Open Access Article
Open Access ArticleSurface-immobilized and self-shaped DNA hydrogels and their application in biosensing†
Xiaoxia
Mao‡
abc,
Guifang
Chen‡
a,
Zihan
Wang
a,
Yuanguang
Zhang
d,
Xiaoli
Zhu
*a and
Genxi
Li
 *ae
*ae
aCenter for Molecular Recognition and Biosensing, School of Life Sciences, Shanghai University, Shanghai 200444, China. E-mail: xiaolizhu@shu.edu.cn
bSchool of Life Sciences, Anqing Normal University, Anqing 246011, China
cInstitute of Biomedical Engineering, School of Communication and Information Engineering, Shanghai University, Shanghai 200444, China
dAnhui Key Laboratory of Functional Coordination Compounds, School of Chemistry and Engineering, Anqing Normal University, Anqing 246011, China
eState Key Laboratory of Pharmaceutical Biotechnology, Collaborative Innovation Center of Chemistry for Life Sciences, Department of Biochemistry, Nanjing University, Nanjing 210093, China. E-mail: genxili@nju.edu.cn
First published on 22nd November 2017
Abstract
Hydrogels are of great interest in the field of biosensing for their good biocompatibility, plasticity, and capability of providing 3D scaffolds. Nevertheless, the application of hydrogels has not been linked with broad surface biosensing systems yet. To overcome the limitations, here for the first time, surface-immobilized pure DNA hydrogels were synthesized using a surficial primer-induced strategy and adopted for biosensing applications. The DNA hydrogel 3D scaffold is successfully constructed on a transparent ITO electrode, which facilitates both colourimetric and electrochemical measurements. Results show that the hydrogel is able to wrap enzymes solidly and exhibits favourable stability under different conditions. Owing to the free diffusion of the micromolecular targets throughout the hydrogel, while isolating the enzymes from the macromolecular interferences outside the hydrogel, the direct colourimetric and electrochemical detection of hydrogen peroxide and bilirubin in serum is achieved. The detection limit of hydrogen peroxide in serum is 22 nM by colourimetric analysis and 13 nM by electrochemical measurement. The detection limit of bilirubin is 32 nM, a favourable limit that could be used in jaundice diagnosis. In addition, the enzyme@hydrogel can be easily regenerated and the catalytic activity is retained for a few cycles, thus allowing the recycling of the hydrogel-based biosensing system. The successful integration of DNA hydrogels with surface biosensing systems will greatly expand the applications of hydrogels for diagnostic and environmental monitoring purposes.
Introduction
Hydrogels consisting of crosslinked three-dimensional (3D) networks of hydrophilic polymers have received enormous attention and have been widely applied as scaffold materials in tissue engineering, regenerative medicine, and drug delivery due to their unique properties, including biocompatibility, great transparency, and high water content.1–7 Unlike two-dimensional materials, hydrogels can absorb and retain large volumes of water or biological fluid relative to their own mass, therefore maintaining their 3D structure and exhibiting plasticity similar to that of the microstructure of natural tissues. Hydrogels thus provide facile encapsulation of bioactive molecules (e.g. nucleotides, proteins, antibodies, and drugs) and retain their biological activities. Moreover, due to their great biocompatibility and huge swelling capacity, hydrogels are believed to be desirable scaffold materials and have attracted growing attention for the fabrication of biosensors in recent years.8,9Hydrogels, working as scaffold materials, include hard hydrogels and soft hydrogels.10 Hard hydrogels have remarkable properties in their dry states, but often lose these properties upon swelling in water, which restricts their applications in various liquid– or solid–liquid interface-based biomedical assays. For instance, the compression modulus of gelatine/alginate hydrogels loaded with hydroxyl apatite is as high as 270 MPa when dried, but decreases sharply to 2 MPa when fully hydrated.11 As for soft hydrogels, they have the advantage of extraordinary water capacity, making it possible for them to be utilized for bioanalysis in liquids.12 However, it is still a great pity that at present these hydrogels are difficult to integrate into diverse interfacial bio-analytical systems, such as enzyme linked immunosorbent assays (ELISAs), electrochemistry, biosensor chips, surface plasmon resonance (SPR), paper-based point-of-care testing (POCT) etc., meaning they are still a minority in bioanalysis. The unavailable bonding between hydrogels and solid interfaces, as well as the difficulty of shaping the soft hydrogels to desirable 3D scaffolds, are the main contributions to the bottleneck.
Adhesion of dry hydrogels to solid interfaces has been reported,13,14 but is not applicable to soft hydrogels that contain significant amounts of water. Therefore, it remains a critical demand and central challenge to develop methods for the design and fabrication of stable hydrogel 3D scaffolds on liquid–solid interfaces, which will be an important way to extend the application of hydrogels in bioanalysis. Herein, we develop a surficial primer-induced (SPI) strategy to immobilize a soft pure DNA hydrogel onto the surface of a solid and transparent electrode i.e. an indium-tin oxide (ITO) electrode. The bump-like shape of the immobilized hydrogel can be formed automatically without using moulds, its base area and height can be well controlled through regulation of the sample dose of the surficial primer and the amount of the hydrogel applied, respectively. Furthermore, the immobilized DNA hydrogel can work as a 3D scaffold to enwrap enzymes, and thus provide a 3D catalytic system for either electrochemical or colourimetric analysis. Some interesting results and unique advantages are thereby obtained.
Results and discussion
Strategy for the synthesis of surface-immobilized enzyme@DNA hydrogels
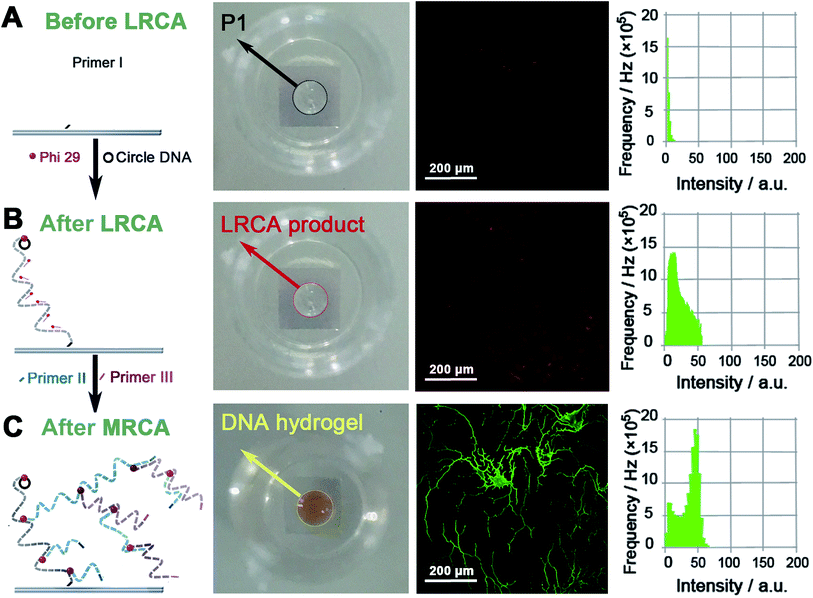
The SPI strategy for the synthesis of the proposed surface-immobilized enzyme@DNA hydrogel is shown in Scheme 1. As illustrated, three steps are involved: (i) formation of a long linear single-stranded DNA (ssDNA) on the surface of an ITO electrode through linear rolling circle amplification (LRCA) of a surface-immobilized primer (primer I). This long linear ssDNA can work as a gripper to immobilize the DNA hydrogel in the next step. (ii) Multi-primed rolling circle amplification (MRCA) launched from the surface-immobilized long linear ssDNA using another two primers (primer II and primer III). In this step, a large number of condensed hyper-branched double-stranded DNA (dsDNA) is produced on the surface of the ITO electrode, this will appear as a hydrogel. Here, the surface tension, gravitational force, and base-paring allow the formation of the hydrogel and shaping to give a bump appearance on the solid–liquid interface. The underside of the bump-like hydrogel relies on the area of the spot of primer I, while the height of the hydrogel depends on the total amount of DNA that is produced through MRCA. (iii) To enwrap the protein enzyme, the hydrogel is dissolved under heating and mixed with proteins in the solution. After cooling down, the dissolved DNA will reassemble slowly around the surface-immobilized long linear ssDNA, during which the crosslinked 3D network of the DNA is expected to enwrap the enzyme and thus form a surface-immobilized and enzyme-enwrapped DNA hydrogel (enzyme@DNA hydrogel for short). In this architecture, a protein enzyme is enwrapped by soft DNA materials instead of being immobilized through traditional chemical bonds or biological affinities. In addition to the superiority of 3D immobilization over 2D immobilization, it is also expected that the enzyme is capable of interfacial bioanalysis, and in the meantime, retains its own independence, which is important for recovering the enzyme.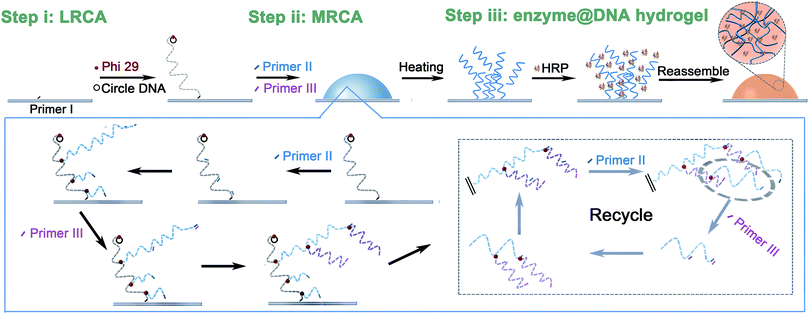 | ||
| Scheme 1 Schematic representation of the fabrication of the surface-immobilized enzyme@DNA hydrogel. | ||
Characterization of the surface-immobilized DNA hydrogel
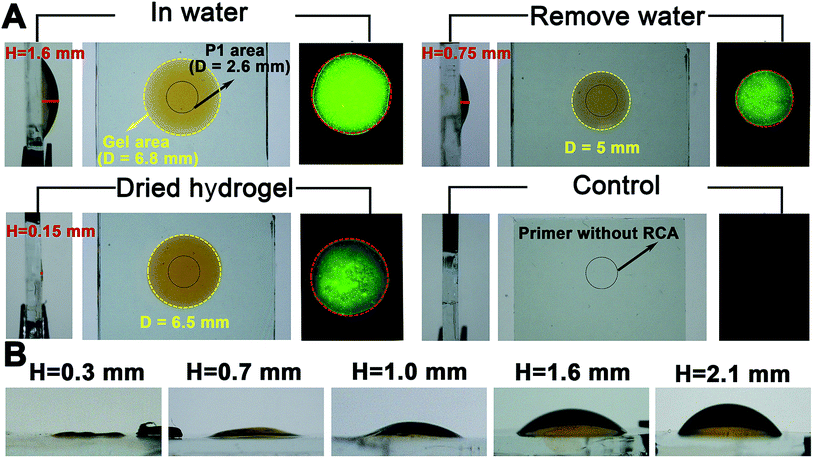
Proof-of-concept experiments were carried out to verify the process in Scheme 1. To establish the success of the LRCA and MRCA reactions as designed, gel electrophoresis is first adopted to characterize the products of the amplification. As shown in Fig. S1,†long segments of LRCA and MRCA products are observed, which are too large to enter into the gel (>2000 bp). In addition, the DNA products increase sharply with the MRCA time, suggesting progressive DNA amplification during the LRCA and MRCA reactions as expected. We next demonstrate that the surface-immobilized primer I induces the LRCA reaction on the surface of the ITO electrode; this is the basis of the formation of the surface-immobilized DNA hydrogel. A fluorophore (Cy3)-labelled DNA probe, complimentary to the LRCA-produced long linear ssDNA but not the primer itself, is adopted to show whether the LRCA has begun. As shown in Fig. 1, primer I is spotted on the centre of an ITO electrode with a spotting area of about 5.3 mm2. Laser confocal microscopic imaging of this spotting region shows that red fluorescence is obtained after the LRCA products bind with a complimentary fluorophore-labelled probe (Fig. 1B), suggesting the expected extension of the primer. In contrast, if a non-complimentary probe is adopted, no obvious signal is observed, demonstrating that the red fluorescence signal is specific to the LRCA products (Fig. S2†). On the basis of the successful LRCA on the surface, MRCA is conducted, producing a self-shaped pure DNA hydrogel that can be observed directly by the naked eye. Because the hydrogel is transparent, SYBR green I, a commonly used nucleic acid dye, is adopted to profile the hydrogel with a brown appearance in daylight and green fluorescence under UV irradiation. Experiments have been conducted to characterize the surface-immobilized DNA hydrogel, including its 3D network, plasticity, flexibility, stability, and swelling behaviour. As shown in Fig. 1C, a clear hyper-branched network of the SYBR green I-intercalated hydrogel can be observed from the confocal microscopic image. If non-complimentary primer II and primer III are adopted for the MRCA reaction as controls, no network is observed (Fig. S3†), confirming that the hydrogel is produced from MRCA and contains a favourable 3D network structure consisting of crosslinked long DNA strands. A macroscopic view of the hydrogel is shown in Fig. 2. Bump-like and round appearances are observed from the side view and top view, respectively. It should be noted again that the brown colour in daylight and green fluorescence under UV irradiation are from the SYBR green I, not the hydrogel itself. It is interesting that when an aqueous solution is supplied, a highly swollen state of the DNA hydrogel is observed with a height of 1.6 mm and a diameter of 6.8 mm. In the cases of removing water or total drying, the hydrogel shrinks to different degrees. The process can be reversed when water is re-added, suggesting good swelling/de-swelling properties.Because the hydrogel is expected to be applied in liquid–solid interfacial biosensing, all the results, hereinafter unless specified, are obtained by immersing the surface-immobilized hydrogel in aqueous solution. That is, the hydrogel is fully swollen. Fig. 2B shows that the size (mainly the height) of the hydrogel depends strongly on the MRCA time. The longer the MRCA time is, the bigger the hydrogel will be. So, though the bump-like shape of the hydrogel cannot be changed, the height and base area can be well controlled, making the hydrogel plastic.
The flexibility of the hydrogel is studied by stretch experiments (Fig. S4A†). The water-removed hydrogel, while its top side is attached to a glass slide, can even be stretched to 5 cm without fracture, a result which is superior to some reported hydrogels.15,16 But in water, a distinct phenomenon is observed (Fig. S4B†). The hydrogel is no longer sticky. Instead, after squeezing the hydrogel between the ITO electrode and a glass slide, it reforms to its original shape quickly. This property allows the hydrogel to retain its shape while facing mechanical disturbance in some liquid–solid interfacial applications. In addition, the Young’s modulus of the surface-immobilized DNA hydrogel is 34.8067 kPa (Fig. S5A†). This value is of the same order of magnitude as other kinds of soft hydrogel.17 The hydrogel is also stable. No remarkable changes are observed if the hydrogel is allowed to stand in water for over 24 h (data not shown).
Loading capacity and stability of the surface-immobilized enzyme@DNA hydrogel
The as-prepared DNA hydrogel is adopted for the encapsulation of enzymes and thereby for biosensing applications. Horseradish peroxidase (HRP), which has an absorbance at 403 nm ascribed to its heme group, and whose catalysis can be monitored via either colourimetric or electrochemical methods,18,19 is adopted as a model. The loading capacity of HRP in the DNA hydrogel is first studied. A high concentration of HRP (1 mM in 100 μL) is mixed with the dissolved hydrogel, and then is co-assembled with the long DNA strands to reform the hydrogel again (Fig. S6A†). Fig. S6B and S7† show that ca. 91.1% HRP (9.7 nmol mm−3) is enwrapped in the hydrogel, and can be retrieved by dissolving the hydrogel using a biomedical method. Compared with the conventional immobilization of HRP on 2D interfaces, which gives a density usually at a level of 1.2 pmol mm−2, here 10.0 nmol HRP per square millimetre of ITO electrode is achieved, 8333 times higher than the conventional 2D immobilization method.20 The results suggest that the hydrogel is an ideal 3D scaffold for the encapsulation of enzymes with ultrahigh efficiency.The stability of the fully-loaded surface-immobilized HRP@DNA hydrogel is further studied under different conditions including buffer solution, metal ions, and temperatures. As shown in Fig. S6C–E,†with an increase of the standing time of the HRP@DNA hydrogel in solution, concentration of some commonly used metal ions, or temperature, different degrees of leakage of HRP are observed. Nevertheless, an acceptable stability (leakage < 5%) can be achieved under conditions with a standing time of less than 1 h, concentration of metal ions less than 10 mM, and temperature lower than 40 °C. For the following biosensing applications, the HRP@DNA hydrogel is applied in 1 mM PBS solution (Na+: 1 mM) at room temperature (25 °C) to guarantee the stability. Generally, DNA hydrogels that depend on hybridization tend to shrink under high ionic strength and melt under low ionic strength. However, here the formation of the soft DNA hydrogel mainly depends on the crosslinking of the long DNA strands under surface tension and gravitational force rather than hybridization between DNA strands.21 As shown in Fig. S8,†little leakage of the enzyme is observed at low salt concentrations. The swelling behaviour of the DNA hydrogel under different concentrations of salt also confirms that the hydrogel maintains its shape well under different ionic strengths (Fig. S8†). This is an important advantage for encapsulation and relevant applications.
Biosensing applications of the surface-immobilized enzyme@DNA hydrogel
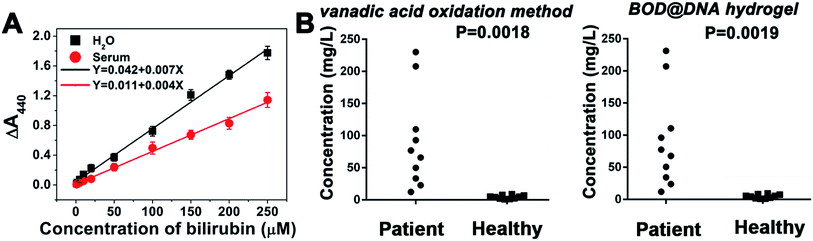
Detection of hydrogen peroxide (H2O2), by adopting the as-prepared HRP@DNA hydrogel, is first conducted. Colourimetric and electrochemical assays are carried out to show the availability of the immobilized hydrogel for different interfacial biosensing systems. For the former, the detection is based on a HRP-catalysed redox reaction whose product, 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS), is green coloured with a maximum absorption at 414 nm (Fig. S9A†). From the results shown in Fig. S9C–E,†a positive relationship is obtained between the spectroscopic signal and the logarithm of the concentration of the target H2O2, this can be fitted well by a linear regressive curve in a range from 50 nM to 10 μM. The linear equation is Y = −1.868 + 1.329X (X is the logarithm of the H2O2 concentration and Y is the value of absorbance, R2 = 0.99). The detection limit reaches 20 nM. Detection of H2O2 in serum is also achieved with a detection range of 50 nM to 10 μM and a detection limit of 22 nM. In the case of the electrochemical assay, an electrochemical active mediator thionine is employed to give electrochemical signals using cyclic voltammetry (Fig. S9B†). From Fig. S9F and S9G,†the catalytic peak currents appear to rise along with the increased concentration of H2O2, either in PBS or in serum samples. As shown in Fig. S9H,†a linear relationship is also observed between the electrochemical signal (current at −0.25 V) and the concentration of H2O2 in both PBS and serum samples. The detection range is 30 nM to 100 μM in PBS and serum. The detection limit is 10 nM in PBS and 13 nM in serum.To further demonstrate the universality of the biosensing applications of the surface-immobilized enzyme@DNA hydrogel, another enzyme, bilirubin oxidase (BOD), is adopted to be enwrapped into the DNA hydrogel and is applied for the detection of bilirubin, a biomarker for jaundice.22 Colourimetric assays are conducted based on a BOD-catalysed reaction:
| 2Bilirubin (maximum absorption: 440 nm) + O2 → 2dehydrobilirubin + 2H2O |
As shown in Fig. 3A, in both PBS and serum samples, a positive linear relationship between the spectroscopic signal and the concentration of bilirubin can be obtained in a wide range from 0 to 250 μM. The detection range covers well the physiological bilirubin level (<17 μM),23 indicating the potential applications for clinical purposes. The detection limit of bilirubin is 32 nM. To further evaluate the diagnostic feasibility, human blood plasma samples, voluntarily collected from healthy people and jaundice patients, were tested. In addition to our hydrogel-based method, the conventional vanadic acid oxidation method is also adopted to work as a standard. The relevant data are supplied in Fig. 3B and Table S2.†Results show that there is favourable consistency between these two methods with an average relative deviation of 2.4%. The healthy group (serum bilirubin 1.69–15.11 μM) and patient group (including mild: 17.1–171 μM, moderate: 171–342 μM, and severe: >342 μM serum bilirubin) can also be well discriminated.
Regenerability of the surface-immobilized enzyme@DNA hydrogel
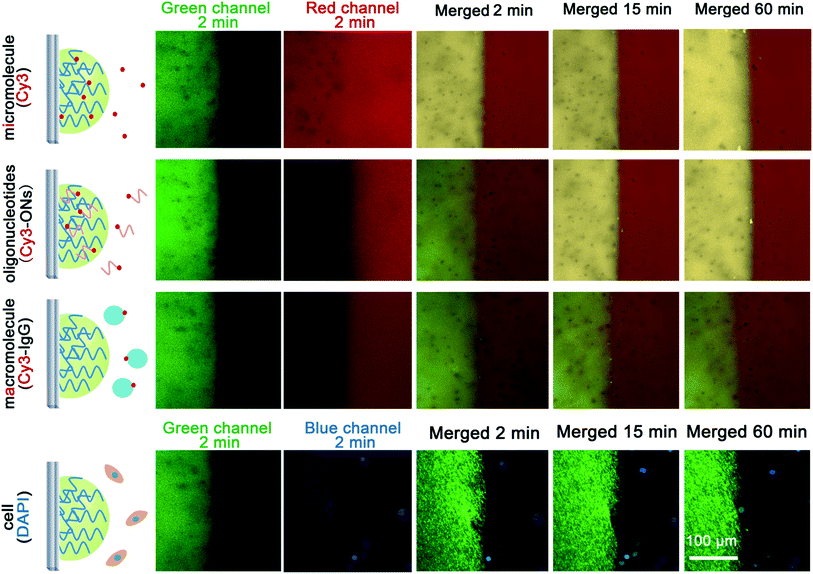
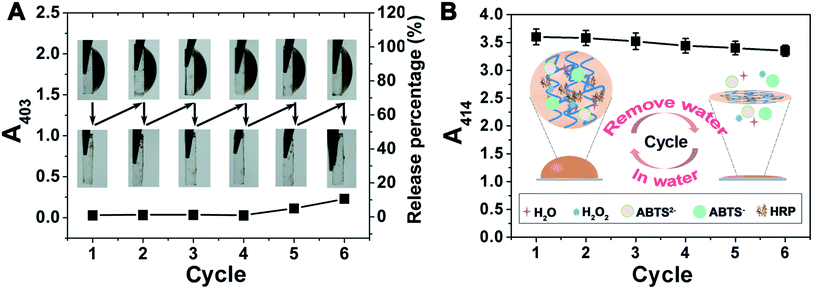
Because there are many interfering constituents in serum and other physiological samples, it is better that they have little affect on the assays, and after tests they can be removed easily to allow for the regeneration of the hydrogel-based biosensing system. We labelled four kinds of typical interfering constituents in physiological samples with fluorophores to study their diffusion from the outside to the inside of the DNA hydrogel (Fig. 4). The results show that micromolecules diffuse quickly into the hydrogel, enabling highly efficient catalysis to be achieved. Oligonucleotides can also diffuse into the hydrogel with a relatively slower diffusion rate. But for macromolecules and cells, it is observed that the DNA hydrogel prevents their diffusion from the outside. It is a favourable result that the hydrogel is working as simple molecular sieve, allowing the substrate to be involved in the catalysis-based biosensing, but avoiding many other possible interfering constituents, mainly macromolecules and cells. It is also possible to regenerate the enzyme@DNA hydrogel by washing it gently after a measurement to remove the outside interfering constituents and to ensure the residues of the catalytic substrates and products diffuse back into the washing buffer. In view of regenerability, together with stability and portability, a dry state is wanted for the enzyme@DNA hydrogel when it is not in use. So after an assay, the ITO electrode together with the surface-immobilized HRP@DNA hydrogel is washed and dried in air. Results show that the HRP@DNA hydrogel can be quickly regenerated after putting the ITO electrode into buffer solution again. Solid detection of H2O2 in serum for at least 6 cycles with little passivation (loss of enzymatic activity < 8%) is thereby achieved (Fig. 5). Compared with some other scaffolds for biosensing, the reusability of the surface-immobilized DNA hydrogel is much easier.24Discussion
Finally, we would like to give a short discussion. The pure DNA-based soft hydrogel has some advantages, e.g. favourable biocompatibility, mild preparation conditions, and high stability in physiological environments. While being immobilized and adopted for 3D scaffolds, we further extend its advantages: (i) it makes it possible for the soft gel to be applied in liquid–solid interfacial biosensing; (ii) the surface-immobilized DNA hydrogel has self-controllable shape and size, making it suitable for quantitative measurements; (iii) the 3D scaffold allows for the encapsulation of large amounts of unlabelled enzymes, which is favourable for improving the performance; (iv) the DNA hydrogel has the ability to be recycled for bioanalysis. In the future, efforts to improve the complex, sophisticated and expensive preparation process of the DNA hydrogel should be made. In addition, owing to the facile modification of DNA as well as its functions such as working as a DNAzyme and aptamer, there is extensive scope for the application of this kind of DNA hydrogel, especially after being immobilized.Conclusions
In summary, we have successfully synthesized surface-immobilized and self-shaped soft pure DNA hydrogels on ITO electrodes. Some interesting properties including plasticity, flexibility, stability, and swelling behaviour are thereby revealed, showing it can be a superior material for some specific interfacial applications. In this study, we show that the surface-immobilized hydrogel is able to work as a 3D scaffold to enwrap a large number of protein enzymes, and therefore can be applied for biosensing. Sensitive detection of H2O2 and bilirubin using a HRP@DNA hydrogel and BOD@DNA hydrogel respectively is achieved. Owing to the molecular sieving effect, as well as the good swelling/de-swelling properties, the enzyme@DNA hydrogel allows for the direct detection of the targets in serum samples and the quick regeneration in swelling-drying cycles. This hydrogel-based biosensing system is also available for both colourimetric and electrochemical assays. The combination of DNA hydrogels with interfacial bio-analytical systems not only extends the scope for the application of DNA hydrogels, but also opens a new avenue to upgrading 2D interfacial biosensing systems to 3D systems. The potential application of the surface-immobilized DNA hydrogel as a toolbox is also expected in the future.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Natural Science Foundation of China (Grant No. 21575088 and 21235003) and the Natural Science Foundation of Shanghai (14ZR1416500).References
- C. B. Ma, Y. Shi, D. A. Pena, L. L. Peng and G. H. Yu, Angew. Chem., Int. Ed., 2015, 127, 7484–7488 CrossRef.
- G. Eke, N. Mangir, N. Hasirci, S. MacNeil and V. Hasirci, Biomaterials, 2017, 129, 188–198 CrossRef CAS PubMed.
- P. Lee and S. Konst, Adv. Mater., 2014, 26, 3415–3419 CrossRef PubMed.
- K. Peng, H. S. Yu, H. Y. Yang, X. Hao, A. Yasin and X. Y. Zhang, Soft Matter, 2017, 13, 2135–2140 RSC.
- J. Li, L. t. Mo, C. H. Lu, T. Fu, H. H. Yang and W. H. Tan, Chem. Soc. Rev., 2016, 45, 1410–1413 RSC.
- H. M. Meng, H. Liu, H. L. Kuai, R. Z. Peng, L. T. Mo and X. B. Zhang, Chem. Soc. Rev., 2016, 45, 2583–2602 RSC.
- J. W. Liu, Soft Matter, 2011, 7, 6757–6767 RSC.
- Q. Wu, J. J. Wei, B. Xu, X. H. Liu, H. B. Wang, W. Wang, Q. G. Wang and W. G. Liu, Sci. Rep., 2017, 7, 41566 CrossRef PubMed.
- X. L. Zhu, X. X. Mao, Z. H. Wang, C. Feng, G. F. Chen and G. X. Li, Nano Res., 2017, 10, 959–970 CrossRef CAS.
- D. Wirthl, R. Pichler, M. Drack, G. Kettlguber, R. Moser, R. Gerstmayr, F. Hartmann, E. Bradt, R. Kaltseis, C. M. Siket, S. E. Schausberger, S. Hild, S. Bauer and M. Kaltenbrunner, Sci. Adv., 2017, 3, e1700053 CrossRef PubMed.
- Y. X. Luo, A. Lode, A. R. Akkineni and M. Gelinsky, RSC Adv., 2015, 5, 43480–43488 RSC.
- H. J. Ding, M. J. Zhong, Y. J. Kim, P. Pholpabu, A. Balasubramanian, C. M. Hui, H. K. He, H. Yang, K. Matyjaszewski and C. J. Bettinger, ACS Nano, 2014, 8, 4348–4357 CrossRef CAS PubMed.
- H. Yuk, T. Zhang, S. T. Lin, G. A. Parada and X. H. Zhao, Nat. Mater., 2016, 15, 190–196 CrossRef CAS PubMed.
- K. Hu, N. Z. Zhou, Y. Li, S. Y. Ma, Z. B. Guo, M. Gao, Q. Y. Zhang, J. F. Sun, T. Z. Zhang and N. Gu, ACS Appl. Mater. Interfaces, 2016, 8, 15113–15119 CAS.
- T. C. Suekama, J. Hu, T. Kurokawa, J. P. Gong and S. H. Gehrke, ACS Macro Lett., 2013, 2, 137–140 CrossRef CAS.
- X. K. Lu, C. Y. Chan, K. I. Lee, P. F. Ng, B. Fei, J. H. Xin and J. Fu, J. Mater. Chem. B, 2014, 2, 7631–7638 RSC.
- K. M. Mabry, R. L. Lawrence and K. S. Anseth, Biomaterials, 2015, 49, 47–56 CrossRef CAS PubMed.
- L. Zhou, N. Sun, L. J. Xu, X. Chen, H. Cheng, J. Wang and R. J. Pei, RSC Adv., 2016, 6, 114500–114504 RSC.
- Y. Zhao, Y. Q. Zheng, R. M. Kong, L. Xia and F. L. Qu, Biosens. Bioelectron., 2016, 75, 383–388 CrossRef CAS PubMed.
- Q. Wang, A. Kromka, J. Houdkova, O. Babchenko, B. Rezek, M. S. Li, R. Boukherroub and S. Szunerits, Langmuir, 2011, 28, 587–592 CrossRef PubMed.
- J. B. Lee, S. M. Peng, D. Y. Yang, Y. H. Roh, H. Funabashi, N. Park, E. J. Rice, L. W. Chen, R. Long, M. M. Wu and D. Luo, Nat. Nanotechnol., 2012, 7, 816–820 CrossRef CAS PubMed.
- G. Garcea, W. Ngu, C. P. Neal, A. R. Dennison and D. P. Berry, HPB, 2011, 13, 426–430 CrossRef PubMed.
- G. Monaghan, A. McLellan, A. McGeehan, S. L. Volti, F. Mollica, I. Salemi, Z. Din, A. Cassidy and R. Hume, J. Pediatr., 1999, 134, 441–446 CrossRef CAS PubMed.
- V. M. Esquerdo, T. R. S. Cadaval Jr, G. L. Dotto and L. A. A. Pinto, J. Colloid Interface Sci., 2014, 424, 7–15 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7sc03716c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |