Open Access Article
Open Access ArticleCrystal structure and functional analysis of large-terpene synthases belonging to a newly found subclass†
Masahiro
Fujihashi
 *a,
Tsutomu
Sato
*a,
Tsutomu
Sato
 *b,
Yuma
Tanaka
a,
Daisuke
Yamamoto
a,
Tomoyuki
Nishi
b,
Daijiro
Ueda
*b,
Yuma
Tanaka
a,
Daisuke
Yamamoto
a,
Tomoyuki
Nishi
b,
Daijiro
Ueda
 b,
Mizuki
Murakami
b,
Yoko
Yasuno
c,
Ai
Sekihara
c,
Kazuma
Fuku
c,
Tetsuro
Shinada
b,
Mizuki
Murakami
b,
Yoko
Yasuno
c,
Ai
Sekihara
c,
Kazuma
Fuku
c,
Tetsuro
Shinada
 c and
Kunio
Miki
c and
Kunio
Miki
 *a
*a
aDepartment of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan. E-mail: mfuji@kuchem.kyoto-u.ac.jp; miki@kuchem.kyoto-u.ac.jp
bDepartment of Applied Biological Chemistry, Faculty of Agriculture, Graduate School of Science and Technology, Niigata University, 8050 Ikarashi-2, Niigata 950-2181, Japan. E-mail: satot@agr.niigata-u.ac.jp
cGraduate School of Science, Osaka City University, 3-3-138 Sugimoto, Sumiyoshi, Osaka 558-8585, Japan
First published on 16th March 2018
Abstract
Thousands of terpenes have been identified to date. However, only two classes of enzymes are known to be involved in their biosynthesis, and each class has characteristic amino-acid motifs. We recently identified a novel large-terpene (C25/C30/C35) synthase, which shares no motifs with known enzymes. To elucidate the molecular mechanism of this enzyme, we determined the crystal structure of a large-β-prene synthase from B. alcalophilus (BalTS). Surprisingly, the overall structure of BalTS is similar to that of the α-domain of class I terpene synthases although their primary structures are totally different from each other. Two novel aspartate-rich motifs, DYLDNLxD and DY(F,L,W)IDxxED, are identified, and mutations of any one of the aspartates eliminate its enzymatic activity. The present work leads us to propose a new subclass of terpene synthases, class IB, which is probably responsible for large-terpene biosynthesis.
Introduction
Terpenes are one of the most diverse compound groups in nature with more than 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 terpenes identified thus far.1 One of the major origins of their diversity is reorganization of their carbon skeletons. The reorganization is mostly initiated in two ways: by the leaving of the pyrophosphate or by the protonation of a carbon–carbon double bond.2–4 The former and the latter reactions are catalyzed by class I and class II terpene synthases, respectively.
000 terpenes identified thus far.1 One of the major origins of their diversity is reorganization of their carbon skeletons. The reorganization is mostly initiated in two ways: by the leaving of the pyrophosphate or by the protonation of a carbon–carbon double bond.2–4 The former and the latter reactions are catalyzed by class I and class II terpene synthases, respectively.
Class I enzymes have DDxx(D,E) and NSE/DTE motifs.3–5 These motifs are located at the entrance of the ligand-binding pocket, and the inner part of the pocket is mainly hydrophobic. The pyrophosphate part of the substrate binds to the two motifs through magnesium ions, and its carbon chain part is accommodated in the hydrophobic pocket. The reaction is triggered by the dissociation of the pyrophosphate group. The dissociation generates an allyl cation, and the subsequent consecutive cyclization, alkyl group shifts and attachment to other nucleophilic groups, generates a diverse range of terpene skeletons. Class II enzymes have a DxDD motif,4,8 which is involved in the protonation of a double bond. After the formation of a carbocation on the substrate, the reaction proceeds in the same manner as that for class I enzymes. These characteristic motifs [DDxx(D,E) and NSE/DTE in class I as well as DxDD in class II] are well conserved, and dominant for their functions.3–5,8 To date, many hemi-(C5) to di-(C20) terpenes are known to be synthesized from prenyl-pyrophosphates by both Class I and II enzymes.10 In contrast, most of the tri-(C30) and tetra-(C40) terpenes are synthesized by only class II enzymes from “head-to-head” type precursors. A few class I terpene synthases catalyzing the formation of large “head-to-tail” type terpenes such as C25, C30 and C35 terpenes are identified.
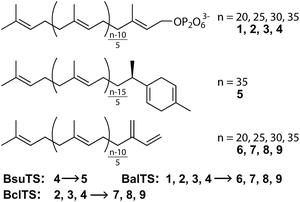
Recently, we revealed a new large-terpene synthase,‡ tetraprenyl β-curcumene synthase, from Bacillus subtilis (BsuTS).11 This enzyme is suggested to be related to the resistance of B. subtilis against an antibiotic bacitracin.12 BsuTS catalyzes the dissociation of the pyrophosphate from the large “head-to-tail” type precursor 4 and forms one ring at one end of the carbon chain (5) (Scheme 1).11 The BsuTS reaction is categorized into class I. However, no signature sequence motifs (DDxx(D,E) and NSE/DTE) of class I enzymes are found in this enzyme, and no sequence similarity was detected between BsuTS and any class I enzymes.11 A BLAST search of BsuTS revealed many homologous enzymes whose functions were unknown, and thus the enzyme probably belongs to a new class of terpene synthases that are involved in the large (around C35) “head-to-tail” type terpene biosynthesis.11 To date, the crystal structure of any enzymes in the new class has never been determined. Investigations of relationships between the structure and the functions of the enzymes are expected to extend the world of terpenes and their biosynthesis, and to provide seeds for novel valuable natural compounds.
Results and discussion
We first look for the target enzyme which is suitable for both the determination of the crystal structure and the analysis of functions. We cloned, over-expressed, purified and attempted to crystallize BsuTS and its homologous proteins and found that one of the proteins from Bacillus alcalophilus (BalTS) successfully formed crystals. Crystals of other homologous proteins and BsuTS have never been obtained. Thus, we decided to investigate BalTS as a representative enzyme in this new class.BalTS possesses 351 amino-acid residues and shows 49% similarity (e-value 2e−130) to BsuTS. The high similarity suggests that both enzymes belong to the same class. The product of the BalTS enzyme was analyzed using 1–4 as substrates. GC-MS analyses showed that these prenyl-pyrophosphates were converted into their corresponding β-prenes (6–9, Fig. 1). GC-MS analysis also confirmed that B. alcalophilus produced 7–9 in the cells as natural products (Fig. 1). These analyses indicate that BalTS is a β-C25/C30/C35-prene synthase. The catalytic reaction is categorized into the same group as BsuTS—i.e., both reactions are probably triggered by the dissociation of the pyrophosphate group from a prenyl pyrophosphate, and terminated by deprotonation to neutralize the carbocation.
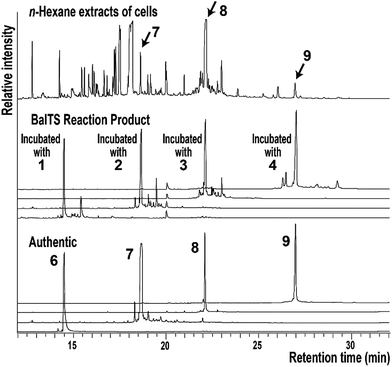 | ||
| Fig. 1 GC-MS analysis of the n-hexane extract of B. alcalophilus and the reaction products of BalTS. | ||
The crystal structure of BalTS is determined at 1.64 Å resolution by a single-wavelength anomalous dispersion method using a seleno-methionine derivative (Fig. 2a and Table S1†). The nomenclature of the secondary structure is shown in Fig. S1.† Although the primary structure of BalTS is completely different from those of any other terpene synthases including the signature motifs, its structure surprisingly belongs to the α-domain folding of terpene synthases (Fig. 2b). The domain is found in various class I terpene synthases and is also found in prenyltransferases, which catalyze the chain length elongation of prenyl pyrophosphates. Thus, BalTS is classified into a newly defined subclass of class I terpene synthases, which we named class IB. The differences between class I and IB enzymes are summarized in Fig. S2.†
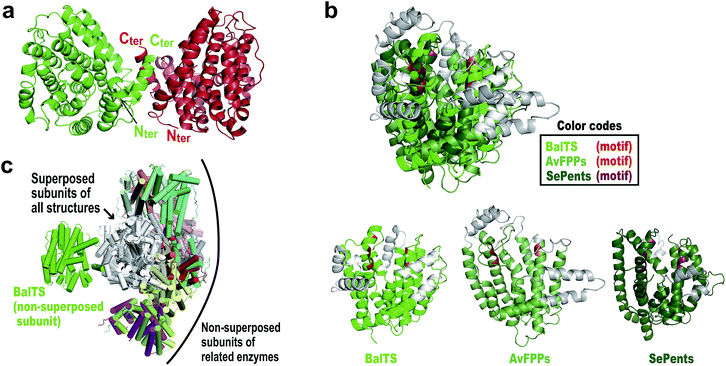 | ||
| Fig. 2 Overall structures of BalTS. (a) Overall dimeric form, subunits A and B are colored green and red, respectively. Nter and Cter represent the N- and C-terminal positions in each subunit of the determined structure. (b) Superposition of farnesyl pyrophosphate synthase from Avian (AvFPPs, a prenyltransferase)6 and pentalenene synthase from Streptomyces exfoliates (SePents, a class I terpene synthase)7 on BalTS. (Potential) pyrophosphate binding motifs are highlighted in red, orange, and red purple. Non-superposed parts are represented in gray. (c) Superpositions of one subunit of various dimeric terpene synthases on one of the BalTS subunits. The superposed subunits are drawn in white. The non-superposed BalTS subunits are drawn in green. Names of superposed molecules are given in the ESI.†Fig. 2 and 3 were prepared using the program PyMOL. | ||
BalTS forms a dimer in solution (Fig. S3†). The geometry of the dimer is distinct from those of the related known enzymes (Fig. 2c). A large cavity is found in each subunit. The cavities are constructed by conserved hydrophobic residues, and the two cavities are fused at the dimer interface (Fig. 3a and Movie S1†). In addition, the cavities have holes that penetrate to the other side of the entrance, which is a very characteristic feature of BalTS that is not seen in the other related enzymes (Fig. 3a and Movie S1†). These properties are very suitable to hold long-chain prenyl-pyrophosphates as a substrate.
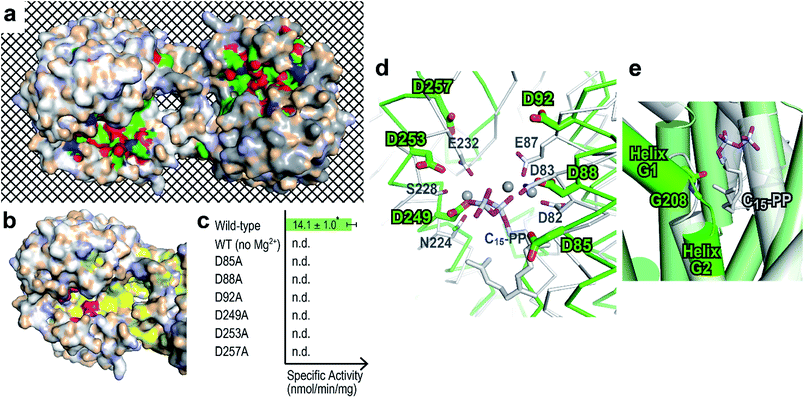 | ||
| Fig. 3 Prediction of the ligand-binding site. (a) Surface representation showing cleft and conserved residues in the physiological dimer of BalTS. White and gray molecules indicate the respective subunits. Carbons, nitrogens, and oxygens of highly conserved residues in homologous proteins are colored green, blue, and red, respectively. The mesh background visualizes the penetrating hole on the BalTS structure. Movie S1† shows the rotation around the x axis of this surface model. (b) Six characteristic aspartates (magenta) and hydrophobic residues conserved in homologous enzymes (yellow) on the BalTS surface. (c) Specific activity of BalTS. n.d.: not detectable. *: standard deviation. (d) Assumption of ligand binding. Selinadiene synthase from S. pristinaespiralis bound with the ligand-analogue molecule (PDB: 4OKM, white)9 is superposed on the BalTS (green) structure. NSE/DTE and DDxx(D,E) motifs on selinadiene synthase, and the six characteristic aspartates of BalTS are represented. Gray spheres represent the Mg2+ cation found between the pyrophosphate part of the ligand-analogue and selinadiene synthase. (e) G-helix kink. The color code is the same as in panel (d). | ||
Around the entrance of the cavity, six characteristic aspartate residues are located (magenta in Fig. 3b and Movie S2†). These 6 aspartate residues are conserved among almost all BalTS homologous proteins and divided into two motifs, DYLDNLxD and DY(F,L,W)IDxxED, on the primary structure (Fig. S4,†13 the 2nd and 5th panels). Mutations of any one of these aspartate residues and removing the Mg2+ cation from the reaction buffer result in an absence of enzymatic activity (Fig. 3c). The non-aspartate conserved residues in the motifs mostly either form the wall of the cleft or come into contact with other conserved residues inside the enzyme. The two motif sequences may be a signature of class IB terpene synthases and could be used to look for other members of this class.
A DALI14 search suggests that the enzyme most structurally similar to BalTS is selinadiene synthase from Streptomyces pristinaespiralis,9 which is a typical bacterial class I sesquiterpene synthase. Superposition of the structure of ligand-bound selinadiene synthase on that of the unliganded BalTS suggests the ligand-binding model of BalTS (Fig. 3d). The NSE/DTE and DDxx(D,E) motifs of selinadiene synthase, which are the signature motifs of class I terpene synthases, correspond to the six characteristic aspartate residues of BalTS. The hydrophobic part of the ligand is accommodated inside the cavity. The C1 atom of the substrate is located close to the carbonyl oxygen of G208 (Fig. 3e). The G-helix is clearly kinked around this residue (Fig. 3e and S1†), and the corresponding glycine in selinadiene synthase is considered to stabilize the positive charge of the allyl cation of the substrate.9 This ligand-binding model is consistent with the biochemical and structural analyses of BalTS, and supports that DYLDNLxD and DY(F,L,W)IDxxED motifs on BalTS are the signature motifs of class IB enzymes.
Next, we try to elucidate the mechanism of how class IB enzymes form complex chemical structures. BsuTS forms one ring in the reaction product, whereas BalTS produces no ring (Scheme 1). However, the residues comprising the interior of the ligand-binding cavity are very well conserved between these two enzymes (Movie S3 and Fig. S5†). This implies that the key residues of large-terpene synthases (class IB terpene synthases) for the formation of new carbon bonds are not located inside the cavity. One of the distinct differences between these two enzymes is the 16 residue C-terminal extension in BsuTS (Fig. S5a†). The C-terminal residue of BalTS, K351, corresponds to Q351 in BsuTS. Thus, we made five BsuTS C-terminal truncation mutants around this residue, which ended at K349, F350, Q351, K352, and M353 (named K349ter, F350ter, Q351ter, K352ter, and M353ter, respectively). The products of the BsuTS mutants were analyzed by GC-MS as shown in Fig. 4 and S6.† As the chain length of the enzyme becomes shorter by the C-terminal truncation, the level of 5 formation is reduced. In contrast, the amounts of 9 formation were basically the same in all truncated and standard enzymes. A related large-terpene synthase from Bacillus clausii (BclTS) catalyzes the conversion of 2–4 into the corresponding β-prenes (7–9).15,16 BclTS does not possess the C-terminal extension as BalTS. These analyses indicate that the C-terminal residues of BsuTS play a dominant role in the cyclization. The location of the BalTS C-terminal residue implies that the BsuTS C-terminal extension might cover the ligand-binding cavity. The coverage of the cavity by the C-terminal residues might be related to curcumene ring formation.
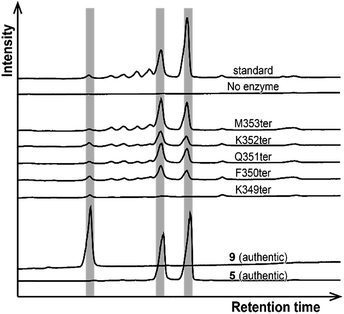 | ||
| Fig. 4 Product analyses of BsuTS variants by GC-MS. The vertical gray bars highlight the retention times of the authentic 5 and 9. | ||
Conclusions
Class IB terpene synthases (“head-to-tail” type large-terpene synthases) are suggested to be widely distributed over various bacterial species, rather than being Bacillus-specific. Most of the BalTS homologous proteins conserve two motifs DYLDNLxD and DY(F,L,W)IDxxED (Fig. S4†). The wall of the putative ligand-binding cavity is composed of conserved hydrophobic residues (Movie S2†). The origins of the homologue genes are distributed across phylum Firmicutes including Bacillus, proteobacteria and cyanobacteria (Fig. S4 and Table S2†). These uncharacterized enzymes—particularly those from non-Bacillus organisms—would have good potential as targets for the identification of new natural compounds. Since only a few large-terpenes synthesized from large prenyl-pyrophosphates have been identified thus far, newly identified compounds related to enzymes belonging to this class promise to expand the chemical space that is important for chemical genomics including drug design.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful to Mr Ryuhei Nagata for the valuable discussions and to the staff members at the beamlines of the Photon Factory and SPring-8 for their help with data collection. This work was supported in part by JSPS KAKENHI (Grant# 24570130 and 17H05439 to MF, 25450149 to T. Sa, and 15K12758 and 17H05448 to T. Sh) as well as by Life Sciences fellowships from the Takeda Science Foundation (to MF). The use of beamlines at the Photon Factory and SPring-8 was approved by the Photon Factory Advisory Committee (2015G645 and 2017G696) and by the Japan Synchrotron Radiation Research Institute (JASRI) (2015B1043 and 2016B2723).Notes and references
- G. G. Harris, P. M. Lombardi, T. A. Pemberton, T. Matsui, T. M. Weiss, K. E. Cole, M. Koksal, F. V. t. Murphy, L. S. Vedula, W. K. Chou, D. E. Cane and D. W. Christianson, Biochemistry, 2015, 54, 7142–7155 CrossRef CAS PubMed.
- M. Baunach, J. Franke and C. Hertweck, Angew. Chem., Int. Ed., 2015, 54, 2604–2626 CrossRef CAS PubMed.
- D. W. Christianson, Chem. Rev., 2006, 106, 3412–3442 CrossRef CAS PubMed.
- Y. Gao, R. B. Honzatko and R. J. Peters, Nat. Prod. Rep., 2012, 29, 1153–1175 RSC.
- D. E. Cane and I. Kang, Arch. Biochem. Biophys., 2000, 376, 354–364 CrossRef CAS PubMed.
- L. C. Tarshis, M. Yan, C. D. Poulter and J. C. Sacchettini, Biochemistry, 1994, 33, 10871–10877 CrossRef CAS PubMed.
- C. A. Lesburg, G. Zhai, D. E. Cane and D. W. Christianson, Science, 1997, 277, 1820–1824 CrossRef CAS PubMed.
- E. Oldfield and F. Y. Lin, Angew. Chem., Int. Ed. Engl., 2012, 51, 1124–1137 CrossRef CAS PubMed.
- P. Baer, P. Rabe, K. Fischer, C. A. Citron, T. A. Klapschinski, M. Groll and J. S. Dickschat, Angew. Chem., Int. Ed. Engl., 2014, 53, 7652–7656 CrossRef CAS PubMed.
- D. W. Christianson, Science, 2007, 316, 60–61 CrossRef PubMed.
- T. Sato, S. Yoshida, H. Hoshino, M. Tanno, M. Nakajima and T. Hoshino, J. Am. Chem. Soc., 2011, 133, 9734–9737 CrossRef CAS PubMed.
- A. W. Kingston, H. Zhao, G. M. Cook and J. D. Helmann, Mol. Microbiol., 2014, 93, 37–49 CrossRef CAS.
- X. Robert and P. Gouet, Nucleic Acids Res., 2014, 42, W320–W324 CrossRef CAS PubMed.
- L. Holm and P. Rosenstrom, Nucleic Acids Res., 2010, 38, W545–W549 CrossRef CAS PubMed.
- T. Sato, H. Yamaga, S. Kashima, Y. Murata, T. Shinada, C. Nakano and T. Hoshino, ChemBioChem, 2013, 14, 822–825 CrossRef CAS PubMed.
- D. Ueda, H. Yamaga, M. Murakami, Y. Totsuka, T. Shinada and T. Sato, ChemBioChem, 2015, 16, 1371–1377 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: One PDF document and three movie files. See DOI: 10.1039/c8sc00289d |
| ‡ Throughout this paper, a “large-terpene” means a terpene that is derived from ≥C25 all-trans head-to-tail prenyl-pyrophosphates. Terpenes derived from head-to-head precursors, which include most of the tri(C30)- and tetra(C40)-terpenes, are excluded from “large-terpenes” in this paper. The molecular size of large-terpenes defined above is apparently larger than those of typical terpenes derived from all-trans head-to-tail prenyl-pyrophosphates. |
| This journal is © The Royal Society of Chemistry 2018 |