Open Access Article
Open Access ArticleUp-regulation of exosomal miR-125a in pneumoconiosis inhibits lung cancer development by suppressing expressions of EZH2 and hnRNPK†
Lin Zhang ab,
Jiangfeng Lic,
Changfu Haob,
Wei Guod,
Di Wangb,
Jianhui Zhangb,
Youliang Zhaob,
Shuyin Duanb and
Wu Yao*b
ab,
Jiangfeng Lic,
Changfu Haob,
Wei Guod,
Di Wangb,
Jianhui Zhangb,
Youliang Zhaob,
Shuyin Duanb and
Wu Yao*b
aDepartment of Reproductive Medical Center, Third Affiliated Hospital of Zhengzhou University, 7 Kangfuqian Road, Zhengzhou, 450001, China
bDepartment of Occupational and Environmental Health, School of Public Health, Zhengzhou University, 100 Science Avenue, Zhengzhou, 450001, China. E-mail: yaowu@zzu.edu.cn; Tel: +86-371-67781922
cSchool of Basic Medicine, Zhengzhou University, 100 Science Avenue, Zhengzhou, 450001, China
dDepartment of Occupational Disease, Henan Provincial Institute of Occupational Health, Middle Street of Kangfu, Zhengzhou, 450052, China
First published on 25th July 2018
Abstract
Exposure to nanoparticles may lead to pneumoconiosis and lung cancer; however, whether patients suffering from pneumoconiosis also face a high risk of lung cancer has been under debate for decades. Recently, exosomes have been found to play critical roles in many diseases via intercellular cargo transportation, which has provided a new insight into the mechanistic investigation of nanoparticle-induced respiratory disorders. Herein, we isolated exosomes from the venous blood of patients with pneumoconiosis and healthy controls and then, we profiled the expression signatures of exosomal miRNAs using high-throughput sequencing technology. A total of 14 aberrantly expressed miRNAs were identified and used to process target gene prediction and functional annotation. Specially, miR-125a along with its target genes EZH2 and hnRNPK was found to play a significant role in the development of lung cancer. We then adopted a series of cellular experiments to validate the role of miR-125a in lung cancer. From the results obtained, we found that the suppression of EZH2 and hnRNPK by high levels of miR-125a inhibited the development of nanoparticle-induced lung adenocarcinoma, which contributed to the clarification of the relation between pneumoconiosis and lung cancer.
Introduction
Long-term or acute exposure to nanoparticles may lead to a variety of pulmonary disorders.1 Crystalline silica dust, one of the most common respirable hazards in both working and living conditions, has been identified as an inducer of pneumoconiosis, silicosis, and lung cancer.2 To date, there are more than 2 million workers being exposed to silica dust in America and 3.2 million workers with the same conditions in Europe.3 This phenomenon is also widely observed in developing or under-developed countries. For example, it has been reported that more than 33 million workers have been exposed to dust in China and India.4 The adverse effects on human health caused by inhaling silica dust have reached a broad consensus globally; in many countries such as America, measures on lowering the concentration of productive dust have been adopted, and the mortality rates of silicosis and lung cancer have reduced to approximately one-half of the predicted rates via re-limiting silica exposure concentration from 0.1 mg m−3 to 0.05 mg m−3 2. Nevertheless, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 new cases and more than 24
000 new cases and more than 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths are annually reported due to silicosis in China.3
000 deaths are annually reported due to silicosis in China.3
Given that pneumoconiosis and lung cancer both could be induced by inhaling silica dust, the association between pneumoconiosis and lung cancer has drawn considerable attention and has also been the subject of debate for decades since silica was defined as a human lung carcinogen by the International Agency for Research on Cancer (IARC) in 1997.5 However, abundant epidemiological evidence is available on this topic, but it is ambiguous and conflicting; therefore, it is still uncertain whether lung cancer occurs exclusively among workers with silicosis. A pooled analysis, which integrated data from 10 large silica-exposed cohorts with over 1000 deaths due to lung cancer, indicated significant positive exposure–response relationship between cumulative silica exposure and lung cancer mortality.6 Some meta-analyses also found a similar phenomenon after excluding confounding factors such as smoking.7 Typically, a 44 years cohort study of 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018 workers indicated an elevated risk of cancer occurrence with the increasing dust exposure concentration, and the hazard ratios of different silica dust exposure categories (0.01–1.2, 1.12–2.91, 2.91–6.22, and ≥6.22 mg m−3 per year vs. non-exposure controls) were 1.26, 1.54, 1.68, and 1.70, respectively. This study also specified that the joint effect of silica exposure and smoking was close to multiplicative rather than additive.3 On the other hand, a recent meta-analysis revealed an elevated risk level of lung cancer in both silicotics and non-silicotics, and the pooled standardized mortality ratio (SMR) values were 2.32 and 1.78 respectively,8 making it more confusing to understand the association between pneumoconiosis and lung cancer. Since current cohort studies could not draw identical conclusions, it is necessary to attempt a new way to investigate the association between pneumoconiosis and lung cancer from the point of pathogenesis.
018 workers indicated an elevated risk of cancer occurrence with the increasing dust exposure concentration, and the hazard ratios of different silica dust exposure categories (0.01–1.2, 1.12–2.91, 2.91–6.22, and ≥6.22 mg m−3 per year vs. non-exposure controls) were 1.26, 1.54, 1.68, and 1.70, respectively. This study also specified that the joint effect of silica exposure and smoking was close to multiplicative rather than additive.3 On the other hand, a recent meta-analysis revealed an elevated risk level of lung cancer in both silicotics and non-silicotics, and the pooled standardized mortality ratio (SMR) values were 2.32 and 1.78 respectively,8 making it more confusing to understand the association between pneumoconiosis and lung cancer. Since current cohort studies could not draw identical conclusions, it is necessary to attempt a new way to investigate the association between pneumoconiosis and lung cancer from the point of pathogenesis.
Mechanistically, inhalation of crystalline silica dust can induce strong inflammatory response, which is an important factor for the occurrence and development of pneumoconiosis and lung cancer.9 Pulmonary inflammation is initiated by macrophages and dendritic cells after being exposed to nanoparticles, and a variety of substances such as cytokines and chemokines can not only lead to the formation of a persistent inflammatory micro-environment in the local lung tissue but also cause epithelial cell injury and proliferation.10 Besides, under such conditions, pulmonary fibroblasts transdifferentiated into myofibroblasts cause collagen oversynthesis and secretion, eventually resulting in pulmonary fibrosis and pneumoconiosis.11 As to lung cancer, persistent inflammation of the lung tissue can cause genotoxic damage to the lung epithelium,12 which is the initial stage of lung cancer. Specially, loss of gap junctional intercellular communication (GJIC) has also been indicated as a typical feature of lung cancer,13 and exosomes have been suggested to play critical roles in cell–cell communication via transporting biomolecules such as protein, miRNAs, lipid, or other signaling molecules.14,15 Classical studies have confirmed that miRNAs transported from the host cell can change the expression of genes in the target cell, which is consistent with the regulatory mechanism of cancer occurrence and metastasis.16 Thus, it would be of great significance to investigate the role of exosomal miRNAs in the development of lung cancer, which would also contribute to the elucidation of the association between pneumoconiosis and lung cancer.
This study was conducted to determine the distinctive exosomal miRNAs involved in pneumoconiosis and further confirm their role in lung cancer. We isolated exosomes from the venous blood of patients with pneumoconiosis and healthy controls and then profiled the expression signatures of all exosomal miRNAs using next-generation sequencing technology. The aberrantly expressed miRNAs were identified and used to conduct target gene prediction and functional annotation. Specially, miR-125a along with its target genes that were detected to be involved in lung cancer development were selected for downstream analysis, and the relationship between the expressions of these target genes and the survival times of different subtypes of lung cancer was investigated. Finally, the regulatory mechanisms of miR-125a on its target genes EZH2 and hnRNPK were validated using cellular experiments in lung cancer.
Materials and methods
Study approval
All subjects involved in this study were well informed about the purpose of this study, and their confirmation was obtained. Clinical analyses were performed with the approval of Henan Provincial Institute of Occupational Health and in accordance with the legal and ethical standards; the whole research protocol was approved by the Institutional Review Board of Zhengzhou University.Study subjects
A total of 154 study subjects were recruited from Henan Provincial Institute of Occupational Health, China, including 54 patients with pneumoconiosis and 100 healthy controls. The National Diagnostic Criteria for Pneumoconiosis of China (GBZ 70-2002) was used to distinguish the patients from the healthy controls. Subjects in the control group were selected from new workers who were undertaking pre-service physical examinations, and it was confirmed that they did not have any occupational dust particle exposure experience or physical disorders that may affect exosome excretion, extraction or transportation. Samples of approximately 5 mL venous blood were obtained from every subject, and exosomes were isolated from the serum mixture of the patients or the healthy controls.Exosome isolation and characterization
The standard exosome isolation and characterization method has been described previously.17 In brief, 5 mL serum mixture was first diluted with 20-fold volume of PBS and centrifuged at 2000g at 4 °C for 30 min. The supernatant was obtained and centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 45 min to exclude the pellet. The supernatant was then centrifuged at 110
000g at 4 °C for 45 min to exclude the pellet. The supernatant was then centrifuged at 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 2 hours (70 Ti centrifuge rotor, Beckman Coulter Inc.), and the raw exosomes at the bottom of the ultracentrifuge tube were collected and diluted with 20 mL PBS. After filtering using a 0.22 μm filter, the suspension was centrifuged for 70 min at 110
000g for 2 hours (70 Ti centrifuge rotor, Beckman Coulter Inc.), and the raw exosomes at the bottom of the ultracentrifuge tube were collected and diluted with 20 mL PBS. After filtering using a 0.22 μm filter, the suspension was centrifuged for 70 min at 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, and the precipitate was obtained. This step was repeated one more time, and the pellet of exosomes at the bottom of centrifuge tube was resuspended in 100 μL PBS. For morphological characterization and counting of exosomes, we adopted transmission electron microscopy (TEM) (HT7700, Hitachi, Tokyo, Japan), scanning and nanoparticle tracking analysis (NTA) (NanoSight® LM10, Malvern, United Kingdom).
000g, and the precipitate was obtained. This step was repeated one more time, and the pellet of exosomes at the bottom of centrifuge tube was resuspended in 100 μL PBS. For morphological characterization and counting of exosomes, we adopted transmission electron microscopy (TEM) (HT7700, Hitachi, Tokyo, Japan), scanning and nanoparticle tracking analysis (NTA) (NanoSight® LM10, Malvern, United Kingdom).
miRNA isolation and high-throughput sequencing
Exosomal miRNAs were isolated using Trizol reagent (Invitrogen/Thermo Fisher Scientific, Waltham, MA, USA) and qualified using Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA). miRNAs were first ligated to their 5′ and 3′ adapters using miRNAs first-strand cDNA synthesis (Tailing Reaction) kit (Biotech (Shanghai) Co., Ltd., Shanghai, China) before constructing miRNA library, and the expressions of total miRNAs were detected using Illumina Hiseq2500 SBS (Illumina, Inc, San Diego, CA, USA). The sequencing raw data were preprocessed using the R software with Bioconductor packages.Identification of differentially expressed miRNAs
The raw data of miRNA expression matrix were first transformed into transcript per million (TPM) format and then normalized. To obtain more conceivable differentially expressed miRNAs, DESeq and orthogonal partial least-squares regression and discriminant analysis (OPLS-DA) method were used to compare miRNA expression values between the samples obtained from the patients and the controls. The conditions used to identify differentially expressed miRNAs were pre-set as follows: DESeq comparing method, P < 0.05, false discovery rate (FDR) < 0.01, fold change (FC) ≥ 2.0 or ≤ 0.5.miRNA target gene prediction and functional annotation
Target genes of miR-125a were predicted using the MR-microT method (available online at http://diana.imis.athena-innovation.gr/DianaTools), which provided near-real-time predictions for custom miRNA sequences, and those target genes with a miTG score ≥0.8 were selected for further investigation. For functional annotation of miRNAs and target genes, Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were conducted using DAVID 6.8 online (https://david.ncifcrf.gov/), ClueGo v2.3.5, and CluePedia v1.3.5 (thresholds account ≥2 and EASE score ≤0.1). Those genes associated with cancers were identified and used for the downstream investigation.Cross-cancer alteration of target genes related to miR-125a
To investigate the alterations of 14 target genes related to miR-125a in lung cancer, we explored their expressions in lung adenocarcinoma, lung squamous carcinoma, and small cell lung cancer using TCGA database. Four types of genomic alterations such as mutation, deletion, amplification, and multiple alterations were recorded, and the expression levels of these target genes in lung adenocarcinoma and lung squamous carcinoma were obtained using heat map.Kaplan–Meier and Cox hazards regression survival analysis
To determine the association between the 14 target genes and the survival times of patients diagnosed with lung cancer, we adopted Kaplan–Meier and Cox hazards regression survival analysis using an online database named KM plotter for lung cancer, which integrated 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 675 genes on survival using 10
675 genes on survival using 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 461 cancer samples including 2437 lung cancer patients with a mean follow-up time of 49 months. Additionally, to examine the regulatory mechanisms of the 14 target genes in different subtypes of lung cancer, the survival analysis was processed using patients who were diagnosed with lung adenocarcinoma or lung squamous carcinoma.
461 cancer samples including 2437 lung cancer patients with a mean follow-up time of 49 months. Additionally, to examine the regulatory mechanisms of the 14 target genes in different subtypes of lung cancer, the survival analysis was processed using patients who were diagnosed with lung adenocarcinoma or lung squamous carcinoma.
Expression detection of target genes related to miR-125a
To validate the hypothesis proposed in this study, we retrieved the expression values of these 14 target genes using Oncomine database premium version, and the expression levels of these genes were also investigated among tissues derived from lung adenocarcinoma, lung squamous carcinoma, large cell lung carcinoma and normal adjacent control samples.Cell culture and transfection
A549 pulmonary epithelial cells were obtained from ATCC Company (ATCC, Manassas, VA, USA) and maintained in high-glucose complete Dulbecco's modified Eagle medium (DMEM) (Hyclone, GE Healthcare Life Sciences, Utah, America) in a humidified atmosphere at 37 °C and 5% CO2. DMEM was supplemented with 10% fetal bovine serum, 100 μg mL−1 streptomycin, 100 U mL−1 penicillin and 2 mM glutamine. After being cultured for three passages, the A549 cells obtained from the fourth generation were harvested and used for downstream experiments. When the confluence of A549 cells reached 70% of a 60 mm dish, the artificially synthesized mimic or inhibitor miR-125a was added to the serum-free cell culture medium. After 48 hours, the A549 cells were harvested for cellular viability detection or protein immunoblotting.Cellular viability detection using CCK8 assay
After the cells were transfected with the miRNA mimic or inhibitor, the viability of the A549 cells was assessed by the CCK8 assay (Dojindo Molecular Technologies, Kumamoto, Japan) according to the manufacturer's instructions. In brief, the A549 cells were plated in 96-well plates at a density of 4 × 104 cells per well and maintained for 24 hours. At this point, the mimic or inhibitor miR-125a was introduced separately. After incubating for 48 hours, the medium was removed, and the cell was washed with PBS. Then, 200 microlitres of the CCK-8 solution was added to each well and kept for 1 hour at 37 °C. The absorbance value of each plate was detected at 450 nm using a microplate reader (SpectraMax M2/M2e, Molecular Devices, Sunnyvale, CA, USA). The cells that were incubated without perturbation were used as controls, and all experiments were repeated with 10 samples in parallel. The cellular viability was expressed as a percentage of test/control (%).Protein quantification using immunoblotting
Total protein extracted from different treatment groups was quantified using immunoblotting. First, 40 μg protein was diluted in 20 μL PBS, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto PVDF membranes (Millipore, Billerica, MA); then, they were incubated in 5% nonfat milk at 4 °C with primary antibodies of hnRNPK, EZH2, or β-action (Abcam, Cambridge, MA, USA). Twelve hours later, they were washed with TBST for three times, 10 min each, and then exposed to goat anti-rabbit secondary antibodies (Abcam, Cambridge, MA, USA). After being washed for three times (10 min per time) with TBST, they were subjected to the Fluor Chem HD2Gel imaging system (Proteinsimple, San Jose, CA, USA) for signal measurement. All experiments conducted in this step were repeated 5 times in parallel.Statistical analysis
All data were analyzed using SAS version 9.2 for Windows. Continuous variables with normal distribution were expressed as mean ± standard deviation. The ages of the patients and healthy controls were compared using the pooled variance t-test. The general linear model was constructed to adjust the interference of age on the expression of miR-125a between different groups, and the pairwise comparison between any two groups was adjusted using the Bonferroni method. A P-value less than 0.05 was considered as statistically significant unless otherwise indicated.Results
Baseline characteristics of study subjects
A total of 154 subjects, including 54 patients with pneumoconiosis and 100 healthy controls, were recruited, among which 37 patients were diagnosed with stage I pneumoconiosis, 12 patients were in stage II, and 5 patients were in stage III. The subjects in the control group were coal workers who undertook pre-service physical examinations and were confirmed to have no occupational dust exposure experience. The average age of the new participating workers in the control group was lower than the average age of the patients (30.1 ± 4.1 vs. 45.5 ± 6.2, t = 16.32, P < 0.0001), which may affect the excretion and the contents of exosomes.Presentation of exosomes isolated from the venous blood of all study subjects
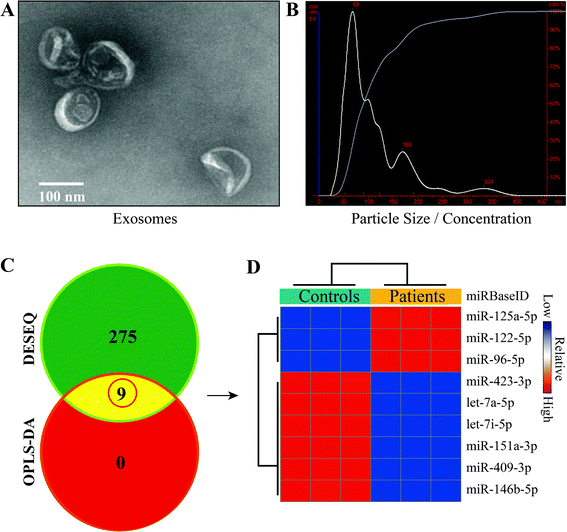
Total exosomes were isolated from the venous blood of all subjects, and their characterization and quantification were conducted using TEM and NTA (Fig. 1). Morphologically, particles looking like a saucer but with smaller diameters between 30 and 150 nm and a clear bilayer plasmalemma structure were identified as exosomes. The average diameter of exosomes detected by NTA in pneumoconiosis patients was 114 ± 65 nm (vs. 102 ± 73 nm in the healthy controls), and exosomes with diameters less than 141 nm (vs. 126 nm) accounted for 90% of total exosomes; the exosome concentration was 2.05 × 108 particles per mL (vs. 5.77 × 108 particles per mL), indicating that the exosomes isolated in this study were highly purified.Identification of differentially expressed miRNAs
To obtain miRNAs of interest in pneumoconiosis, we used two algorithms (DESEQ and OPLS-DA) to identify differentially expressed miRNAs. The algorithms integrated univariate and multivariate statistical analyses and eliminated bias that usually occurs in a single algorithm. As a result, a total of 9 miRNAs were determined, among which 3 miRNAs were up-regulated and 6 miRNAs were down-regulated (Fig. 1). We then focused on miR-125a that was differentially expressed with the highest value of fold change.Prediction and functional annotation of target genes related to miR-125a
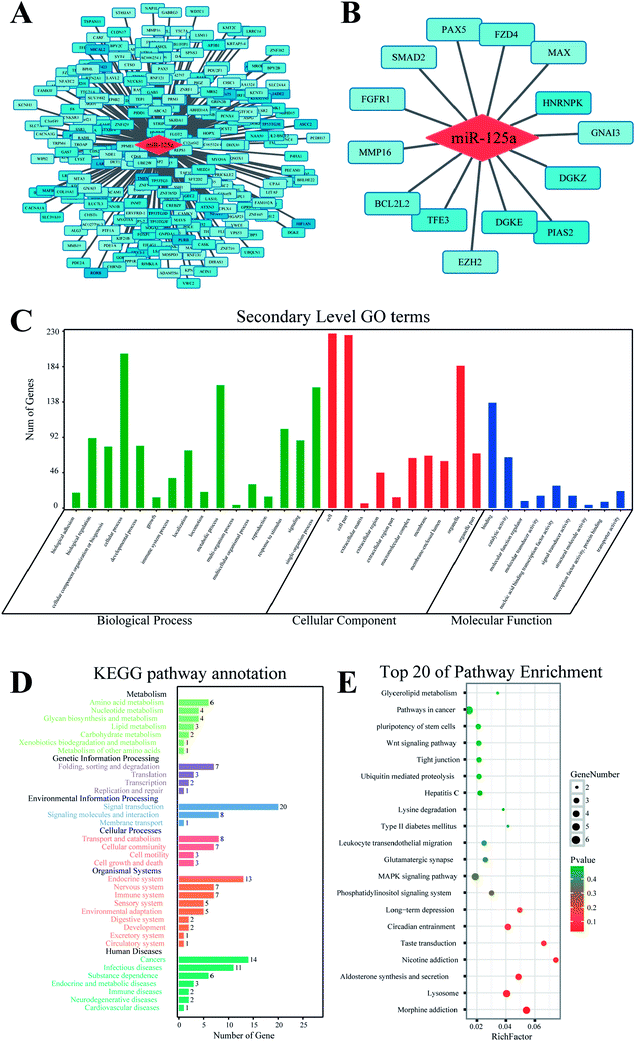
The target gene of miR-125a was predicted using MR-microT database based on a MapReduce-based implementation target prediction method, and a set of 266 target genes was determined (ESI Table 1†). The functional annotation of miR-125a was conducted using databases of GO and KEGG from four aspects of biological process, molecular function, cellular component, and signaling pathway (Fig. 2). The results of GO analysis showed that 35 secondary GO items were enriched, among which some GO items such as chromosome organization (GO: 0051276) and transmembrane transport (GO: 0055085) in biological process, nucleic acid binding transcription factor activity (GO: 0001071) and mRNA binding (GO: 0003729) in molecular function, and nucleoplasm (GO:0005654) and intracellular part (GO:0044424) in cellular component indicated that the main function of miR-125a is the regulation of molecular or cellular binding processes. The KEGG enrichment analysis showed that 20 target genes related to miR-125a were involved in the signal transduction item, which was highly consistent with the widely accepted miRNA-target gene regulatory mechanisms. In addition, 14 target genes of miR-125a were enriched in cancers, accounting for the majority of target genes included in human diseases. Hence, we subsequently focused on the roles of miR-125a and its corresponding 14 target genes related to cancers.Expression of 14 target genes in lung cancer
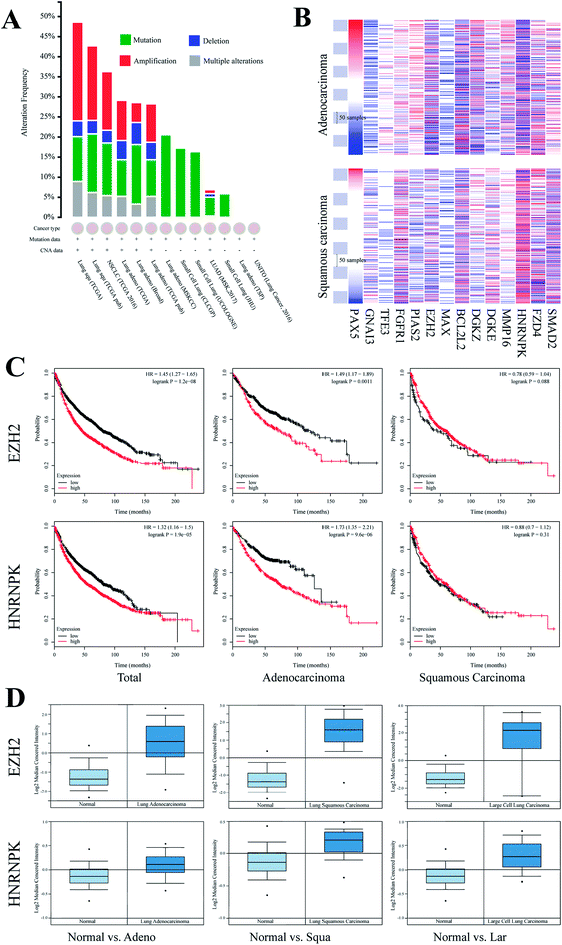
To elucidate the function of miR-125a via regulating 14 cancer-related target genes, we investigated their expressions in lung cancers of different subtypes. First, we recruited all samples that were labeled as lung cancer in TCGA database and found that genomic mutation was widely distributed in all subtypes of lung cancer, whereas genomic amplification was mainly found in lung squamous carcinoma and lung adenocarcinoma. We then investigated the expressions of these genes in these two subtypes of lung cancers (Fig. 3). Moreover, we compared the expressions of these genes in tissues of carcinoma vs. the adjacent normal tissue using the Oncomine database; of these, EZH2 and hnRNPK were both differentially expressed in lung squamous carcinoma, lung adenocarcinoma, and large cell lung carcinoma, whereas the changes in the other 12 target genes were not statistically significant (Table 1). To investigate the association of the expressions of EZH2 and hnRNPK with the survival times of lung cancer patients, we processed Kaplan-Meier and Cox hazards survival regression analysis. For patients diagnosed with lung adenocarcinoma, the lower the expression of EZH2 or hnRNPK, the longer the survival time of lung cancer patients. However, the trends of survival time were conversely shown in patients diagnosed with lung squamous carcinoma, but there was no association between the expression of hnRNPK and the patient survival time, which indicated that the regulation of miR-125a on EZH2 and hnRNPK was mainly involved in lung adenocarcinoma.| Subtype (vs. normal) | EZH2 | hnRNPK | ||||
|---|---|---|---|---|---|---|
| Fold change | t | p | Fold change | t | p | |
| Adenocarcinoma | 3.19 | 9.08 | 2.29 × 10−13 | 1.16 | 4.65 | 5.85 × 10−6 |
| Squamous carcinoma | 6.47 | 13.66 | 4.45 × 10−16 | 1.22 | 5.81 | 1.93 × 10−7 |
| Large-cell lung cancer | 6.42 | 6.44 | 1.65 × 10−6 | 1.32 | 5.68 | 3.22 × 10−6 |
Functional annotation of 14 target genes and investigation of their roles in lung cancer
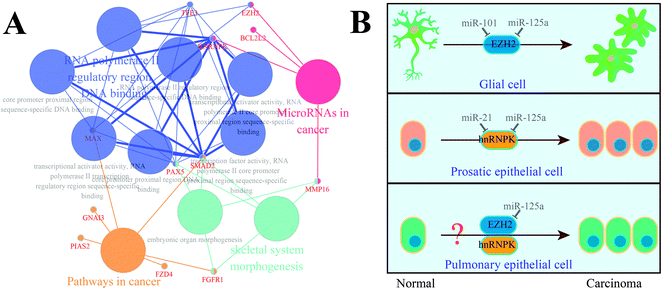
In parallel with the detection and comparison of 14 cancer-related target genes, we also performed functional re-annotation for these genes (Fig. 4). A total of 4 functional groups were enriched, among which four genes (MMP16, BCL2L2, hnRNPK, and EZH2) were involved in miRNAs in cancer signaling pathway. Considering the relationship between the gene expression and the patient survival time, we localized hnRNPK and EZH2 on the signaling pathway map of miRNAs in cancer and found that EZH2 and hnRNPK were mainly involved in the carcinogenesis of glial cells and prostatic epithelial cells, respectively; these two biological processes could be regulated by miR-101 and miR-21, respectively. Similarly, miR-125a was predicted to be a regulator of hnRNPK and EZH2, but it has never been investigated whether miR-125a can regulate the carcinogenesis of pulmonary epithelial cells. Hence, we assumed that the up-regulation of miR-125a in pneumoconiosis could suppress the expressions of hnRNPK and EZH2 and inhibit the carcinogenesis of pulmonary epithelial cells.Validation of the suppression of EZH2 and hnRNPK by miR-125a in lung cancer
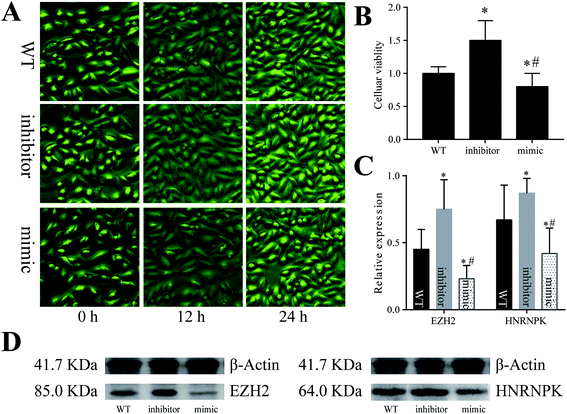
To validate the assumptions proposed above, we adopted cellular experiments using the A549 cell line derived from the pulmonary epithelial cells of lung adenocarcinoma (Fig. 5). After the cells were co-cultured with artificially synthesized miR-125a mimic or inhibitor, the number and viability of A549 cells both decreased in the miR-125a mimic group but increased in the miR-125a inhibitor group (compared with those of the wild-type cells). Moreover, the protein expression levels of hnRNPK and EZH2 were up-regulated when the cells were treated with the miR-125a inhibitor, but they were down-regulated after being co-cultured with the miR-125a mimic, indicating that miR-125a could directly suppress the expressions of hnRNPK and EZH2 and further negatively affect the carcinogenesis of pulmonary epithelial cells.Discussion
Due to the wide applications of nanomaterials, the chance for exposure of humans to nano dust particles has been increasing substantially. Crystalline silica dust, one of the main causes of pneumoconiosis in coal workers, has been identified as group I carcinogen with sufficient evidence of carcinogenicity by IARC;5 however, the relationship between pneumoconiosis and lung cancer, both of which can be induced by silica dust, is still under debate. To reveal their associations, we profiled the genome-wide miRNA expression signatures in exosomes isolated from pneumoconiosis patients.A total of 14 differentially expressed miRNAs were identified using DESeq and OPLS-DA analyses; of these, 3 miRNAs were up-regulated, and 6 miRNAs were down-regulated. Interestingly, miRNAs that were aberrantly expressed in pneumoconiosis also contributed to the development of different types of cancers. miR-125a, for example, an up-regulated miRNA analyzed in this study, has been found to possess lung cancer-suppressive features by targeting KRAS in the NF-κB signaling pathway.18 Meanwhile, this miRNA also participates in inhibiting cell proliferation19,20 and cancer metastasis.21 Another two up-regulated miRNAs, namely, miR-122 and miR-96 are seldom discussed. Available studies on miR-122 are mainly involved in hepatosteatosis.22 Additionally, current studies on the down-regulated miRNAs such as miR-146b and let-7i have indicated their roles in regulating papillary thyroid cancer,23 ulcerative colitis,24 and breast cancer.25 The down-regulation of miR-146b has been suggested to promote cancer development, whereas the up-regulation of let-7i has been predicted to improve the survival time of breast cancer. Thus, based on the similar regulatory mechanisms of miRNAs, we may conclude that the patients diagnosed with pneumoconiosis are more likely to suffer from cancer compared with the normal controls.
Specially, the role of miR-125a in lung cancer and pneumoconiosis was proposed and investigated in this study. We investigated the expression signatures of miR-125a in pneumoconiosis and explored its functions related to carcinogenesis. A total of 14 target genes including PAX5, GNAI3, TFE3, FGFR1, PIAS2, EZH2, MAX, BCL2L2, DGKZ, DGKE, MMP16, hnRNPK, FZD4, and SMAD2 were identified in cancer-related signaling pathways via KEGG enrichment analysis; the biological functions of these 14 target genes were re-annotated in the downstream analysis, for which signaling pathways involving miRNAs were selected for further investigations. Similar to previously reported results, SMAD2 was found both in pneumoconiosis and in lung cancer.26,27 However, the other 13 target genes were found to be responsible for cancers only;28–31 they have not been reported to be responsible for pneumoconiosis.
For better understanding with respect to clinical conditions, we investigated the relationship between the expressions of these 14 target genes and the survival times of patients diagnosed with lung cancer. After searching for the Oncomine database, we found that EZH2 and hnRNPK were differentially expressed between groups of disease and normal. Interestingly, the up-regulated EZH2 and hnRNPK could clearly decrease the survival probability of patients diagnosed with lung cancer (EZH2, hazard ratio (HR) (95% CI): 1.45 (1.27–1.65), P < 0.001; hnRNPK, HR (95% CI): 1.32 (1.16–1.50), P < 0.001). Besides, a similar expression pattern was also found for lung adenocarcinoma (EZH2, HR (95% CI): 1.49 (1.17–1.89), P < 0.001; hnRNPK, HR (95% CI): 1.73 (1.35–2.21), P < 0.001), but it was not found for lung squamous carcinoma (EZH2, HR (95% CI): 0.78 (0.59–1.04), P = 0.09; hnRNPK, HR (95% CI): 0.88 (0.70–1.12), P = 0.31), indicating that the regulatory mechanism of miR-125a on EZH2 and hnRNPK was mainly involved in lung adenocarcinoma, whereas the occurrence and the development of lung squamous carcinoma may not be affected by miR-125a and its target genes. Moreover, the expressions of EZH2 and hnRNPK in different subtypes of lung cancer were also investigated and compared with those of the normal adjacent tissue. Consistent with the survival analysis, the expression levels of EZH2 and hnRNPK in all subtypes of lung cancer were all up-regulated. The roles of EZH2 and hnRNPK have also been proved in many previously published studies. For example, the inhibition of EZH2 by miR-4465 in non-small-cell lung cancer has been suggested to suppress tumor proliferation and metastasis,32 and three available studies also demonstrated the role of hnRNPK in lung cancer resistance via inhibiting GSK3β-Ser9 phosphorylation.33–35
Considering the functional re-annotation results of 14 cancer-related target genes and the survival analysis of miRNA-related genes, we selected EZH2 and hnRNPK for further examination. First, we localized these two genes on the map of the signaling pathway of microRNAs in cancer and then, diseases such as prostate cancer and glioblastoma, which are related to EZH2 and hnRNPK, were identified. In detail, EZH2 could be down-regulated by an excessive amount of miR-101 in the prostatic epithelial cells,36 whereas hnRNPK was up-regulated by an insufficient amount of miR-21 in the glial cells;37 these two genes were predicted to be regulated by miR-125a. However, should miR-125a along with its target genes EZH2 and hnRNPK also play roles in lung cancer? To answer this question, we assumed that miR-125a along with its target genes EZH2 and hnRNPK regulated the carcinogenesis of pulmonary epithelial cells, which can affect the occurrence and development of lung adenocarcinoma.
To validate our assumption, the A549 cell line derived from lung adenocarcinoma was co-cultured with an artificially synthesized miR-125a mimic or inhibitor. Surprisingly, the number of A549 cells increased after incubation with the miR-125a inhibitor, but it decreased in the miR-125a mimic group, and the cellular viability detected by the CCK8 assay showed a similar trend, specifying the role for miR-125a in suppressing A549 cell proliferation in lung adenocarcinoma. To investigate regulatory mechanisms involved, the protein expression levels of EZH2 and hnRNPK were detected using western blot; both of these two proteins were down-regulated in the miR-125a mimic group, whereas they were up-regulated in the miR-125a inhibitor group, indicating that miR-125a could inhibit A549 cell proliferation via down-regulating the expressions of EZH2 and hnRNPK. Similar to miR-101, miR-125a excessively expressed and contributed to the suppression of EZH2 and hnRNPK in pneumoconiosis. However, a large amount of miR-125a is not beneficial for the proliferation of cancer cells. Some studies have indicated that an elevated expression level of EZH2 might be potentially used as a biomarker for the diagnosis of prostate cancer.38,39 Typically, the expression of EZH2 has been detected to be up-regulated in non-small-cell lung cancer,40,41 which was consistent with the findings in this study.
However, there were some inadequacies in this study. For example, the unbalanced distribution of baseline characteristics such as age, body mass index, and other confounding factors may affect the excretion and composition of exosomes. Although no available evidence has been found for the concerns we proposed here, the roles of these confounding factors should be highlighted in further investigations.
In conclusion, the exosomal miR-125a was identified to be up-regulated in nanoparticle-induced pneumoconiosis, and the suppression of EZH2 and hnRNPK by miR-125a was then determined in lung adenocarcinoma. Thus, we may conclude that miR-125a is a bridge for connecting pneumoconiosis and lung adenocarcinoma.
Conflicts of interest
The authors have declared that no conflict of interest exists.Acknowledgements
We thank the members of the basic medical college, Zhengzhou University for their comments and suggestions and also the reviewers who made valuable suggestions that improved the quality of the presentation. This work was supported by the Natural Science Foundation of China [81273039, 81472954, and 81773404].References
- S. H. Lee, D. S. Kim, Y. W. Kim, M. P. Chung, S. T. Uh, C. S. Park, S. H. Jeong, Y. B. Park, H. L. Lee, J. S. Song, J. W. Shin, N. S. Yoo, E. J. Lee, J. H. Lee, Y. Jegal, H. K. Lee and M. S. Park, Chest, 2015, 147, 465–474 CrossRef PubMed.
- K. Steenland and E. Ward, Ca-Cancer J. Clin., 2014, 64, 63–69 CrossRef PubMed.
- Y. Liu, K. Steenland, Y. Rong, E. Hnizdo, X. Huang, H. Zhang, T. Shi, Y. Sun, T. Wu and W. Chen, Am. J. Epidemiol., 2013, 178, 1424–1433 CrossRef PubMed.
- J. Y. W. Chan, J. C. C. Tsui, P. T. W. Law, W. K. W. So, D. Y. P. Leung, M. M. K. Sham, S. K. W. Tsui and C. W. H. Chan, J. Appl. Toxicol., 2017, 37, 1162–1173 CrossRef PubMed.
- I. W. G. o. t. E. o. C. R. t. Humans, I. A. f. R. o. Cancer and W. H. Organization, Silica, some silicates, coal dust and para-aramid fibrils, World Health Organization, 1997 Search PubMed.
- K. Steenland, A. Mannetje, P. Boffetta, L. Stayner, M. Attfield, J. Chen, M. Dosemeci, N. DeKlerk, E. Hnizdo, R. Koskela and H. Checkoway, International Agency for Research on, Cancer Causes Control, 2001, vol. 12, pp. 773–784 Search PubMed.
- I. W. G. o. t. E. o. C. R. t. Humans, IARC Monogr. Eval. Carcinog. Risks Hum., 2012, 100, 11–465 Search PubMed.
- S. Poinen-Rughooputh, M. S. Rughooputh, Y. Guo, Y. Rong and W. Chen, BMC Public Health, 2016, 16, 1137 CrossRef PubMed.
- B. A. Muzembo, Y. Deguchi, N. R. Ngatu, M. Eitoku, R. Hirota and N. Suganuma, Environ. Int., 2015, 77, 16–24 CrossRef PubMed.
- N. Esmaeil, M. Gharagozloo, A. Rezaei and G. Grunig, Am. J. Clin. Exp. Immunol., 2014, 3, 20–29 Search PubMed.
- C. Li, Y. Lu, S. Du, S. Li, Y. Zhang, F. Liu, Y. Chen, D. Weng and J. Chen, Theranostics, 2017, 7, 4255–4275 CrossRef PubMed.
- F. Doerr, J. George, A. Schmitt, F. Beleggia, T. Rehkamper, S. Hermann, V. Walter, J. P. Weber, R. K. Thomas, M. Wittersheim, R. Buttner, T. Persigehl and H. C. Reinhardt, Sci. Rep., 2017, 7, 15511 CrossRef PubMed.
- M. K. Elzarrad, A. Haroon, K. Willecke, R. Dobrowolski, M. N. Gillespie and A. B. Al-Mehdi, BMC Med., 2008, 6, 20 CrossRef PubMed.
- L. Xu, B. F. Yang and J. Ai, J. Cell. Physiol., 2013, 228, 1713–1719 CrossRef PubMed.
- K. Boriachek, M. N. Islam, A. Moller, C. Salomon, N. T. Nguyen, M. S. A. Hossain, Y. Yamauchi and M. J. A. Shiddiky, Small, 2018, 14, 21 CrossRef PubMed.
- A. Montecalvo, A. T. Larregina, W. J. Shufesky, D. B. Stolz, M. L. Sullivan, J. M. Karlsson, C. J. Baty, G. A. Gibson, G. Erdos, Z. Wang, J. Milosevic, O. A. Tkacheva, S. J. Divito, R. Jordan, J. Lyons-Weiler, S. C. Watkins and A. E. Morelli, Blood, 2012, 119, 756–766 CrossRef PubMed.
- L. Zhang, C. Hao, S. Yao, R. Tang, W. Guo, H. Cong, J. Li, L. Bao, D. Wang, Y. Li, X. Yu, S. Duan and W. Yao, Small, 2018, 14, e1704008 CrossRef PubMed.
- S. Naidu, L. Shi, P. Magee, J. D. Middleton, A. Lagana, S. Sahoo, H. S. Leong, M. Galvin, K. Frese, C. Dive, V. Guzzardo, M. Fassan and M. Garofalo, Sci. Rep., 2017, 7, 15441 CrossRef PubMed.
- X. D. Yao, P. Li and J. S. Wang, Exp. Ther. Med., 2017, 14, 1699–1705 CrossRef PubMed.
- H. Zheng, J. Wu, J. Shi, C. Lu, Y. Wang, Q. Sun, G. Zhang and G. Zhao, Oncol. Rep., 2017, 38, 1790–1796 CrossRef PubMed.
- D. Chen, Q. Sun, L. Zhang, X. Zhou, X. Cheng, D. Zhou, F. Ye, J. Lin and W. Wang, Oncotarget, 2017, 8, 70642–70652 Search PubMed.
- M. Naderi, A. Pazouki, E. Arefian, S. M. Hashemi, F. Jamshidi-Adegani, O. Gholamalamdari, S. Soudi, K. Azadmanesh, S. Mirab Samiee, S. Merat, M. Gholami Fesharaki, M. Mondanizadeh, M. Vasei and M. Soleimani, Arch. Iran. Med., 2017, 20, 165–171 Search PubMed.
- T. Stokowy, D. Gawel and B. Wojtas, Int. J. Endocrinol., 2016, 2016, 1427042 Search PubMed.
- R. Ranjha, S. Aggarwal, S. Bopanna, V. Ahuja and J. Paul, PLoS One, 2015, 10, e0142869 CrossRef PubMed.
- A. Carmona, F. Guerrero, P. Buendia, T. Obrero, P. Aljama and J. Carracedo, Front. Physiol., 2017, 8, 666 CrossRef PubMed.
- X. Ji, B. Wu, J. Fan, R. Han, C. Luo, T. Wang, J. Yang, L. Han, B. Zhu, D. Wei, J. Chen and C. Ni, Sci. Rep., 2015, 5, 14131 CrossRef PubMed.
- A. Wang, C. Lu, Z. Ning, W. Gao, Y. Xie, N. Zhang, J. Liang, F. S. Abbasi, Q. Yan and J. Liu, Oncotarget, 2017, 8, 28247–28259 Search PubMed.
- H. Greschik, D. Duteil, N. Messaddeq, D. Willmann, L. Arrigoni, M. Sum, M. Jung, D. Metzger, T. Manke, T. Gunther and R. Schule, Cell Death Dis., 2017, 8, e3173 CrossRef PubMed.
- C. Fumarola, D. Cretella, S. La Monica, M. A. Bonelli, R. Alfieri, C. Caffarra, F. Quaini, D. Madeddu, A. Falco, A. Cavazzoni, G. Digiacomo, G. Mazzaschi, V. Vivo, E. Barocelli, M. Tiseo, P. G. Petronini and A. Ardizzoni, Oncotarget, 2017, 8, 91841–91859 CrossRef PubMed.
- T. Januario, X. Ye, R. Bainer, B. Alicke, T. Smith, B. Haley, Z. Modrusan, S. Gould and R. L. Yauch, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 12249–12254 CrossRef PubMed.
- E. Arbajian, F. Puls, C. R. Antonescu, F. Amary, R. Sciot, M. Debiec-Rychter, V. P. Sumathi, M. Jaras, L. Magnusson, J. Nilsson, J. Hofvander and F. Mertens, Clin. Cancer Res., 2017, 23, 7426–7434 CrossRef PubMed.
- J. Sun, X. Tian, S. Q. Lu and H. B. Hu, Biomed. Pharmacother., 2017, 96, 1358–1362 CrossRef PubMed.
- X. Gao, J. Feng, Y. He, F. Xu, X. Fan, W. Huang, H. Xiong, Q. Liu, W. Liu, X. Liu, X. Sun, Q. Y. He, Q. Zhang and L. Liu, Sci. Rep., 2016, 6, 22999 CrossRef PubMed.
- L. P. Li, C. H. Lu, Z. P. Chen, F. Ge, T. Wang, W. Wang, C. L. Xiao, X. F. Yin, L. Liu, J. X. He and Q. Y. He, Proteomics, 2011, 11, 429–439 CrossRef PubMed.
- A. M. Leopoldino, F. Carregaro, C. H. Silva, O. Feitosa, U. M. Mancini, J. M. Freitas and E. H. Tajara, Genome, 2007, 50, 451–462 CrossRef PubMed.
- X. Wu, X. Dinglin, X. Wang, W. Luo, Q. Shen, Y. Li, L. Gu, Q. Zhou, H. Zhu, Y. Li, C. Tan, X. Yang and Z. Zhang, Oncotarget, 2017, 8, 76015–76028 Search PubMed.
- Y. M. Park, S. J. Hwang, K. Masuda, K. M. Choi, M. R. Jeong, D. H. Nam, M. Gorospe and H. H. Kim, Mol. Cell. Biol., 2012, 32, 4237–4244 CrossRef PubMed.
- Z. Ling, X. Wang, T. Tao, L. Zhang, H. Guan, Z. You, K. Lu, G. Zhang, S. Chen, J. Wu, J. Qian, H. Liu, B. Xu and M. Chen, J. Exp. Clin. Cancer Res., 2017, 36, 159 CrossRef PubMed.
- D. P. Labbe, C. J. Sweeney, M. Brown, P. Galbo, S. Rosario, K. M. Wadosky, S. Y. Ku, M. Sjostrom, M. Alshalalfa, N. Erho, E. Davicioni, R. J. Karnes, E. M. Schaeffer, R. B. Jenkins, R. B. Den, A. E. Ross, M. Bowden, Y. Huang, K. P. Gray, F. Y. Feng, D. E. Spratt, D. W. Goodrich, K. H. Eng and L. Ellis, Clin. Cancer Res., 2017, 23, 7072–7083 CrossRef PubMed.
- Z. Q. Yuan, W. L. Chen, B. G. You, Y. Liu, S. D. Yang, J. Z. Li, W. J. Zhu, X. F. Zhou, C. Liu and X. N. Zhang, J. Control. Release, 2017, 268, 198–211 CrossRef PubMed.
- C. C. Sun, S. J. Li, G. Li, R. X. Hua, X. H. Zhou and D. J. Li, Mol. Ther.–Nucleic Acids, 2016, 5, e385 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra03081b |
| This journal is © The Royal Society of Chemistry 2018 |