Open Access Article
Open Access ArticleIn silico identification of protein targets for chemical neurotoxins using ToxCast in vitro data and read-across within the QSAR toolbox†
Y. G.
Chushak
 *a,
H. W.
Shows
b,
J. M.
Gearhart
a and
H. A.
Pangburn
c
*a,
H. W.
Shows
b,
J. M.
Gearhart
a and
H. A.
Pangburn
c
aHenry M Jackson Foundation for the Advancement of Military Medicine, Wright-Patterson AFB, Ohio 45433, USA. E-mail: Chushak@yahoo.com
bBiological Sciences Department, Wright State University, Dayton, Ohio 45435, USA
cUnited States Air Force School of Aerospace Medicine, Aeromedical Research Department, Force Health Protection, Wright-Patterson AFB, Ohio 45433, USA
First published on 12th March 2018
Abstract
There are many mechanisms of neurotoxicity that are initiated by the interaction of chemicals with different neurological targets. Under the U.S. Environmental Protection Agency's ToxCast program, the biological activity of thousands of chemicals was screened in biochemical and cell-based assays in a high-throughput manner. Two hundred sixteen assays in the ToxCast screening database were identified as targeting a total of 123 proteins having neurological functions according to the Gene Ontology database. Data from these assays were imported into the Organization for Economic Co-operation and Development QSAR Toolbox and used to predict neurological targets for chemical neurotoxins. Two sets of data were generated: one set was used to classify compounds as active or inactive and another set, composed of AC50s for only active compounds, was used to predict AC50 values for unknown chemicals. Chemical grouping and read-across within the QSAR Toolbox were used to identify neurologic targets and predict interactions for pyrethroids, a class of compounds known to elicit neurotoxic effects in humans. The classification prediction results showed 79% accuracy while AC50 predictions demonstrated mixed accuracy compared with the ToxCast screening data.
1 Introduction
Every day humans are exposed to thousands of manufactured chemicals. Some of these chemicals, such as organic solvents or pesticides, can interact with neurological proteins in the brain and cause neurotoxic effects leading to headaches, altered sensation or motor skills, impaired memory and cognitive functions, behavioural problems, even paralysis and death. The neurotoxicity of chemicals greatly depends on their interactions with neurological targets. The recently introduced adverse outcome pathway1 (AOP) framework links these molecular interactions (Molecular Initiating Event) with a series of key events on different biological levels that result in an adverse outcome effect. Within the AOP framework, neurotoxicity can be defined as an adverse effect on the functioning of the nervous system.2With the recent advances made in the field of in vitro high-throughput screening (HTS), it is now possible to screen the biological activity of large chemical libraries in a cost efficient and timely manner. In 2006, the U.S. Environmental Protection Agency (EPA) initiated the ToxCast program to develop and evaluate in vitro biochemical and cell-based assays for screening thousands of chemicals at multiple concentrations in the high-throughput mode.3 In Phase II of the program, approximately 1800 compounds were tested in ∼900 HTS assays. The chemicals in the library included pesticides, commercial compounds, and some failed pharmaceuticals. In 2008, the ToxCast program was merged with a large multiagency Tox21 collaboration. Under this new program, ∼8400 chemicals were screened in ∼70 HTS assays.4 These in vitro screenings generated an enormous volume of data, which are publicly available at https://www.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data. Although ToxCast and other in vitro screening programs provide a significant amount of information about the biological activities of thousands of chemicals, a great deal of information for millions of chemicals remains missing. Computational methods together with the HTS data offer a great opportunity to partially address this data gap and to identify molecular targets and other endpoints for chemical toxins of interest.
According to the European Chemical Agency (ECHA) guidance on information requirements and chemical safety assessment,5 two computational methods – (quantitative) structure–activity relationship [(Q)SAR] and grouping of chemicals with read-across – can be used for evaluating the intrinsic properties of chemicals. Both methods are based on the similarity principle, i.e., similar molecules have similar properties and biological activities are defined by the molecular structure.6 (Q)SAR methods are statistical in nature, as they try to correlate the molecular descriptors of chemicals with their properties. Furthermore, these methods are global in their scope, as they build models for all chemicals in the training dataset and make predictions for a wide range of chemicals within the applicability domain. QSAR modelling was applied to develop predictive models based on ToxCast HTS data. Some of the models were successful,7–9 while other QSAR models yielded low predictive performance.10,11
Grouping of chemicals into a category and read-across is another important technique for data gap filling in chemical hazard assessment. This approach is local in scope, as its predictions are based on the properties of a small set of similar chemicals. The OECD Guidance on Grouping of Chemicals defines a chemical category as a group of chemicals whose physico-chemical and toxicological properties are similar or follow a regular pattern as a result of structural similarity.12 The similarities may be based on common functional groups, common modes or mechanisms of action, common constituents or chemical classes, etc. Read-across is a technique to predict the unknown properties of chemicals of interest based on the known properties of chemicals in the same chemical group.5 Grouping of chemicals and the read-across technique are implemented in several freely available tools such as QSAR Toolbox,13 Toxmatch,14 and ToxRead.15 QSAR Toolbox is a software platform developed by the Organisation for Economic Co-operation and Development (OECD) in collaboration with the European Chemical Agency, intended to be used to fill data gaps in the hazard assessment of chemicals. QSAR Toolbox v.3.5 has a database with about 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 chemicals provided by governmental and commercial institutions. Furthermore, it allows access to import custom databases and use data for hazard assessment. The main aim of the present study was to explore the application of the QSAR Toolbox and data from ToxCast HTS assays to identify and predict the molecular interactions of chemical neurotoxins with their targets.
000 chemicals provided by governmental and commercial institutions. Furthermore, it allows access to import custom databases and use data for hazard assessment. The main aim of the present study was to explore the application of the QSAR Toolbox and data from ToxCast HTS assays to identify and predict the molecular interactions of chemical neurotoxins with their targets.
Recently, activities of 86 compounds from the ToxCast library were tested in neuronal cultures on multi-well microelectrode arrays (MEAs).16 Activities of these compounds on MEAs were compared with their activities on 20 ToxCast binding assays that measured the interaction of chemicals with 8 different ion channels. In our approach, we identified 123 proteins from ToxCast HTS assays that are related to neurological functions. This set of proteins includes ion channels, G-protein coupled receptors, nuclear receptors, transporters, and enzymes as potential neurological targets. The developed approach was evaluated by predicting neurological targets for pyrethroids and comparing the predicted results with ToxCast screening data.
2 Materials and methods
2.1 ToxCast compound dataset
The Tox21/ToxCast dataset released in October 2015 consists of 9076 chemicals tested in 1193 cellular and biochemical assays.17 These assays were developed across multiple human and animal cell lines by several providers including Attagene Inc. (marked as ATG), BioSeek (BSK), NIH Chemical Genomics Center (Tox21), and NovaScreen (NVS), among others. However, not all chemicals were tested in all of the assays. The majority of biochemical assays related to the activity of neurological proteins, such as ligand-gated ion channels and G-protein coupled receptors, were screened in the NovaScreen assay platform. Therefore, for our analysis we selected a subset of 1077 chemicals that were all screened in NVS assays. Furthermore, we reduced this subset by eliminating mixtures and compounds without the molecular description of their structure in the SMILES (Simplified Molecular Input Line-Entry System) format. As a result, the final subset contained 1050 chemicals that were screened in 656 ToxCast HTS assays.2.2 Bioactivity data associated with neurotoxicity
The Tox21/ToxCast HTS assays targeted 342 different proteins. Using the Gene Ontology (GO) database, we have identified that 123 of these proteins have neurological functions. To identify proteins that are related to neuronal functions, we used three terms in the GO search: “neurological”, “synapse”, and “axon”. This search identified 2499 unique proteins related to neurological functions, and 123 of these proteins were screened in 216 ToxCast assays. Data from these assays were imported into the QSAR Toolbox and used in further analysis. The chemical concentration at half maximum efficacy AC50 (in μM) was used to identify chemical-assay activities. Two sets of data were generated: one set coded with a “1” for active compounds and a “0” for inactive compounds was used for classification, while the second dataset containing AC50s for only active compounds was used for prediction of AC50 values for unknown chemicals of interest.2.3 Performance evaluation
To evaluate the performance of ToxCast HTS assays on chemical neurotoxins, compounds with known protein interactions from two databases were used: DrugBank (DB) (https://www.drugbank.ca/) and the Ki database from the Psychoactive Drug Screening Program (PDSP) (https://kidbdev.med.unc.edu/databases/kidb.php).The DB database combines detailed drug data with comprehensive drug–target information18 containing 8261 drugs and 4338 non-redundant proteins that are linked to these drug entries. Twenty-nine chemicals from the DB database were screened in selected ToxCast assays and were used to evaluate the activity of neurological proteins in ToxCast screening.
The PDSP Ki database, which is funded by the U.S. National Institute of Mental Health Psychoactive Drug Screening Program, serves as a data warehouse for published and internally derived Ki, or affinity, values for a large number of drugs and drug candidates at an expanding number of G-protein coupled receptors, ion channels, transporters, and enzymes.19 Currently, it has ∼60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Ki values. Seventeen chemicals from this database were tested on their targets in selected ToxCast HTS assays and were used to evaluate the performance of ToxCast assays.
000 Ki values. Seventeen chemicals from this database were tested on their targets in selected ToxCast HTS assays and were used to evaluate the performance of ToxCast assays.
Another database, the Toxin and Toxin Target Database (http://www.t3db.ca/), also provides the mechanisms of toxicity and target proteins for toxins. However, ToxCast HTS data are already included in this database for chemical–protein associations. Therefore, this database information was not used for evaluation of ToxCast screening assays to avoid bias.
2.4 Software
Data processing and management were performed using SQLite v.3 SQL database engine (https://www.sqlite.org/). Grouping of chemicals into a category and read-across was performed within OECD QSAR Toolbox v. 3.5. The compounds were grouped by organic functional groups and by structural similarity.3 Results and discussion
In the presented approach, we used 1050 chemicals from the ToxCast dataset that were screened in 656 ToxCast high-throughput assays. Among the 342 different proteins that were screened in the ToxCast HTS assays, there are 123 proteins with neurological functions identified according to the Gene Ontology database. The distribution of active chemicals for these proteins was exceptionally skewed, with some proteins exhibiting >450 active chemicals, regressing to some with only two (Fig. 1). The top 22 neurological proteins with more than 100 active chemicals are shown in the inset of Fig. 1. Detailed information on these 22 proteins, together with their neurological functions and curated associations with mental and nervous system diseases obtained from the Comparative Toxicogenomics Database (http://ctdbase.org), is presented in ESI Table S1.† The estrogen receptor (ESR1), well-known for its promiscuous interactions with structurally diverse chemicals that can potentially cause a range of adverse outcome effects,20 demonstrated the highest number of interacting chemicals (453). ESR1 is associated with several nervous system diseases including migraine21 and Alzheimer's disease.22 Recent studies indicate that ESR1 antagonists play a crucial role in neuroinflammation and neurodegeneration.23 Two other proteins with more than 300 active chemicals are HLA class II histocompatibility antigen (HLA-DRA) with 414 active chemicals and C–C motif chemokine (CCL2) with 386 active chemicals. HLA-DRA is associated with Parkinson's disease24 and multiple sclerosis,25 while CCL2 is involved in a variety of neuroinflammatory26 and neurodegenerative27 diseases (see ESI Table S1† for more complete information). Furthermore, these three proteins have 184 common active chemicals, e.g., bisphenol B, DDT (dichlorodiphenyltrichloroethane), and PFDA (perfluorodecanoic acid). On the other end of the spectrum, low chemical–protein interaction, 16 neurological proteins have less than 10 active chemicals, which correspond to less than 1% of the screened compounds. Among the proteins with a low number of active chemicals detected by the ToxCast HTS assays are important neuronal proteins such as GABA receptors (GABRA5, GABRA6, GABBR1), glutamate receptors GRIK1 and GRM5, glycine receptor GLRA1, and voltage gated calcium ion channel CACNA1B. The low number of active chemicals for these proteins limits the predictability for these proteins for new unknown chemicals.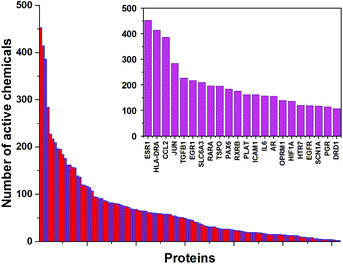 | ||
| Fig. 1 Distribution of the number of active chemicals for neurological proteins. The inset shows 22 proteins with more than 100 active chemicals. | ||
As previously mentioned, neurological proteins were screened in 263 ToxCast high-throughput assays. In the ToxCast screening protocol, initially all chemicals were screened in each assay at a single concentration of 25 μM (10 μM for CYP assays) to identify active chemical-assay combinations in which the mean assay signal differed by at least 30% from the DMSO control signal.28 Next, for active chemical-assay pairs, each chemical was run in eight point serial dilutions starting from a top concentration of 50 μM down to 0.023 μM to estimate the AC50s. We generated two sets of data for neurological proteins from the ToxCast dataset: one set for classification of compounds as active or inactive and another set for prediction of AC50 values which were subsequently imported into the QSAR Toolbox to identify and predict the molecular interactions of chemical neurotoxins with their targets.
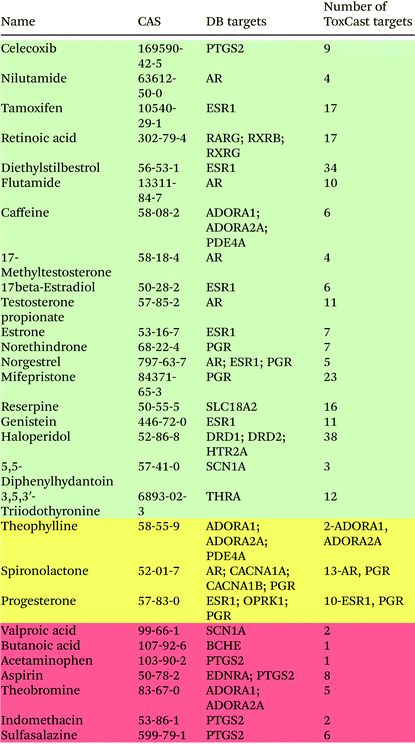
We evaluated the performance of the ToxCast HTS assays on neurological proteins by using chemical neurotoxins with known mechanisms of action from the two databases: DrugBank and Ki databases from the Psychoactive Drug Screening Program. Twenty-nine chemicals from the DB database were screened in ToxCast assays on neurological protein targets. The results of the comparison of drug–target pairs from DB and ToxCast screening are shown in Table 1. Only proteins that were screened in ToxCast assays are presented in this table. Chemicals in green rows were found to have all of their targets from DB represented in the ToxCast database. For chemicals in the yellow rows, only some DB chemical–target pairs were reproduced in ToxCast screening, and for chemicals in the red rows, their targets were not present in the ToxCast screening. Overall, there are 46 chemical–protein interactions between the selected compounds and neurological proteins in the DB database. Thirty-three of these interactions, corresponding to ∼72% of the interactions, were reproduced in the ToxCast HTS screening. ESR1 is a target for seven of the screened drugs in the DB database, and it was identified as a target for all selected chemicals in the ToxCast screening. The androgen receptor (AR) interacts with six drugs in DB, and the progesterone receptor (PGR) interacts with five drugs; all these interactions were also identified in ToxCast screening. Among the 13 missing chemical–protein pairs, 4 of them involve the interactions of prostaglandin-endoperoxide synthase PTGS2 with common analgesics such as aspirin or acetaminophen. Only one interaction of celecoxib with PTGS2 was identified in ToxCast screening.
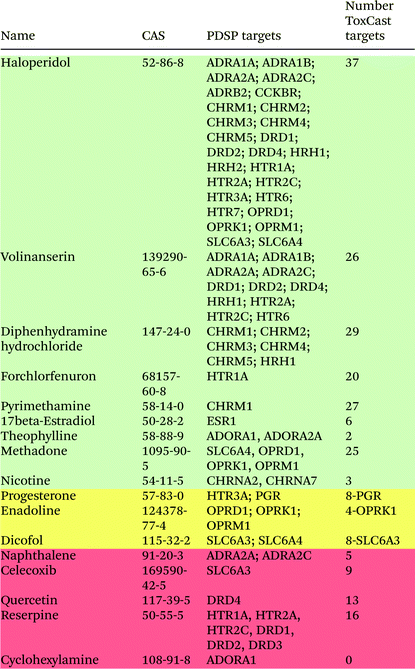
Seventeen chemicals from the Ki database were screened in the ToxCast assays, and their neurological targets are shown in Table 2. The colouring schema in Table 2 is the same as in Table 1. Although only 17 chemicals from the Ki database were screened in the ToxCast assays, there are 74 chemical–protein interactions. The majority of these chemical–protein pairs represent the interactions of two drugs – haloperidol and volinanserin – with their targets. Haloperidol, which is used to treat schizophrenia and other psychoses, has 5 protein targets in the DB database (only 3 of them were used in the ToxCast screening), but it has 27 protein targets in the PDSP database. In the ToxCast screening, haloperidol was active on 37 neurological protein targets, which indicates the very high activity of this drug having potential side effects. Volinanserin was tested in clinical trials as a potential antipsychotic drug, but it was never advanced to the market place. It interacts with 11 protein targets in the PDSP database and has 26 interactions in the ToxCast screening assays. Among the chemicals whose protein interactions were not identified in the ToxCast screening, celecoxib and reserpine are also present in the DB database. The anti-inflammatory drug celecoxib interacts with prostaglandin synthase PTGS2 in DB, and this interaction was captured in ToxCast screening. However, its interaction with the sodium-dependent dopamine transporter SLC6A3 from the Ki database was not observed. The antipsychotic drug reserpine has the synaptic vesicular amine transporter SLC18A2 listed as a target in DB, which was identified in ToxCast, while in PDSP it interacts with serotonin and dopamine receptors that were not observed in the ToxCast screening. Overall, out of 73 chemical–protein interactions between the selected compounds and neurological proteins in the Ki database, 58 chemical–protein pairs that correspond to 80% of interactions were identified in the ToxCast screening assays.
The neuronal activities of 86 compounds from the ToxCast library were recently examined in primary cortical cultures with MEAs.16 The changes in the weighted mean firing rate were used as indicators of chemical activities on neuronal networks. The effects of tested chemicals on spontaneous neuronal activity in MEAs were compared with their activities on 20 ToxCast assays that measured the interaction of chemicals with ion channels. Pyrethroids were among the classes of chemicals that affected neuronal activity in MEAs. The primary mechanism of pyrethroid neurotoxicity is proposed to be via their effects on the sodium channels of nerve cells.29 However, only one of the six studied pyrethroid compounds – allethrin – showed activity in the ToxCast ion channel assays, while all but permethrin were active in MEA measurements.16 This indicates the presence of different mechanisms of toxicity for pyrethroids. We selected chemicals from this group to evaluate the predictive capability of the proposed approach.
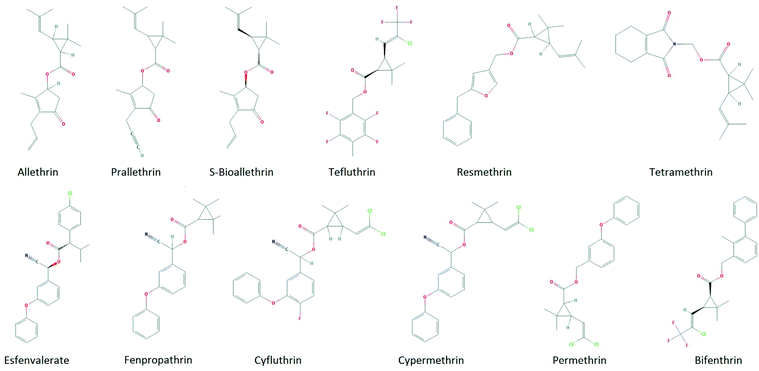
The first step in the read-across approach is to identify a group of chemicals from the ToxCast screening database that belong to the same class as a studied compound. There are several ways to perform a similarity search in the QSAR Toolbox (“Category Definition”). We used two terms, “pyrethroids” and “esters”, to identify pyrethroids in the ToxCast database. The search recognized 12 compounds that are shown in Fig. 2. Two different types of pyrethroids are recognized based on the differences in their structure and the symptoms of poisoning.29 Structures of four of the selected compounds include a cyano group, and these chemicals belong to the Type II pyrethroid group, while the remaining chemicals, which lack a cyano group, belong to the Type I pyrethroid group.
The majority of pyrethroids are active in 10 to 20 ToxCast HTS assays with neurological proteins (Fig. 3). However, allethrin shows very high activity as it is active in 26 assays, while bifenthrin is only active in 4 assays and tefluthrin in 6 assays. This is a clear indication that some compounds in the pyrethroid group have unique structural features that are reflected in their distinctive activities. As for neuronal targets, the ATG_DR5_CIS assay that targeted retinoic acid receptors RARA, RARB, and RARG exhibited activity for 10 of the 12 pyrethroids examined. Another assay that also targeted RARA – ATG_RARa_TRANS – has seven active compounds, indicating that retinoic acid receptors are targeted by pyrethroids.
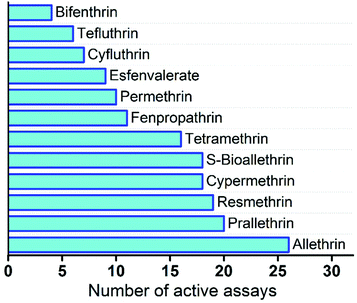 | ||
| Fig. 3 Number of active assays with neurological proteins for pyrethroids from the database of chemicals. | ||
Another potential target of pyrethroid compounds is the dopamine transporter SLC63. Two assays that targeted human and guinea pig transporters – NVS_TR_hDAT and NVS_NVS_gDAT – have nine and five active pyrethroids, respectively. Among the neuronal proteins with only one or two active chemicals are GABA receptor GABRA1, which is affected only by allethrin, and muscarinic cholinergic receptors CHRM1 and CHMR3, which are affected by prallethrin. Since we can only make predictions for protein targets that are affected by a group of chemicals and not just by a single chemical, we selected a set of 14 ToxCast assays with 5 or more active pyrethroids to assess the predictive capability of the proposed approach. The detailed information about these assays is presented in ESI Table S2.† Eight of the studied assays are cell-based assays and the effect of chemicals was investigated by measuring the activity of proteins or changes in the protein expression level. Five of the biochemical assays measured protein–ligand binding, while in one assay enzymatic activity was measured to evaluate the activity of chemicals.
The predictions for pyrethroids were based on the activity of five nearest neighbour chemicals, and data from target chemicals were not used in the prediction. The detailed classification prediction results are presented in ESI Table S3.† For four compounds – allethrin, S-bioallethrin, prallethrin, and fenpropathrin – activity in only one of the studied assays was incorrectly predicted. On the other hand, activities for tetramethrin and for bifenthrin in 7 and 5 assays, respectively, were incorrectly predicted. Tetramethrin is the only compound among the studied pyrethroids that has an imide group that potentially explains the high number of incorrect predictions. Bifenthrin, as mentioned before, showed extremely low activity in the ToxCast screening, as it was active with neurological proteins only in four assays (see Fig. 3). The summary of classification predictions for pyrethroid compounds is presented in Table 3. Overall, there are 168 chemical–protein pairs; 89 are active and 79 are inactive. Eighty three percent of active interactions, 73 of 89 interactions in the ToxCast database, were correctly predicted using our approach. We also correctly predicted 60 inactive chemical–protein pairs, which corresponds to a 76% hit rate.
| ToxCast | Predicted | ||
|---|---|---|---|
| Active | Inactive | Hit rate | |
| Active | 73 | 16 | 83% |
| Inactive | 19 | 60 | 76% |
| Accuracy | 79% | 79% | |
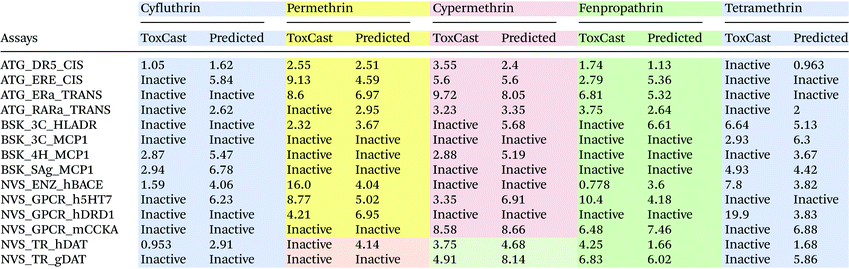
Table 4 shows the predicted AC50 values (in mg L−1) for a set of five pyrethroid compounds in comparison with the ToxCast screening results. The AC50 prediction results for all 12 pyrethroids are presented in ESI Table S4.† For cyfluthrin, we correctly predicted activity in 11 out of 14 assays. The predicted AC50 values for cyfluthrin are in good correlation with those observed experimentally. Activity predictions for permethrin yielded incorrect results for two assays while only a few predicted AC50 values were in agreement with those measured in ToxCast assays. In several in vitro assays, permethrin shows very low activity, with AC50 values in the range of 8.6–16.0 mg L−1. This is in agreement with the low potency of permethrin, which showed no effect on the neuronal network activity measured using the MEA technique.16 The predicted values are based on the values measured for a group of similar chemicals. As a result, the predicted AC50 values for permethrin in these assays are lower compared with the observed. For cypermethrin, only the BSK_3C_HLADR assay activity was incorrectly predicted, while the predicted AC50 values show, in general, a good correlation with the measured ones. Although tetramethrin is active in 16 neurological assays, it is active only in 5 of the assays selected for the evaluation. As mentioned before, classification incorrectly predicted the activity of tetramethrin in seven of the ToxCast assays. The predicted AC50 values also showed mixed accuracy compared with the ToxCast data. These results indicate that it is currently difficult to predict the activity of a compound that has different structural and activity properties compared with the other chemicals in the same group.
4 Conclusions
The neurotoxicity of chemicals greatly depends on their interactions with neurological targets. We used ToxCast in vitro HTS data and the OECD QSAR Toolbox to identify and predict AC50 values for the interaction of chemical neurotoxins with their protein targets. The developed approach was evaluated by predicting chemical–protein interactions for a set of pyrethroid compounds. The classification prediction results showed 79% accuracy while AC50 predictions demonstrated mixed accuracy compared with the ToxCast screening data. Several observed challenges of the proposed approach need to be highlighted.Grouping of chemicals. The first and most critical part in the read-across prediction is finding similar chemicals in the database. There is no unique way for grouping chemicals into a category. Therefore, the same chemical can belong to different groups based on the similarity criteria.30 We used two terms to identify pyrethroids in the ToxCast database, “pyrethroids” and “esters”. However, 12 pyrethroid compounds that were identified showed very different activities in the ToxCast assays. For example, allethrin is active in 26 assays, while bifenthrin is active only in 4 assays and tefluthrin is active in 6 assays. Furthermore, this set of pyrethroids contains eight Type I and four Type II pyrethroids classified according to eliciting different symptomology. Clearly, the difference in the chemical structure of these chemicals resulted in their different activities. One way to improve the similarity is to subcategorize the list of chemicals based on the structural similarity on a chemical-by-chemical basis.
Low activity of chemicals. In some cases, a chemical showed low activity in a ToxCast assay with multiple points above the baseline, but the AC50 value could not be calculated from the dose–response data.28 In such cases, chemicals were marked as “inactive” in the ToxCast database, making it difficult to identify the active/inactive boundary for chemicals with low activity.
The number of active/inactive compounds in the classification dataset should be similar. The ATG_DR5_CIS assay has 10 active pyrethroids out of 12 and several assays have 9 active pyrethroids. It is impossible to predict inactive compounds for ATG_DR5_CIS (like tetramethrin and bifenthrin) when 10 neighbours are active in the assay. The same is true also for assays that have just 1–3 active compounds. Therefore, these assays were not considered in the current approach. In this case, a different grouping schema needs to be applied that will identify a different set of similar chemicals.
Descriptors and applicability domain. The QSAR Toolbox allows the use of a different set of molecular descriptors for read-across prediction. Special attention is needed when selecting descriptors to be sure that they show correlation with the AC50 values and the tested chemical is within the applicability domain for the selected descriptors. Another approach is to use trend analysis, which can improve the predicted results when the chemical is outside the applicability domain for the selected descriptor. However, that approach was not used in the current study.
Our results demonstrate that ToxCast in vitro screening data and the QSAR Toolbox can be used to identify neurological targets and predict AC50 values for chemical neurotoxins that have similar chemicals in the ToxCast database. This approach can be combined with MEA measurements to link molecular interactions to cellular response. Very recently, MEA techniques were used to evaluate the effect of a set of pyrethroids on the spontaneous activity of cortical neuronal networks.31 Besides the weighted mean firing rate, authors also measured other burst parameters such as the mean burst duration, the mean interspike interval in burst, etc. In the present study, we show that pyrethroids affect multiple protein targets that can result in complex neurological responses. Therefore, it is of interest to investigate the correlation of molecular interactions with the multiple endpoints in MEA measurements. Such a combined in vitro/in silico approach would allow us to connect the MIE with the cellular response of chemical neurotoxins.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research was funded by the Defence Health Program under the contract RSAAC 17-003 and by the US Air Force School of Aerospace Medicine. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Air Force, the Department of Defence, or the U.S. Government.References
- M. Vinken, The adverse outcome pathway concept: a pragmatic tool in toxicology, Toxicology, 2013, 312, 158–165 CrossRef CAS PubMed.
- A. Bal-Price, K. M. Crofton, M. Sachana, T. J. Shafer, M. Behl, A. Forsby, A. Hargreaves, B. Landesmann, P. J. Lein, J. Louisse, F. Monnet-Tschudi, A. Paini, A. Rolaki, A. Schrattenholz, C. Suñol, C. van Thriel, M. Whelan and E. Fritsche, Putative adverse outcome pathways relevant to neurotoxicity, Crit. Rev. Toxicol., 2015, 45, 83–91 CrossRef CAS PubMed.
- D. J. Dix, K. A. Houck, M. T. Martin, A. M. Richard, R. W. Setzer and R. J. Kavlock, The ToxCast program for prioritizing toxicity testing of environmental chemicals, Toxicol. Sci., 2007, 95, 5–12 CrossRef CAS PubMed.
- R. R. Tice, C. P. Austin, R. J. Kavlock and J. R. Bucher, Improving the human hazard characterization of chemicals: a Tox21 update, Environ. Health Perspect., 2013, 121, 756–765 CrossRef PubMed.
- Guidance on information requirements and chemical safety assessment, Chapter R.6: QSARs and grouping of chemicals, https://echa.europa.eu/guidance-documents/guidance-on-information-requirements-and-chemical-safety-assessment, (accessed January 2016).
- G. Patlewicz and J. M. Fitzpatrick, Current and future perspectives on the development, evaluation, and application of in silico approaches for predicting toxicity, Chem. Res. Toxicol., 2016, 29, 438–451 CrossRef CAS PubMed.
- J. Liu, K. Mansouri, R. S. Judson, M. T. Martin, H. Hong, M. Chen, X. Xu, R. S. Thomas and I. Shah, Predicting hepatotoxicity using ToxCast in vitro bioactivity and chemical structure, Chem. Res. Toxicol., 2015, 28, 738–751 CrossRef CAS PubMed.
- K. Mansouri and R. S. Judson, in In Silico Methods for Predicting Drug Toxicity, ed. E. Benfenati, Springer, New York, 2016, ch. 16, pp. 361–381 Search PubMed.
- K. Mansouri, A. Abdelaziz, A. Rybacka, A. Roncaglioni, A. Tropsha, A. Varnek, A. Zakharov, A. Worth, A. M. Richard, C. M. Grulke, D. Trisciuzzi, D. Fourches, D. Horvath, E. Benfenati, E. Muratov, E. B. Wedebye, F. Grisoni, G. F. Mangiatordi, G. M. Incisivo, H. Hong, H. W. Ng, I. V. Tetko, I. Balabin, J. Kancherla, J. Shen, J. Burton, M. Nicklaus, M. Cassotti, N. G. Nikolov, O. Nicolotti, P. L. Andersson, Q. Zang, R. Politi, R. D. Beger, R. Todeschini, R. Huang, S. Farag, S. A. Rosenberg, S. Slavov, X. Hu and R. S. Judson, CERAPP: Collaborative Estrogen Receptor Activity Prediction Project, Environ. Health Perspect., 2016, 124, 1023–1033 CrossRef PubMed.
- S. Novotarskyi, A. Abdelaziz, Y. Sushko, R. Körner, J. Vogt and I. V. Tetko, ToxCast EPA in vitro to in vivo challenge: insight into the Rank-I model, Chem. Res. Toxicol., 2016, 29, 768–775 CrossRef CAS PubMed.
- R. S. Thomas, M. B. Black, L. Li, E. Healy, T. M. Chu, W. Bao, M. E. Andersen and R. D. Wolfinger, A comprehensive statistical analysis of predicting in vivo hazard using high-throughput in vitro screening, Toxicol. Sci., 2012, 128, 398–417 CrossRef CAS PubMed.
- Organization for Economic Co-operation and Development, Guidance on Grouping of Chemicals, Organization for Economic Co-operation and Development, Paris, 2nd edn, 2014, OECD Environment, Health and Safety Publications Series on Testing and Assessment No. 194, ENV/JM/MONO(2014)4 Search PubMed.
- S. D. Dimitrov, R. Diderich, T. Sobanski, T. S. Pavlov, G. V. Chankov, A. S. Chapkanov, Y. H. Karakolev, S. G. Temelkov, R. A. Vasilev, K. D. Gerova, C. D. Kuseva, N. D. Todorova, A. M. Mehmed, M. Rasenberg and O. G. Mekenyan, QSAR Toolbox – workflow and major functionalities, SAR QSAR Environ. Res., 2016, 27, 203–219 CrossRef CAS PubMed.
- A. Gallegos-Saliner, A. Poater, N. Jeliazkova, G. Patlewicz and A. P. Worth, Toxmatch - a chemical classification and activity prediction tool based on similarity measures, Regul. Toxicol. Pharmacol., 2008, 52, 77–84 CrossRef CAS PubMed.
- G. Gini, A. M. Franchi, A. Manganaro, A. Golbamaki and E. Benfenati, ToxRead: a tool to assist in read across and its use to assess mutagenicity of chemicals, SAR QSAR Environ. Res., 2014, 25, 999–1011 CrossRef CAS PubMed.
- P. Valdivia, M. Martin, W. R. LeFew, J. Ross, K. A. Houck and T. J. Shafer, Multi-well microelectrode array recordings detect neuroactivity of ToxCast compounds, Neurotoxicology, 2014, 44, 204–217 CrossRef CAS PubMed.
- Toxicity ForeCaster (ToxCast™) data, ToxCast & Tox21 data spreadsheet from invitrodb_v2, http://www2.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data, (accessed January 2016).
- D. S. Wishart, C. Knox, A. C. Guo, D. Cheng, S. Shrivastava, D. Tzur, B. Gautam and M. Hassanali, DrugBank: a knowledgebase for drugs, drug actions and drug targets, Nucleic Acids Res., 2008, 36, D901–D906 CrossRef CAS PubMed.
- B. L. Roth, E. Lopez, S. Patel and W. K. Kroeze, The multiplicity of serotonin receptors: uselessly diverse molecules or an embarrassment of riches?, Neuroscientist, 2000, 6, 252–262 CrossRef CAS.
- H. W. Ng, R. Perkins, W. Tong and H. Hong, Versatility or promiscuity: the estrogen receptors, control of ligand selectivity and an update on subtype selective ligands, Int. J. Environ. Res. Public Health, 2014, 11, 8709–8742 CrossRef CAS PubMed.
- N. C. Chai, B. L. Peterlin and A. H. Calhoun, Migraine and estrogen, Curr. Opin. Neurol., 2014, 27, 315–324 CrossRef CAS PubMed.
- L. Bertram, M. B. McQueen, K. Mullin, D. Blacker and R. E. Tanzi, Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database, Nat. Genet., 2007, 39, 17–23 CrossRef CAS PubMed.
- M. Chakrabarti, A. Haque, N. L. Banik, P. Nagarkatti, M. Nagarkatti and S. K. Ray, Estrogen receptor agonists for attenuation of neuroinflammation and neuro-degeneration, Brain Res. Bull., 2014, 109, 22–31 CrossRef CAS PubMed.
- I. Ahmed, R. Tamouza, M. Delord, R. Krishnamoorthy, C. Tzourio, C. Mulot, M. Nacfer, J. C. Lambert, P. Beaune, P. Laurent-Puig, M. A. Loriot, D. Charron and A. Elbaz, Association between Parkinson's disease and the HLA-DRB1 locus, Mov. Disord., 2012, 27, 1104–1110 CrossRef CAS PubMed.
- International Multiple Sclerosis Genetics Consortium, D. A. Hafler, A. Compston, S. Sawcer, E. S. Lander, M. J. Daly, P. L. De Jager, P. I. de Bakker, S. B. Gabriel, D. B. Mirel, A. J. Ivinson, M. A. Pericak-Vance, S. G. Gregory, J. D. Rioux, J. L. McCauley, J. L. Haines, L. F. Barcellos, B. Cree, J. R. Oksenberg and S. L. Hauser, Risk alleles for multiple sclerosis identified by a genomewide study, N. Engl. J. Med., 2007, 357, 851–862 CrossRef PubMed.
- G. Conductier, N. Blondeau, A. Guyon, J. L. Nahon and C. Rovère, The role of monocyte chemoattractant protein MCP1/CCL2 in neuroinflammatory diseases, J. Neuroimmunol., 2010, 224, 93–100 CrossRef CAS PubMed.
- S. Bose and J. Cho, Role of chemokine CCL2 and its receptor CCR2 in neurodegenerative diseases, Arch. Pharmacal Res., 2013, 36, 1039–1050 CrossRef CAS PubMed.
- N. S. Sipes, M. T. Martin, P. Kothiya, D. M. Reif, R. S. Judson, A. M. Richard, K. A. Houck, D. J. Dix, R. J. Kavlock and T. B. Knudsen, Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signalling assays, Chem. Res. Toxicol., 2013, 26, 878–895 CrossRef CAS PubMed.
- H. P. Vijverberg and J. van den Bercken, Neurotoxicological effects and the mode of action of pyrethroid insecticides, Crit. Rev. Toxicol., 1990, 21, 105–126 CrossRef CAS PubMed.
- I. Shah, J. Liu, R. S. Judson, R. S. Thomas and G. Patlewicz, Systematically evaluating read-across prediction and performance using a local validity approach characterized by chemical structure and bioactivity information, Regul. Toxicol. Pharmacol., 2016, 79, 12–24 CrossRef CAS PubMed.
- B. Mohana Krishnan and B. M. Prakhya, In vitro evaluation of pyrethroid-mediated changes on neuronal burst parameters using microelectrode arrays, Neurotoxicology, 2016, 57, 270–281 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Tables S1–S4. See DOI: 10.1039/c7tx00268h |
| This journal is © The Royal Society of Chemistry 2018 |