90–98 CrossRef CAS.
- A. C. Bhowal, S. Pandit and S. Kundu, J. Phys. D: Appl. Phys., 2019, 52, 015302 CrossRef.
- J. Bornacelli, C. Torres-Torres, H. Silva-Pereyra, G. Labrada-Delgado, A. Crespo-Sosa, J. Cheang-Wong and A. Oliver, Sci. Rep., 2019, 9, 5699 CrossRef CAS PubMed.
- G. S. Yuvasri, N. Goswami and J. Xie, Principles and Applications of Aggregation-Induced Emission, Springer, 2019, pp. 265–289 Search PubMed.
- G. Wang, T. Huang, R. W. Murray, L. Menard and R. G. Nuzzo, J. Am. Chem. Soc., 2005, 127, 812–813 CrossRef CAS PubMed.
- G. Wang, R. Guo, G. Kalyuzhny, J.-P. Choi and R. W. Murray, J. Phys. Chem. B, 2006, 110, 20282–20289 CrossRef CAS PubMed.
- S. Maity, D. Bain and A. Patra, J. Phys. Chem. C, 2019, 123, 2506–2515 CrossRef CAS.
- Y. Wang, Y.-e. Shi, T. Li, H. Wang, Y. Li, Y. Xiong, S. Peng and Z. Wang, Nanoscale Adv., 2019, 1, 834–839 RSC.
- R. Jalili and A. Khataee, Microchim. Acta, 2019, 186, 29 CrossRef PubMed.
- Y. Guo, F. Cao, X. Lei, L. Mang, S. Cheng and J. Song, Nanoscale, 2016, 8, 4852–4863 RSC.
- W. Wei, Y. Lu, W. Chen and S. Chen, J. Am. Chem. Soc., 2011, 133, 2060–2063 CrossRef CAS PubMed.
- Z. Qing, X. He, D. He, K. Wang, F. Xu, T. Qing and X. Yang, Angew. Chem., Int. Ed., 2013, 52, 9719–9722 CrossRef CAS PubMed.
- C. Wang, L. Ling, Y. Yao and Q. Song, Nano Res., 2015, 8, 1975–1986 CrossRef CAS.
- A. W. Cook and T. W. Hayton, Acc. Chem. Res., 2018, 51, 2456–2464 CrossRef CAS PubMed.
- H.-H. Deng, K.-L. Li, Q.-Q. Zhuang, H.-P. Peng, Q.-Q. Zhuang, A.-L. Liu, X.-H. Xia and W. Chen, Nanoscale, 2018, 10, 6467–6473 RSC.
- Z. Wang, B. Chen, A. S. Susha, W. Wang, C. J. Reckmeier, R. Chen, H. Zhong and A. L. Rogach, Adv. Sci., 2016, 3, 1600182 CrossRef PubMed.
- Z. Wu, H. Liu, T. Li, J. Liu, J. Yin, O. F. Mohammed, O. M. Bakr, Y. Liu, B. Yang and H. Zhang, J. Am. Chem. Soc., 2017, 139, 4318–4321 CrossRef CAS PubMed.
- K. D. M. Weerawardene, H. Häkkinen and C. M. Aikens, Annu. Rev. Phys. Chem., 2018, 69, 205–229 CrossRef CAS PubMed.
- Y. Du, H. Sheng, D. Astruc and M. Zhu, Chem. Rev., 2019 DOI:10.1021/acs.chemrev.8b00726.
- Y. Huang, H. Feng, W. Liu, S. Zhang, C. Tang, J. Chen and Z. Qian, J. Mater. Chem. B, 2017, 5, 5120–5127 RSC.
- C. Wang, H. Cheng, Y. Huang, Z. Xu, H. Lin and C. Zhang, Analyst, 2015, 140, 5634–5639 RSC.
- N. K. Das, S. Ghosh, A. Priya, S. Datta and S. Mukherjee, J. Phys. Chem. C, 2015, 119, 24657–24664 CrossRef CAS.
- H. Huang, H. Li, J.-J. Feng, H. Feng, A.-J. Wang and Z. Qian, Sens. Actuators, B, 2017, 241, 292–297 CrossRef CAS.
- J. Liu, Q. M. Zhang, Y. Feng, Z. Zhou and K. Shih, ChemPhysChem, 2016, 17, 225–231 CrossRef CAS PubMed.
- A. Baghdasaryan, R. Grillo, S. Roy Bhattacharya, M. Sharma, E. Reginato, H. Theraulaz, I. Dolamic, M. Dadras, S. Rudaz and E. Varesio, ACS Appl. Nano Mater., 2018, 1, 4258–4267 CrossRef CAS.
- B. Han, X. Hu, M. Yu, T. Peng, Y. Li and G. He, RSC Adv., 2018, 8, 22748–22754 RSC.
- Y. Huang, W. Liu, H. Feng, Y. Ye, C. Tang, H. Ao, M. Zhao, G. Chen, J. Chen and Z. Qian, Anal. Chem., 2016, 88, 7429–7434 CrossRef CAS PubMed.
- H.-Y. Huang, K.-B. Cai, M. J. Talite, W.-C. Chou, P.-W. Chen and C.-T. Yuan, Sci. Rep., 2019, 9, 4053 CrossRef PubMed.
- A. Ganguly, I. Chakraborty, T. Udayabhaskararao and T. Pradeep, J. Nanopart. Res., 2013, 15, 1522 CrossRef.
- X. Liu, W. Ding, Y. Wu, C. Zeng, Z. Luo and H. Fu, Nanoscale, 2017, 9, 3986–3994 RSC.
- S. M. Lin, S. Geng, N. Li, N. B. Li and H. Q. Luo, Talanta, 2016, 151, 106–113 CrossRef CAS PubMed.
- Y. Du, J. Fang, H. Wang and Y. Yang, ACS Appl. Mater. Interfaces, 2017, 9, 11035–11044 CrossRef CAS PubMed.
- Y.-S. Borghei, M. Hosseini, M. Khoobi and M. R. Ganjali, J. Fluoresc., 2017, 27, 529–536 CrossRef CAS PubMed.
- Y. S. Borghei, M. Hosseini, M. Khoobi and M. R. Ganjali, Luminescence, 2017, 32, 1045–1050 CrossRef CAS PubMed.
- N. Goswami, K. Zheng and J. Xie, Nanoscale, 2014, 6, 13328–13347 RSC.
- X. Su and J. Liu, ACS Appl. Mater. Interfaces, 2017, 9, 3902–3910 CrossRef CAS PubMed.
- K. Khonkayan, S. Sansuk, S. Srijaranai, T. Tuntulani, C. Saiyasombat, W. Busayaporn and W. Ngeontae, Microchim. Acta, 2017, 184, 2965–2974 CrossRef CAS.
- S. Sharma, K. K. Chakrahari, J.-Y. Saillard and C. Liu, Acc. Chem. Res., 2018, 51, 2475–2483 CrossRef CAS PubMed.
- M. Cui, G. Song, C. Wang and Q. Song, Microchim. Acta, 2015, 182, 1371–1377 CrossRef CAS.
- T. Zhou, W. Xu, Q. Yao, T. Zhao and X. Chen, Methods Appl. Fluoresc., 2015, 3, 044002 CrossRef PubMed.
- T. Zhou, Q. Yao, T. Zhao and X. Chen, Talanta, 2015, 141, 80–85 CrossRef CAS PubMed.
- Y.-J. Lin, P.-C. Chen, Z. Yuan, J.-Y. Ma and H.-T. Chang, Chem. Commun., 2015, 51, 11983–11986 RSC.
- S. Zhou, Y. Li, F. Wang and C. Wang, RSC Adv., 2016, 6, 38897–38905 RSC.
- K. T. Prakash, N. Singh and V. Venkatesh, Chem. Commun., 2019, 55, 322–325 RSC.
- J.-S. Shen, Y.-L. Chen, Q.-P. Wang, T. Yu, X.-Y. Huang, Y. Yang and H.-W. Zhang, J. Mater. Chem. C, 2013, 1, 2092–2096 RSC.
- X.-J. Zheng, R.-P. Liang, Z.-J. Li, L. Zhang and J.-D. Qiu, Sens. Actuators, B, 2016, 230, 314–319 CrossRef CAS.
- A. Rotaru, S. Dutta, E. Jentzsch, K. Gothelf and A. Mokhir, Angew. Chem., Int. Ed., 2010, 49, 5665–5667 CrossRef CAS PubMed.
- B. Han, R. Xiang, X. Hou, M. Yu, T. Peng, Y. Li and G. He, Anal. Methods, 2017, 9, 2590–2595 RSC.
- F. Zhou, X. Cui, A. Shang, J. Lian, L. Yang, Y. Jin and B. Li, Microchim. Acta, 2017, 184, 773–779 CrossRef CAS.
- Q. Song, Y. Shi, D. He, S. Xu and J. Ouyang, Chem. – Eur. J., 2015, 21, 2417–2422 CrossRef CAS PubMed.
- G. Liu, Y. Shao, J. Peng, W. Dai, L. Liu, S. Xu, F. Wu and X. Wu, Nanotechnology, 2013, 24, 345502 CrossRef PubMed.
- Y. Ling, N. Zhang, F. Qu, T. Wen, Z. F. Gao, N. B. Li and H. Q. Luo, Spectrochim. Acta, Part A, 2014, 118, 315–320 CrossRef CAS PubMed.
- J. Feng, Y. Ju, J. Liu, H. Zhang and X. Chen, Anal. Chim. Acta, 2015, 854, 153–160 CrossRef CAS PubMed.
- M. Zhao, L. Sun and R. M. Crooks, J. Am. Chem. Soc., 1998, 120, 4877–4878 CrossRef CAS.
- N. Vilar-Vidal, J. Rivas and M. A. Lopez-Quintela, ACS Catal., 2012, 2, 1693–1697 CrossRef CAS.
- L. Balogh and D. A. Tomalia, J. Am. Chem. Soc., 1998, 120, 7355–7356 CrossRef CAS.
- P. N. Floriano, C. O. Noble IV, J. M. Schoonmaker, E. D. Poliakoff and R. L. McCarley, J. Am. Chem. Soc., 2001, 123, 10545–10553 CrossRef CAS PubMed.
- R. Ghosh, U. Goswami, S. S. Ghosh, A. Paul and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2014, 7, 209–222 CrossRef PubMed.
- Z. Wang, A. S. Susha, B. Chen, C. Reckmeier, O. Tomanec, R. Zboril, H. Zhong and A. L. Rogach, Nanoscale, 2016, 8, 7197–7202 RSC.
- H. Zhang, X. Huang, L. Li, G. Zhang, I. Hussain, Z. Li and B. Tan, Chem. Commun., 2012, 48, 567–569 RSC.
- Z. Yan, Q. Niu, M. Mou, Y. Wu, X. Liu and S. Liao, J. Nanopart. Res., 2017, 19, 235 CrossRef.
- L. Xiaoqing, L. Ruiyi, L. Zaijun, S. Xiulan, W. Zhouping and L. Junkang, New J. Chem., 2015, 39, 5240–5248 RSC.
- N. Goswami, A. Giri, M. Bootharaju, P. L. Xavier, T. Pradeep and S. K. Pal, Anal. Chem., 2011, 83, 9676–9680 CrossRef CAS PubMed.
- M. Chen, W. Li, H. Xiong, W. Wen, X. Zhang and S. Wang, Microchim. Acta, 2017, 1–8 Search PubMed.
- L. Zhao and Z. Ma, Sens. Actuators, B, 2017, 241, 849–854 CrossRef CAS.
- H. Miao, D. Zhong, Z. Zhou and X. Yang, Nanoscale, 2015, 7, 19066–19072 RSC.
- R. Ghosh, A. K. Sahoo, S. S. Ghosh, A. Paul and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2014, 6, 3822–3828 CrossRef CAS PubMed.
- L. Jin, Z. Zhang, A. Tang, C. Li and Y. Shen, Biosens. Bioelectron., 2016, 79, 108–113 CrossRef CAS PubMed.
- Y. Wang, Y. Cui, R. Liu, Y. Wei, X. Jiang, H. Zhu, L. Gao, Y. Zhao, Z. Chai and X. Gao, Chem. Commun., 2013, 49, 10724–10726 RSC.
- W. Wang, F. Leng, L. Zhan, Y. Chang, X. X. Yang, J. Lan and C. Z. Huang, Analyst, 2014, 139, 2990–2993 RSC.
- Y. Ding, X. Li, C. Chen, J. Ling, W. Li, Y. Guo, J. Yan, L. Zha and J. Cai, Sci. Rep., 2017, 7, 9638 CrossRef PubMed.
- Y. Qiao, T. Xu, Y. Zhang, C. Zhang, L. Shi, G. Zhang, S. Shuang and C. Dong, Sens. Actuators, B, 2015, 220, 1064–1069 CrossRef CAS.
- X. Hu, X. Mao, X. Zhang and Y. Huang, Sens. Actuators, B, 2017, 247, 312–318 CrossRef CAS.
- S. Biswas, J. T. Miller, Y. Li, K. Nandakumar and C. S. Kumar, Small, 2012, 8, 688–698 CrossRef CAS PubMed.
- M. T. Reetz and W. Helbig, J. Am. Chem. Soc., 1994, 116, 7401–7402 CrossRef CAS.
- N. Vilar-Vidal, M. C. Blanco, M. A. López-Quintela, J. Rivas and C. Serra, J. Phys. Chem. C, 2010, 114, 15924–15930 CrossRef CAS.
- S. Huseyinova, J. Blanco, F. l. G. Requejo, J. M. Ramallo-López, M. C. Blanco, D. Buceta and M. A. López-Quintela, J. Phys. Chem. C, 2016, 120, 15902–15908 CrossRef CAS.
- N. Vilar-Vidal, J. R. Rey and M. A. López Quintela, Small, 2014, 10, 3632–3636 CrossRef CAS PubMed.
- X. Jia, J. Li and E. Wang, Small, 2013, 9, 3873–3879 CrossRef CAS PubMed.
- Z. Li, S. Guo and C. Lu, Analyst, 2015, 140, 2719–2725 RSC.
- X. Yuan, Z. Luo, Q. Zhang, X. Zhang, Y. Zheng, J. Y. Lee and J. Xie, ACS Nano, 2011, 5, 8800–8808 CrossRef CAS.
- C. Vázquez-Vázquez, M. Bañobre-López, A. Mitra, M. A. López-Quintela and J. Rivas, Langmuir, 2009, 25, 8208–8216 CrossRef.
- R. K. Koninti, S. Satpathi and P. Hazra, J. Phys. Chem. C, 2018, 122, 5742–5752 CrossRef CAS.
- H. Kawasaki, Y. Kosaka, Y. Myoujin, T. Narushima, T. Yonezawa and R. Arakawa, Chem. Commun., 2011, 47, 7740–7742 RSC.
- C. Toh, X. Liu, P. Ho and J. Chen, IEEE Trans. Magn., 2011, 47, 4003–4006 CAS.
- H. Haberland, M. Karrais, M. Mall and Y. Thurner, J. Vac. Sci. Technol., A, 1992, 10, 3266–3271 CrossRef CAS.
- J. Chen, C. Tan, S. Chow, B. Liu and G. Chow, J. Appl. Phys., 2005, 98, 064306 CrossRef.
- S. Pan, X. Zhang, W. Lu and S. F. Yu, J. Mater. Chem. A, 2018, 6, 18687–18693 RSC.
- M. Hyotanishi, Y. Isomura, H. Yamamoto, H. Kawasaki and Y. Obora, Chem. Commun., 2011, 47, 5750–5752 RSC.
- X. Nie, H. Qian, Q. Ge, H. Xu and R. Jin, ACS Nano, 2012, 6, 6014–6022 CrossRef CAS PubMed.
- S. Jalili, C. Mochani, M. Akhavan and J. Schofield, Mol. Phys., 2012, 110, 267–276 CrossRef CAS.
- M. Liu, X. Qiu, K. Hashimoto and M. Miyauchi, J. Mater. Chem. A, 2014, 2, 13571–13579 RSC.
- M. P. de Lara-Castells, A. W. Hauser, J. M. Ramallo-López, D. Buceta, L. J. Giovanetti, M. A. Lopez-Quintela and F. Requejo, J. Mater. Chem. A, 2019, 7, 7489–7500 RSC.
- B. Beguin, B. Denise and R. Sneeden, J. Organomet. Chem., 1981, 208, C18–C20 CrossRef CAS.
- Q. Tang, Y. Lee, D.-Y. Li, W. Choi, C. W. Liu, D. Lee and D.-e. Jiang, J. Am. Chem. Soc., 2017, 139, 9728–9736 CrossRef CAS PubMed.
- J. Fang, B. Zhang, Q. Yao, Y. Yang, J. Xie and N. Yan, Coord. Chem. Rev., 2016, 322, 1–29 CrossRef CAS.
- C. Sun, N. Mammen, S. Kaappa, P. Yuan, G. Deng, C. Zhao, J. Yan, S. Malola, K. Honkala and H. Häkkinen, ACS Nano, 2019, 13, 5975–5986 CrossRef CAS PubMed.
- B. Zhang, A. Sels, G. Salassa, S. Pollitt, V. Truttmann, C. Rameshan, J. Llorca, W. Olszewski, G. Rupprechter and T. Bürgi, ChemCatChem, 2018, 10, 5372–5376 CrossRef CAS.
- O. Lopez-Acevedo, K. A. Kacprzak, J. Akola and H. Häkkinen, Nat. Chem., 2010, 2, 329–334 CrossRef CAS.
- W. Chen and S. Chen, Angew. Chem., Int. Ed., 2009, 48, 4386–4389 CrossRef CAS.
- Y. Lu and W. Chen, J. Power Sources, 2012, 197, 107–110 CrossRef CAS.
- Y. Lu, W. Wei and W. Chen, Chin. Sci. Bull., 2012, 57, 41–47 CrossRef CAS.
- W. Wei and W. Chen, Int. J. Smart Nano Mater., 2013, 4, 62–71 CrossRef CAS.
- Q.-Y. Zhang, Q.-F. Zhao, X.-M. Liang, X.-L. Wang, F.-X. Ma, B.-B. Suo, W.-L. Zou, H.-X. Han, Q. Song and Q. Wu, Int. J. Hydrogen Energy, 2018, 43, 9935–9942 CrossRef CAS.
- M. Takahashi, H. Koizumi, W.-J. Chun, M. Kori, T. Imaoka and K. Yamamoto, Sci. Adv., 2017, 3, e1700101 CrossRef PubMed.
- L. He, H. Liu, C.-X. Xiao and Y. Kou, Green Chem., 2008, 10, 619–622 RSC.
- B. Sarkar, C. Pendem, L. N. Sivakumar Konathala, R. Tiwari, T. Sasaki and R. Bal, Chem. Commun., 2014, 50, 9707–9710 RSC.
- A. A. Athawale and S. V. Bhagwat, J. Appl. Polym. Sci., 2003, 89, 2412–2417 CrossRef CAS.
- L. Hu, Y. Yuan, L. Zhang, J. Zhao, S. Majeed and G. Xu, Anal. Chim. Acta, 2013, 762, 83–86 CrossRef CAS PubMed.
- D. Tang and J. Zhang, RSC Adv., 2013, 3, 15225–15236 RSC.
- R. S. Dhayal, H. P. Chen, J. H. Liao, W. E. van Zyl and C. Liu, ChemistrySelect, 2018, 3, 3603–3610 CrossRef CAS.
- S. S. Tafreshi, A. Roldan and N. H. de Leeuw, J. Phys. Chem. C, 2014, 118, 26103–26114 CrossRef CAS.
- R. A. Hoyt, M. M. Montemore and E. Kaxiras, J. Phys. Chem. Lett., 2018, 9, 5339–5343 CrossRef CAS.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., 2002, 114, 2708–2711 CrossRef.
- B.-H. Lee, C.-C. Wu, X. Fang, C. Liu and J.-L. Zhu, Catal. Lett., 2013, 143, 572–577 CrossRef CAS.
- Y. Zhong, Q. Wang, Y. He, Y. Ge and G. Song, Sens. Actuators, B, 2015, 209, 147–153 CrossRef CAS.
- J. Liu, Z. Wu, Y. Tian, Y. Li, L. Ai, T. Li, H. Zou, Y. Liu, X. Zhang and H. Zhang, ACS Appl. Mater. Interfaces, 2017, 9, 24899–24907 CrossRef CAS.
- G. Zhang, R. Wang, L. Shi, C. Zhang, Y. Zhang, Y. Zhou, C. Dong, G. Li and S. Shuang, Sens. Actuators, B, 2019, 279, 361–368 CrossRef CAS.
- H. Cao, Z. Chen and Y. Huang, Talanta, 2015, 143, 450–456 CrossRef CAS.
- L. Ruiyi, W. Huiying, Z. Xiaoyan, L. Xiaoqing, S. Xiulan and L. Zaijun, New J. Chem., 2016, 40, 732–739 RSC.
- X. Hu, W. Wang and Y. Huang, Talanta, 2016, 154, 409–415 CrossRef CAS.
- X. Yang, Y. Feng, S. Zhu, Y. Luo, Y. Zhuo and Y. Dou, Anal. Chim. Acta, 2014, 847, 49–54 CrossRef CAS PubMed.
- D. Li, Z. Chen, Z. Wan, T. Yang, H. Wang and X. Mei, RSC Adv., 2016, 6, 34090–34095 RSC.
- L. Lin, Y. Hu, L. Zhang, Y. Huang and S. Zhao, Biosens. Bioelectron., 2017, 94, 523–529 CrossRef CAS PubMed.
- S. M. Lin, S. Geng, N. Li, S. G. Liu, N. B. Li and H. Q. Luo, Sens. Actuators, B, 2017, 252, 912–918 CrossRef CAS.
- H. Y. Zou, J. Lan and C. Z. Huang, RSC Adv., 2015, 5, 55832–55838 RSC.
- N. K. Das, S. Ghosh, A. Priya, S. Datta and S. Mukherjee, J. Phys. Chem. C, 2015, 119, 24657–24664 CrossRef CAS.
- Z. Shojaeifard, N. Heidari and B. Hemmateenejad, Spectrochim. Acta, Part A, 2019, 209, 202–208 CrossRef CAS.
- J. Liu, B. Wang, M. Xu, L. Wang and Z. Zhou, J. Lumin., 2017, 185, 258–262 CrossRef CAS.
- D. Q. Feng, W. Zhu, G. Liu and W. Wang, RSC Adv., 2016, 6, 96729–96734 RSC.
- L. Kong, X. Chu, W. Liu, Y. Yao, P. Zhu and X. Ling, New J. Chem., 2016, 40, 4744–4750 RSC.
- Y. S. Lin, T. C. Chiu and C. C. Hu, RSC Adv., 2019, 9, 9228–9234 RSC.
- Z.-C. Liu, J.-W. Qi, C. Hu, L. Zhang, W. Song, R.-P. Liang and J.-D. Qiu, Anal. Chim. Acta, 2015, 895, 95–103 CrossRef CAS.
- D. Li, B. Li and S. I. Yang, Anal. Methods, 2015, 7, 2278–2282 RSC.
- Z. Shen, C. Zhang, X. Yu, J. Li, B. Liu and Z. Zhang, Microchem. J., 2019, 145, 517–522 CrossRef CAS.
- Y. Huang, W. Liu, H. Feng, Y. Ye, C. Tang, H. Ao, M. Zhao, G. Chen, J. Chen and Z. Qian, Anal. Chem., 2016, 88, 7429–7434 CrossRef CAS.
- P.-C. Chen, Y.-C. Li, J.-Y. Ma, J.-Y. Huang, C.-F. Chen and H.-T. Chang, Sci. Rep., 2016, 6, 24882 CrossRef CAS PubMed.
- J. Y. Ma, P. C. Chen and H. T. Chang, Nanotechnology, 2014, 25, 195502 CrossRef PubMed.
- U. Sivasankaran, J. Radecki, H. Radecka and K. Girish Kumar, Luminescence, 2019, 34, 243–248 CrossRef CAS PubMed.
- Z. Wang, C. C. Zhang, J. Gao and Q. Wang, J. Lumin., 2017, 190, 115–122 CrossRef CAS.
- R. S. Aparna, J. S. Anjali Devi, P. Sachidanandan and S. George, Sens. Actuators, B, 2018, 254, 811–819 CrossRef CAS.
- Z. Wang, R. Chen, Y. Xiong, K. Cepe, J. Schneider, R. Zboril, C. S. Lee and A. L. Rogach, Part. Part. Syst. Charact., 2017, 34, 1700029 CrossRef.
- H. Li, J. Chang, T. Hou, L. Ge and F. Li, Talanta, 2016, 160, 475–480 CrossRef CAS.
- H.-W. Zhu, W.-X. Dai, X.-D. Yu, J.-J. Xu and H.-Y. Chen, Talanta, 2015, 144, 642–647 CrossRef CAS PubMed.
- Y. Nerthigan, A. K. Sharma, S. Pandey and H. F. Wu, Microchim. Acta, 2019, 186(3), 130 CrossRef PubMed.
- X. Zhang, Q. Liu, Y. Jin and B. Li, ChemistrySelect, 2019, 4, 2398–2403 CrossRef CAS.
- R. Liu, L. Zuo, X. Huang, S. Liu, G. Yang, S. Li and C. Lv, Microchim. Acta, 2019, 186, 250 CrossRef.
- H. Jiang and X.-M. Wang, Chin. J. Anal. Chem., 2017, 45, 1776–1785 Search PubMed.
- Q. Song, Y. Shi, D. He, S. Xu and J. Ouyang, Chem. – Eur. J., 2015, 21, 2417–2422 CrossRef CAS PubMed.
- L. Zhao and Z. Ma, Sens. Actuators, B, 2017, 241, 849–854 CrossRef CAS.
- C. Wang, S. Shu, Y. Yao and Q. Song, RSC Adv., 2015, 5, 101599–101606 RSC.
- Y. Zhou, H. Wang, H. Zhang, Y. Chai and R. Yuan, Anal. Chem., 2018, 90, 3543–3549 CrossRef CAS.
- C. Muñoz-Bustos, A. Tirado-Guízar, F. Paraguay-Delgado and G. Pina-Luis, Sens. Actuators, B, 2017, 244, 922–927 CrossRef.
- Z. Gao, R. Su, W. Qi, L. Wang and Z. He, Sens. Actuators, B, 2014, 195, 359–364 CrossRef CAS.
- N. Zhang, F. Qu, H. Q. Luo and N. B. Li, Anal. Chim. Acta, 2013, 791, 46–50 CrossRef CAS PubMed.
- S. Xu, Y. Wang, D. Zhou, M. Kuang, D. Fang, W. Yang, S. Wei and L. Ma, Sci. Rep., 2016, 6, 39157 CrossRef CAS PubMed.
- W. Li, W. Li, Y. Hu, Y. Xia, Q. Shen, Z. Nie, Y. Huang and S. Yao, Biosens. Bioelectron., 2013, 47, 345–349 CrossRef CAS.
- Y. Hu, Q. Zhang, L. Xu, J. Wang, J. Rao, Z. Guo and S. Wang, Anal. Bioanal. Chem., 2017, 409, 6677–6688 CrossRef CAS.
- L. Wang, M. Wang, F. Shi, Z. Liu and X. Su, Sens. Actuators, B, 2017, 252, 209–214 CrossRef CAS.
- H. Zhao, J. Dong, F. Zhou and B. Li, Sens. Actuators, B, 2017, 238, 828–833 CrossRef CAS.
- Y. Zhang, Y. Li, C. Zhang, Q. Zhang, X. Huang, M. Yang, S. A. Shahzad, K. K.-W. Lo, C. Yu and S. Jiang, Anal. Bioanal. Chem., 2017, 409, 4771–4778 CrossRef CAS PubMed.
- Y. Hong, J. W. Y. Lam and B. Z. Tang, Chem. Soc. Rev., 2011, 40, 5361–5388 RSC.
- T. Qing, C. Long, X. Wang, K. Zhang, P. Zhang and B. Feng, Microchim. Acta, 2019, 186, 248 CrossRef PubMed.
- M. Ye, Y. Yu, B. Lin, Y. Cai, Y. Cao, M. Guo and D. Zhu, Sens. Actuators, B, 2019, 284, 36–44 CrossRef CAS.
- Q. Liu, Q. Lai, N. Li and X. Su, Microchim. Acta, 2018, 185, 182 CrossRef PubMed.
- Y. Huang, H. Feng, W. Liu, Y. Zhou, C. Tang, H. Ao, M. Zhao, G. Chen, J. Chen and Z. Qian, Anal. Chem., 2016, 88, 11575–11583 CrossRef CAS PubMed.
- X.-d. Wang, O. S. Wolfbeis and R. J. Meier, Chem. Soc. Rev., 2013, 42, 7834–7869 RSC.
- X. Ma, Y. Wang, T. Zhao, Y. Li, L.-C. Su, Z. Wang, G. Huang, B. D. Sumer and J. Gao, J. Am. Chem. Soc., 2014, 136, 11085–11092 CrossRef CAS PubMed.
- L. Xiaoqing, L. Ruiyi, L. Xiaohuan and L. Zaijun, RSC Adv., 2015, 5, 48835–48841 RSC.
- S. Uchiyama, Y. Matsumura, A. P. de Silva and K. Iwai, Anal. Chem., 2003, 75, 5926–5935 CrossRef CAS PubMed.
- F. H. C. Wong, D. S. Banks, A. Abu-Arish and C. Fradin, J. Am. Chem. Soc., 2007, 129, 10302–10303 CrossRef CAS PubMed.
- L. Wang, H. Miao, D. Zhong and X. Yang, Anal. Methods, 2016, 8, 40–44 RSC.
- T. Zhao, X.-W. He, W.-Y. Li and Y.-K. Zhang, J. Mater. Chem. B, 2015, 3, 2388–2394 RSC.
- M. Karimi, P. Sahandi Zangabad, A. Ghasemi, M. Amiri, M. Bahrami, H. Malekzad, H. Ghahramanzadeh Asl, Z. Mahdieh, M. Bozorgomid and A. Ghasemi, ACS Appl. Mater. Interfaces, 2016, 8, 21107–21133 CrossRef CAS PubMed.
- P. S. Zangabad, S. Mirkiani, S. Shahsavari, B. Masoudi, M. Masroor, H. Hamed, Z. Jafari, Y. D. Taghipour, H. Hashemi and M. Karimi, Nanotechnol. Rev., 2018, 7, 95–122 CAS.
- Z. Liu, X. Jin, S. Zhang and Y. Tian, Anal. Chem., 2019, 91, 2488–2497 CrossRef CAS PubMed.
- S. Shahsavari, J. Nanomed. Res., 2017, 5, 000101 Search PubMed.
- R. Siegel, D. Naishadham and A. Jemal, Ca-Cancer J. Clin., 2013, 63, 11–30 CrossRef PubMed.
- M. Shokeen and C. J. Anderson, Acc. Chem. Res., 2009, 42, 832–841 CrossRef CAS PubMed.
- H. Lyon, Biotech. Histochem., 2002, 77, 57–80 CrossRef CAS PubMed.
- X. Zhu, H. Shi, Y. Shen, B. Zhang, J. Zhao and G. Li, Nano Res., 2015, 8, 2714–2720 CrossRef CAS.
- K. Nakatani, ChemBioChem, 2004, 5, 1623–1633 CrossRef CAS PubMed.
- X. Jia, J. Li, L. Han, J. Ren, X. Yang and E. Wang, ACS Nano, 2012, 6, 3311–3317 CrossRef CAS PubMed.
- A. Abi and E. E. Ferapontova, J. Am. Chem. Soc., 2012, 134, 14499–14507 CrossRef CAS PubMed.
- X. Zhu, S. Liu, J. Cao, X. Mao and G. Li, Sci. Rep., 2016, 6, 19515 CrossRef CAS PubMed.
- K. Zheng, M. I. Setyawati, D. T. Leong and J. Xie, ACS Nano, 2017, 11, 6904–6910 CrossRef CAS.
- S. Wang, Y. Wang, Y. Peng and X. Yang, ACS Appl. Mater. Interfaces, 2019, 11, 8461–8469 CrossRef CAS PubMed.
- A. T. Simon, D. Dutta, A. Chattopadhyay and S. S. Ghosh, ACS Omega, 2019, 4, 4697–4706 CrossRef CAS PubMed.
- S. Kundu and A. Patra, Chem. Rev., 2016, 117, 712–757 CrossRef.
- H. Choi, Y.-S. Chen, K. G. Stamplecoskie and P. V. Kamat, J. Phys. Chem. Lett., 2014, 6, 217–223 CrossRef.
- K. G. Stamplecoskie and P. V. Kamat, J. Am. Chem. Soc., 2014, 136, 11093–11099 CrossRef CAS PubMed.
- S. C. Erwin, L. Zu, M. I. Haftel, A. L. Efros, T. A. Kennedy and D. J. Norris, Nature, 2005, 436, 91–94 CrossRef CAS PubMed.
- Z. Wang, B. Chen, M. Zhu, S. V. Kershaw, C. Zhi, H. Zhong and A. L. Rogach, ACS Appl. Mater. Interfaces, 2016, 8, 33993–33998 CrossRef CAS PubMed.
- Y. Liu, D. Yao and H. Zhang, ACS Appl. Mater. Interfaces, 2017, 10, 12071–12080 CrossRef PubMed.
- H. Wang, Z. Wang, Y. Xiong, S. V. Kershaw, T. Li, Y. Wang, Y. Zhai and A. L. Rogach, Angew. Chem., Int. Ed., 2019, 58, 7040–7044 CrossRef CAS PubMed.
- Z. Wang, Y. Xiong, S. V. Kershaw, B. Chen, X. Yang, N. Goswami, W.-F. Lai, J. Xie and A. L. Rogach, Chem. Mater., 2017, 29, 10206–10211 CrossRef CAS.
- Y. e. Shi, X. Zhuang, L. Cao, S. Gou, Y. Xiong, W. F. Lai, Z. Wang and A. L. Rogach, ChemNanoMat, 2019, 5, 110–115 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © the Partner Organisations 2019 |