Open Access Article
Open Access ArticleComposition and role of the attached and planktonic microbial communities in mesophilic and thermophilic xylose-fed microbial fuel cells†
Paolo Dessì *a,
Estefania Porcab,
Johanna Haavistoa,
Aino-Maija Lakaniemi
*a,
Estefania Porcab,
Johanna Haavistoa,
Aino-Maija Lakaniemi a,
Gavin Collins
a,
Gavin Collins b and
Piet N. L. Lens
b and
Piet N. L. Lens ac
ac
aLaboratory of Chemistry and Bioengineering, Tampere University of Technology, P.O. Box 541, FI-33101 Tampere, Finland. E-mail: paolo.dessi@tut.fi; Tel: +358 417239696
bMicrobial Communities Laboratory, School of Natural Sciences, National University of Ireland Galway, University Road, Galway, H91 TK33, Ireland
cUNESCO-IHE, Institute for Water Education, Westvest 7, 2611AX Delft, The Netherlands
First published on 15th January 2018
Abstract
A mesophilic (37 °C) and a thermophilic (55 °C) two-chamber microbial fuel cell (MFC) were studied and compared for their power production from xylose and the microbial communities involved. The anode-attached, membrane-attached, and planktonic microbial communities, and their respective active subpopulations, were determined by next generation sequencing (Illumina MiSeq), based on the presence and expression of the 16S rRNA gene. Geobacteraceae accounted for 65% of the anode-attached active microbial community in the mesophilic MFC, and were associated to electricity generation likely through direct electron transfer, resulting in the highest power production of 1.1 W m−3. A lower maximum power was generated in the thermophilic MFC (0.2 W m−3), likely due to limited acetate oxidation and the competition for electrons by hydrogen oxidizing bacteria and hydrogenotrophic methanogenic archaea. Aerobic microorganisms, detected among the membrane-attached active community in both the mesophilic and thermophilic MFC, likely acted as a barrier for oxygen flowing from the cathodic chamber through the membrane, favoring the strictly anaerobic exoelectrogenic microorganisms, but competing with them for xylose and its degradation products. This study provides novel information on the active microbial communities populating the anodic chamber of mesophilic and thermophilic xylose-fed MFCs, which may help in developing strategies to favor exoelectrogenic microorganisms at the expenses of competing microorganisms.
1. Introduction
The microbial fuel cell (MFC) is an emerging technology for the direct bioconversion of chemical energy of organic substrates to electrical energy. MFCs consist of two electrodes (anode and cathode) connected through an external electrical circuit. The anode acts as electron acceptor in the bioelectrochemical redox reactions of microbial metabolism, whereas the cathode acts as electron donor for biotic or abiotic reactions. The combination of anodic and cathodic reactions creates a potential difference between the electrodes which drives the electrons to migrate from the anode to the cathode, thus generating electrical current (for a review, see Butti et al.1).Biological electricity production in MFCs requires microorganisms capable to oxidize the substrates and transfer the electrons exogenously to the solid anode electrode. Electrons can be transferred to the anode essentially through three mechanisms: short range, long range, and mediated electron transfer (for reviews, see Kumar et al.2 and Kalathil et al.3). Some microorganisms, such as Geobacter sulfurreducens, can transfer electrons to a surface directly via redox-active proteins present on the outer surface of their cell membrane, such as c-type cytochromes, or via conductive pili called nanowires.4,5 G. sulfurreducens develops multi-layer structured biofilms, in which nanowires connect the different cells, enabling the electron transfer to the anode.6 Mediators, in their oxidized form, penetrate the microbial cell and become reduced during cellular metabolism. They then diffuse out of the cell and release the electrons at the anode, becoming oxidized again and thus reusable.5 Some species, such as Pseudomonas, produce mediators such as pyocyanin endogenously.7 Once mediators are produced, also other microorganisms present in the mixed culture system can use them to transfer the electrons to the anode.8
Pure cultures of electrochemically active microorganisms, such as Geobacter sp.9–11 and Shewanella sp.,12,13 have shown power production from simple substrates such as volatile fatty acids and sugars at mesophilic conditions (25–37 °C) and neutral pH (6.8–7.3). Mixed cultures are more practical for wastewater treatment, as they contain a consortium of hydrolytic, fermentative and electroactive microorganisms able to produce electricity from complex substrates.9 However, due to the competition for electron donor with non-exoelectrogenic microorganisms such as methanogenic archaea,14 power production can remain low, and operational conditions must be optimized to favor exoelectrogenic microorganisms. Catal et al.15 compared electricity production from 12 monosaccharides present in lignocellulosic biomass, including pentoses and hexoses, in a mesophilic (30 °C) MFC inoculated with a mixed culture adapted to acetate. Xylose resulted in the highest potential for electricity production over the other hexoses and pentoses tested.
Thermophilic electricity production could be advantageous because of the high rate of biochemical reactions, and thus high electron production rates, of thermophilic microorganisms.16 MFCs have been operated at temperatures up to 98 °C.17 However, although over 20 species of microorganisms, mainly belonging to the Proteobacteria phylum, have been reported to produce electricity under mesophilic conditions, the number of known thermophilic exoelectrogenic microorganisms is much lower.18 To date, only few species have been reported to produce electricity at thermophilic conditions, including Firmicutes such as Caloramator australicus,18 Thermincola potens,19 Thermincola ferriacetica,20 and Thermoanaerobacter pseudethanolicus,21 as well as Deferribacteres such as Calditerrivibrio nitroreducens.22
Investigating the composition of the active subpopulation, rather than the whole microbial community, is crucial in understanding the role of microorganisms in MFCs. DNA-based methods may drive to erroneous conclusions in the detection of the key species in bioreactors.23 Previously performed microbial community analyses have, nevertheless, mainly targeted the presence of the 16S rRNA gene (DNA) whereas, to our knowledge, only one study19 has also focused on 16S rRNA gene expression (RNA), which is an indicator of the microbial activity.23 Furthermore, especially in studies on thermophilic MFCs, microbial community analyses have mainly focused on the anode-attached microbial community, lacking information on the planktonic microbial community. The latter community could be involved in electricity generation as well, either directly, by performing mediated electron transfer to the anode24 or indirectly, by converting the substrates to compounds readily available for the exoelectrogenic microorganisms.
In addition, the membrane is a suitable surface for the establishment of a biofilm. Although biofouling of the membrane has been reported in MFC studies,14,25 only Lu et al.26 have reported the composition of a membrane-attached microbial community in two brewery wastewater-fed MFCs operated in series at ambient temperature (20–22 °C). However, the microbial community analysis was performed only at DNA level, and the role of the membrane-attached microorganisms detected on the MFC performance was not discussed.26 Although likely not directly involved in electricity generation, membrane-attached microorganisms may have a role in the functioning of MFCs, which must be elucidated. Therefore, the aim of this study was to investigate the microbial communities growing (i) as anodic biofilm, (ii) in suspended form in the anodic solution (planktonic), and (iii) as biofilm on the membrane of a mesophilic (37 °C) and a thermophilic (55 °C) xylose-fed MFC. Both presence and expression of the 16S rRNA gene were determined with the aim to investigate both the composition of the overall microbial community and the active subpopulation. Power production, as well as xylose and metabolite concentration profiles were also analyzed to determine the possible differences in the electricity production pathways at 37 and 55 °C.
2. Experimental
2.1 Source of anodic microorganisms
In order to ensure a large variety of microbial species capable of living under a broad temperature range and degrading xylose, two inocula (15 mL each) were mixed and provided to each MFC. The first one was activated sludge from a municipal wastewater treatment plant (Viinikanlahti, Tampere, Finland), which has shown potential for anaerobic energy production in the form of dark fermentative hydrogen production at temperatures up to 55 °C.27 The second one was anolyte from a xylose-fed MFC operating at 37 °C.28 The volatile solids content was 10.6 (±0.2) and 8.4 (±0.5) g L−1 for the activated sludge and the anolyte, respectively. The mixture of the two inocula was flushed with N2 for 10 min before introducing into the anode chambers of the MFCs.2.2 Anolyte composition
The anolyte was prepared according to Mäkinen et al.,29 but EDTA, yeast extract, and resazurin were not added. The substrate was xylose (0.3 or 1 g L−1, as specified in Section 2.4). The pH was kept at 7 (±0.2) using phosphate buffer. The anolyte conductivity was 14.6 mS cm−1. The composition of the feeding solution was the same as the anolyte, but with a 10-times higher xylose concentration.2.3 MFC configuration
The h-type two-chamber MFCs were constructed by connecting two glass bottles (Adams & Chittenden Scientific Glass, USA) separated by an anion exchange membrane (AMI-7001, Membranes International Inc., USA) with a diameter of 5.2 cm. The total volume of the anodic and the cathodic chamber was 350 mL each. The anode was a carbon brush (5 cm length and 1.5 cm diameter), while the cathode was a carbon cloth (5 × 4 cm) coated with approximately 20 mg of a Pt-based catalyst (20% platinum on Vulcan XC-72R carbon, E-TEK, USA). The two electrodes were connected through an external resistance of 100 Ω. A reference electrode (BASi RE-5B Ag/AgCl) was inserted into the anodic chamber, close to the anode. The anodic chamber was filled with the anolyte (270 mL) and inoculum (30 mL), and its content was continuously mixed by magnetic stirring. The cathodic chamber was filled with milliQ water (300 mL), and oxygen was provided as the terminal electron acceptor by pumping air from outside using an aquarium air pump (Marina 50) at a flow rate of 130 mL min−1. In the mesophilic MFC, the water lost by evaporation (circa 15 mL per day) was replaced manually every 1–2 days. In the thermophilic MFC, due to the faster evaporation (circa 50 mL per day), the water was replaced daily through a pump connected to a timer. Temperature of the mesophilic and thermophilic MFC was kept at 37 (±2) °C and 55 (±2) °C, respectively, by using two incubators (Memmert, Germany).2.4 MFC operation
Both MFCs were operated in fed-batch mode. The feeding steps were done by replacing 30 mL of anolyte (10% of the total volume) with 30 mL of the feeding solution. In order to avoid substrate overload in the start-up phase, the first six fed-batch cycles were conducted with an initial xylose concentration of 0.3 g L−1. The initial xylose concentration was then increased to 1.0 g L−1 for the following eleven fed-batch cycles, referred to as I–XI. In the results section, day 0 refers to the first day of operation with a xylose concentration of 1 g L−1. The MFCs were fed every 5–6 days when the initial xylose concentration was 0.3 g L−1, and every 7–8 days when initial xylose concentration was 1.0 g L−1.2.5 Sampling
Anolyte samples were collected at the beginning and at the end of every feeding step. During the feeding step “IX” (from day 55 to day 61) anolyte samples were collected at various time points to determine the COD, xylose and volatile fatty acids concentration profiles. Biomass samples from both MFCs were collected at the end of the experiment (day 72) from three different sampling points: anodic electrode (anode-attached), membrane (membrane-attached) and anolyte (planktonic). The anodic electrode and the membranes were taken out of the MFC, put into 50 mL sterile Falcon tubes with about 30 mL of autoclaved 0.9% NaCl solution, sonicated for 2 minutes at 50–60 Hz (Finnsonic, Finland) and strongly shaken in order to detach as much biofilm as possible. The two resulting samples, as well as a sample of the anolyte, were concentrated by consecutive centrifugation cycles (10 min, 5000 rpm) in 5 mL sterile Eppendorf tubes using a Sigma 4k145 centrifuge. The resulting pellets were re-suspended in 5 mL autoclaved 0.9% NaCl solution and stored at −85 °C until carrying out the microbial community analyses.2.6 Analytical methods
Voltage and anodic potential were measured with a data logger (Agilent 34970A, Agilent technologies, Canada) at 2 minutes intervals. The anodic potential was measured against the Ag/AgCl reference electrode. Soluble COD was measured using the dichromate method according to the Finnish standard SFS 5504. Anolyte conductivity and pH were measured with a conductivity meter (WTW inoLab, Germany) and a pH meter (WTW pH 330 meter with Hamilton Slimtrode probe), respectively. Dissolved oxygen in the cathodic chamber was measured by a multi-parameter meter (HQ40d) with a standard luminescent/optical dissolved oxygen probe (IntelliCAL). Xylose, volatile fatty acids, and alcohols were measured by a high performance liquid chromatography (HPLC) system equipped with a Rezex RHM-monosaccharide column (Phenomenex, USA) as described earlier by Dessì et al.27 Some of the chromatograms obtained are provided as an example in the ESI (Fig. S1†).2.7 Coulombic efficiency, power and polarization curves
Coulombic efficiency (CE) was calculated according to Oh et al.,30 accounting 20 mol electrons exchanged per mol of xylose, according to the following equation:| C5H10O5 + 5H2O → 5CO2 + 20H+ + 20e− | (1) |
Power and polarization curves were obtained on day 64 and 71 from the mesophilic and thermophilic MFC, respectively. The electrical circuit was kept open for 30 minutes before the analysis to obtain the open circuit voltage (OCV). The circuit was then closed through a resistor box (TENMA 72-7270, Taiwan) and the resistance was decreased stepwise from 15 kΩ to 5 Ω at 30 minute intervals. Voltage was recorded just before switching the resistance. Power density and current density were calculated as P = U2/(R·V) and I = U/(R·V), respectively, where U is the voltage recorded in the data logger, R is the external resistance, and V is the anolyte volume (300 mL).
2.8 Microbial community analyses
Nucleic acids were co-extracted from the biomass samples using the method from Griffiths et al.,31 with the following modifications: 3 M sodium acetate (1/10 of sample volume) and cold (−20 °C) 100% isopropanol (1 sample volume) were added for precipitation instead of polyethelene glycol and nucleic acids were re-suspended in sterile water instead of tris–EDTA buffer. DNA and RNA were quantified by a Nanodrop spectrophotometer (NanoDrop Technologies, Wilmington, USA), and their quality was assessed by measuring the absorbance ratio at 260/280 nm and 260/230 nm wavelength. No further treatment was performed on nucleic acid samples for DNA level analysis. For RNA level analysis, nucleic acid samples were diluted to a final concentration of 25 ng mL−1. DNA was removed by the addition of 1 μL turbo DNase and 2.5 μL turbo DNase buffer (Invitrogen, Thermo Fisher, USA), followed by incubation at 37 °C for 30 minutes. DNase was then inactivated by addition of 2.5 μL DNase inactivator (Invitrogen) and separated from the RNA containing liquid by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 1.5 minutes). The absence of DNA was confirmed by bacterial 16S rRNA gene PCR (primers 338f and 805r) followed by electrophoresis in 1% agarose gel (no bands obtained). Complementary DNA (cDNA) was obtained from RNA using M-MuLV Reverse Transcriptase (New England BioLabs, USA), according to the instructions provided by the supplier. Bacterial 16S rRNA gene PCR was then applied to confirm the success of the reverse transcription (bands appeared).
000 × g, 1.5 minutes). The absence of DNA was confirmed by bacterial 16S rRNA gene PCR (primers 338f and 805r) followed by electrophoresis in 1% agarose gel (no bands obtained). Complementary DNA (cDNA) was obtained from RNA using M-MuLV Reverse Transcriptase (New England BioLabs, USA), according to the instructions provided by the supplier. Bacterial 16S rRNA gene PCR was then applied to confirm the success of the reverse transcription (bands appeared).
Samples of both DNA and cDNA (12 μL) were collected in a 96-well plate and sent to FISABIO (Valencia, Spain) for partial 16S rRNA genes (DNA) or 16S rRNA (cDNA) high-throughput sequencing on an Illumina MiSeq platform. Forward and reverse primers for PCR were 515f and 806r, respectively.32 Sequence screening, alignment to the Silva (v128) database, clustering, chimeras removal and taxonomic classification (97% cutoff) were performed using Mothur v1.39.3,33 following the procedure described by Kozich et al.34 A total of 1,130,353 raw sequences was obtained from 12 samples, and 1,058,675 passed the quality filters. Relative abundance and diversity analyses were performed using the R software.35 The Illumina sequencing data was deposited to the NCBI Sequence Read Archive under BioProject Number PRJNA428321.
3. Results
3.1 Power production in the mesophilic and thermophilic MFC
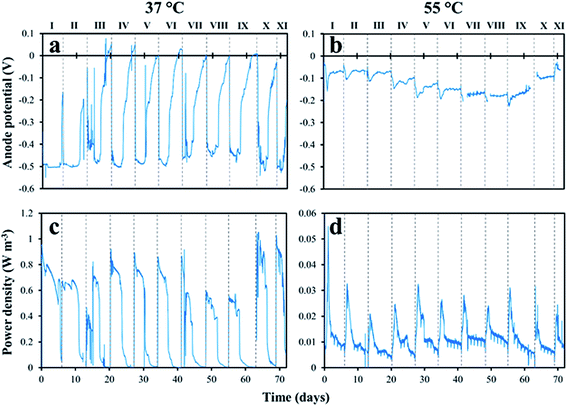
After the start-up period, the anodic potential in the mesophilic MFC was reproducible in all of the eleven fed-batch cycles with a xylose concentration of 1.0 g L−1 (Fig. 1a). It ranged between −450 mV and −520 mV (vs. Ag/AgCl). The anode potential of the thermophilic MFC was stable during the first 20 days of operation, with a minimum of about −100 mV. It then started to decrease, reaching a minimum of −230 mV in cycle IX, but increased again in cycles X and XI (Fig. 1b). A higher power density was obtained from the mesophilic MFC compared to the thermophilic MFC in the eleven fed-batch cycles (Fig. 1c vs. 1d). The maximum power density in the mesophilic MFC ranged between 0.55 W m−3 in cycle IX and 1.0 W m−3 in cycles X and XI, with a maximum CE of about 12%, while the power density in the thermophilic MFC rarely exceeded 0.03 W m−3 with a maximum CE of about 3%.3.2 Substrate degradation
At the end of all the eleven fed-batch cycles, the concentrations of xylose and organic metabolites in the anolyte of the mesophilic MFC were below the detection limit of the HPLC. However, acetate was found in the anolyte of the thermophilic MFC with a maximum concentration of 0.1 g L−1. Substrate consumption and metabolite production were monitored more closely in the fed-batch cycle IX, and compared to the obtained power density (Fig. 2). In the mesophilic MFC, xylose (1.0 g L−1) was consumed within 4 hours after its addition. Acetate and butyrate (0.2 and 0.1 g L−1, respectively) were detected after xylose consumption, but were then consumed within 48 hours. The soluble COD concentration decreased from 1.3 g L−1 to a final concentration of 0.25 g L−1. The power density increased until xylose was completely depleted, reaching a maximum of about 0.50 W m−3, then stabilized for about 80 hours before dropping to <0.05 W m−3 about 30 hours after the complete depletion of acetate and butyrate (Fig. 2).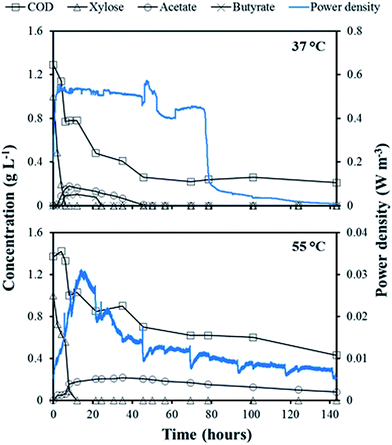 | ||
| Fig. 2 Power density and concentration of soluble COD, xylose, acetate, and butyrate present in the anolyte of the mesophilic (37 °C) and thermophilic (55 °C) MFC during fed-batch cycle IX. | ||
In the thermophilic MFC, xylose was depleted within 12 hours, and the resulting acetate produced reached a maximum of 0.2 g L−1 after 22 hours. The acetate concentration remained stable for about 45 hours after xylose depletion, before starting to be slowly consumed to a final concentration of 0.1 g L−1 after 143 hours (Fig. 2). The soluble COD concentration also slowly decreased to a final concentration of 0.5 g L−1. A power density peak of 0.03 W m−3 was obtained immediately after xylose depletion but, unlike the mesophilic MFC, no sudden power density drop occurred (Fig. 2).
3.3 Power and polarization curve
Based on the polarization data (Fig. 3), the power density was higher in the mesophilic MFC than in the thermophilic MFC regardless of the resistance applied. The maximum power densities of 1.1 and 0.2 W m−3 were obtained applying a resistance of 250 and 1000 Ω to the mesophilic and thermophilic MFC, respectively (Fig. 3a). The open circuit voltage (OCV) was approximately 0.7 and 0.4 V for the mesophilic and thermophilic MFC, respectively (Fig. 3b). Power overshoot (drastic drop of power and current density) occurred in the thermophilic MFC, when a resistance lower than 500 Ω was applied. The internal resistance, measured as the slope of the linear part of the polarization curve, was 270 Ω and 560 Ω for the mesophilic and the thermophilic MFC, respectively.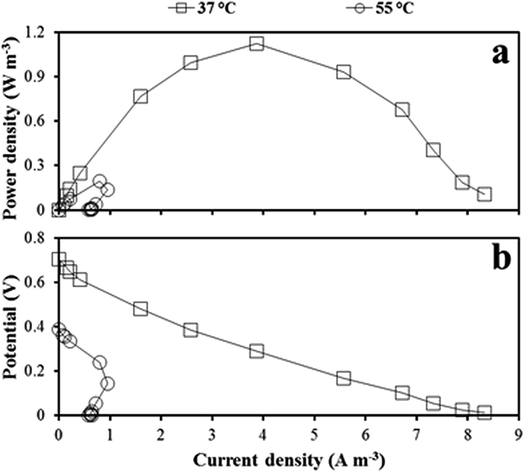 | ||
| Fig. 3 Power (a) and polarization (b) curve obtained from the mesophilic MFC in cycle X, and from the thermophilic MFC in cycle XI. Current and power density were normalized to the anolyte volume. | ||
3.4 Microbial community analysis
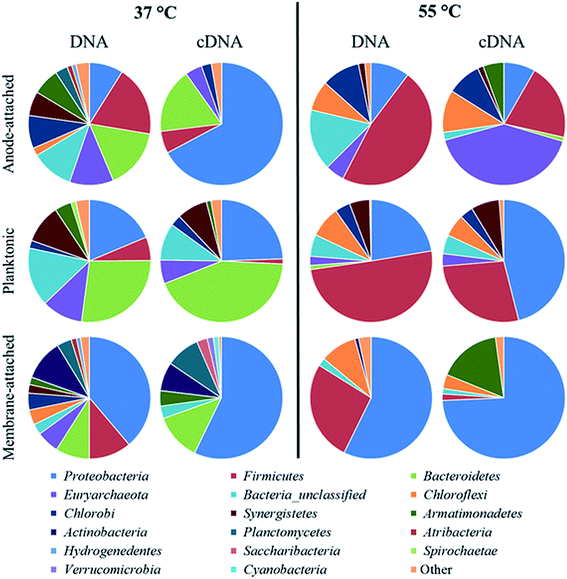
High-throughput Illumina MiSeq sequencing showed a clear difference in the composition of the microbial community not only among the mesophilic and thermophilic MFC, but also among the different sampling points in the same MFC (Fig. 4 and 5). A higher diversity was obtained in the DNA than in the cDNA samples, and in the mesophilic than in the thermophilic MFC, based on the diversity and evenness indexes (Table 1).| Temperature (°C) | Sample type | Microbial community | No. of sequencesa | No. of families | Shannon diversity | Simpson diversity | J′ evenness |
|---|---|---|---|---|---|---|---|
| a Refers to sequences which passed the quality check. All sample sizes were normalized according to the lowest number of sequences (28,349) prior to perform relative abundance and diversity analysis. | |||||||
| 37 | DNA | Anode-attached | 197,036 | 617 | 3.22 | 0.94 | 0.71 |
| Planktonic | 100,125 | 391 | 2.95 | 0.92 | 0.68 | ||
| Membrane-attached | 136,388 | 798 | 3.72 | 0.97 | 0.83 | ||
| cDNA | Anode-attached | 28,349 | 161 | 1.45 | 0.55 | 0.36 | |
| Planktonic | 108,967 | 326 | 2.74 | 0.91 | 0.64 | ||
| Membrane-attached | 68,638 | 341 | 3.26 | 0.93 | 0.80 | ||
| 55 | DNA | Anode-attached | 96,743 | 237 | 2.59 | 0.90 | 0.63 |
| Planktonic | 55,907 | 136 | 2.24 | 0.85 | 0.58 | ||
| Membrane-attached | 68,558 | 141 | 2.40 | 0.87 | 0.56 | ||
| cDNA | Anode-attached | 103,439 | 72 | 2.06 | 0.80 | 0.56 | |
| Planktonic | 33,729 | 98 | 1.75 | 0.72 | 0.46 | ||
| Membrane-attached | 60,796 | 80 | 1.48 | 0.65 | 0.41 | ||
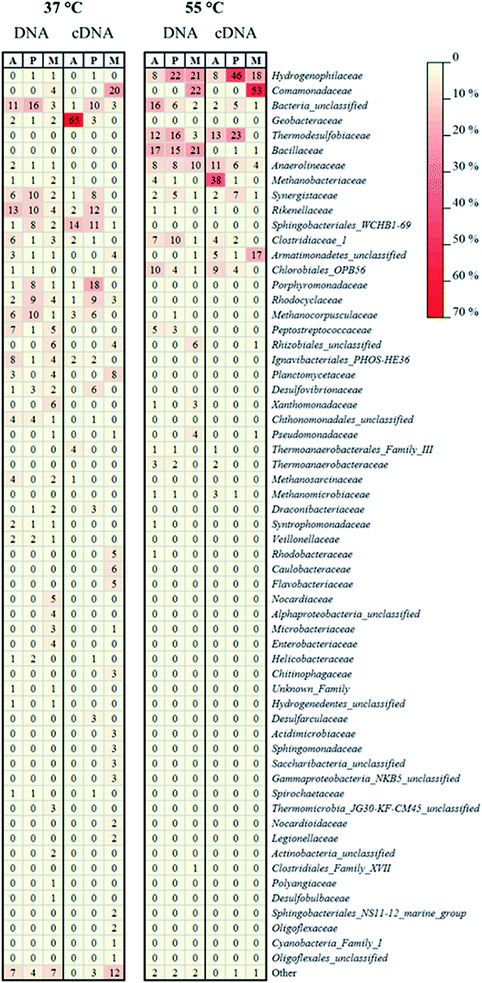
In the mesophilic MFC, the active anode-attached subpopulation was mainly composed of Proteobacteria belonging to the family of Geobacteraceae (65% of the total relative abundance), but Sphingobacteriales (14%) were also found (Fig. 4 and 5). The planktonic active subpopulation was more diverse, with the families of Porphyromonadaceae, Rikenellaceae, and Sphingobacteriales WCHB1-69 all above 10% of the relative abundance. Proteobacteria also dominated the membrane-attached active microbial community in the mesophilic MFC. Comamonadaceae was the most abundant family (20%), followed by a variety of families contributing to <10% of the total relative abundance (Fig. 4 and 5).
In the thermophilic MFC, the anode-attached active subpopulation was composed by Euryarchaeota (Fig. 4 and 5), mainly Methanobacteriaceae (38% of the total relative abundance), Firmicutes such as Thermodesulfobiaceae (13%), and Chloroflexi such as Anaerolineaceae (11%). Thermodesulfobiaceae were found also in the planktonic active subpopulation (23%), together with Hydrogenophylaceae (46%), and other less abundant families. Comamonadaceae was the most abundant active membrane-attached family (53%) of the thermophilic MFC, which included also Hydrogenophylaceae (18%), and an unclassified family belonging to the order of Armatimonadetes (17%) (Fig. 4 and 5).
4. Discussion
4.1 Bioelectricity production and microbial dynamics in the mesophilic MFC
An active microbial community mainly composed of Proteobacteria (Fig. 4) generated a relatively high power density in the mesophilic xylose-fed MFC (Fig. 1c). Indeed, most of the known mesophilic exoelectrogens belong to the phylum Proteobacteria.36 The diversity of the active anode-attached subpopulation (cDNA) was remarkably lower than the diversity of the whole community (DNA) (Table 1), confirming that the presence of microorganisms in a bioreactor does not relate to their activity.23 In particular, Geobacteraceae accounted only for 2% of the anode-attached microbial community, but was the prevalent (65%) active family (Fig. 5), and likely played a major role in power production. In fact, the Geobacteraceae family includes known exoelectrogenic microorganisms which have been widely reported to dominate the anodic microbial community in mesophilic MFCs, regardless of the inoculum source, substrate, and the MFC set-up.28,37–39 For example, Mei et al.40 showed that different microbial communities could develop in mesophilic (30 °C) MFCs started-up with different inocula, but Geobacter was found regardless of the inoculum.In this study, the remarkably higher diversity of the anode-attached community (DNA) than the active subpopulation (cDNA) (Table 1) suggests the presence of inactive or dead microorganisms, which could have hampered the activity of the Geobacteraceae, thus lowering power production.41 The relative abundance of active planktonic Geobacteraceae was only 3% (Fig. 5), suggesting that they were mainly growing attached to the anode. In fact, Geobacter sp. transfers electrons to the anode by direct contact transfer, but is unable to conduct long-range electron transfer.42 This is confirmed by the prompt power increase after the addition of xylose at the beginning of each fed-batch cycle (Fig. 1c), which is common in MFCs dominated by microorganisms performing direct electron transfer.20 Sphingobacteriales, found among both the active anode-attached and planktonic subpopulations in the mesophilic MFC (14 and 11% relative abundance, respectively), have been previously reported as part of the anodic microbial community,39,43 but further studies are required to assess their role in electricity generation.
No dominant family was detected in the active mesophilic planktonic subpopulation, but instead 6–7 families were present with a similar relative abundance (Fig. 5). Among them, both Desulfovibrionaceae44 and Rikenellaceae45 have been reported to produce electricity as pure cultures in MFCs. Rikenella sp. can perform glycolysis and mediated electron transfer to the anode,45 which likely explains its presence among the active mesophilic planktonic microbial community in this study (Fig. 5). The Rhodocyclaceae family includes Fe(III) reducers, such as Ferribacterium, which can be involved in bioelectricity production46 and has also been found in an anodic biofilm of an acetate-fed MFC.47 Porphyromonadeceae, which accounted for 18% of the active mesophilic planktonic subpopulation, have been previously detected both in the anode-attached and planktonic population in a mesophilic MFC treating starch, peptone, and fish extract.48 Although likely not directly involved in bioelectricity production, other microorganisms may also have contributed to the overall performance of the MFC. For example, Synergistaceae (8% of the relative abundance in the mesophilic active planktonic community) may be involved in the recycling of nutrients by quickly digesting the proteins of dead microorganisms.38
The membrane-attached active microbial community in the mesophilic MFC was highly diverse (Table 1). Comamonadaceae, which accounted for 20% of the active population, include facultative anaerobic microorganisms capable of using short chain volatile fatty acids as a source of carbon for their metabolism.49 Species belonging to the Comamonadaceae family, such as Comamonas denitrificans, have been previously found in the anodic biofilm of MFCs, and even shown to produce electricity in the absence of oxygen.50 However, Comamonadaceae were found in this study exclusively on the membrane, suggesting that they had a minor role in bioelectricity generation. Oxygen can flow from the cathodic to the anodic chamber through the AMI-7001 anion exchange membrane with a diffusivity coefficient of 4.3 × 10−6 cm2 s−1,51 thus exposing the anodic microorganisms to oxygen. The aerobic or facultative membrane-attached microorganisms may consume the oxygen crossing the membrane, favoring the strictly anaerobic exoelectrogens, but also competing with them for the substrates. Kim et al.51 estimated that, due to the higher biomass yield of aerobes compared to anaerobes, about 10% of the substrate was consumed through aerobic metabolism, reducing the CE of their acetate-fed (1.2 g L−1) MFCs. However, they did not perform microbial community analysis to confirm their hypothesis. Besides, membrane-attached microorganisms may reduce power output also by forming a thick biofilm which limits proton transfer from the anodic to the cathodic chamber.25
4.2 Bioelectricity production and microbial dynamics in the thermophilic MFC
In the thermophilic MFC, the relatively low number of active anode-attached microbial families (Table 1) suggests the scarcity of thermophilic exoelectrogenic species. The inoculum selected for the experiment, which was not previously enriched for thermophilic electricity production, can be one of the causes hindering the establishment of an active exoelectrogenic community. However, the same activated sludge was successfully used to enrich dark fermentative hydrogen producers at 55 °C in a previous study.27 In addition, 20% of the anode-attached active subpopulation was composed by Firmicutes, which have been previously reported to generate electricity in thermophilic, acetate-fed MFCs.19 About 66% of Firmicutes found in the thermophilic anode-attached community belonged to the family Thermodesulfobiaceae, which includes Coprothermobacter sp., a proteolytic microorganism involved in the fermentation of organic substrates, with production of pyruvate, formate and acetate, and also in syntrophic acetate oxidation (for a review, see Gagliano et al.52). The activity of Coprothermobacter is enhanced by establishing a syntrophy with hydrogenotrophic methanogenic archaea such as Methanothermobacter.53 Methanothermobacter belongs to the family of Methanobacteraceae, which was indeed among the most abundant active anode-attached families in the thermophilic MFC in this study (Fig. 5). Although Coprothermobacter was previously found among the anode-attached microbial community of thermophilic acetate-fed MFCs,19,54 and is thus a possible acetate-utilizing anode respiring bacterium, its electrochemical activity as a pure culture has not yet been investigated. Also microorganisms belonging to the order of Chlorobiales, despite being mainly phototrophs, can perform heterotrophic anaerobic respiration, and have been reported as part of the anodic biofilm in MFCs.46,55 Anaerolineaceae, also found among the thermophilic anode-attached microbial community, is a family of filamentous bacteria involved in the fermentation of various sugars.56 They are also involved in the syntrophic oxidation of butyrate, and, similarly to Coprothermobacter, grow better in the presence of H2-consuming microorganisms, such as methanogenic archaea.57The lower power production in the thermophilic MFC is likely due to the lack of effective exoelectrogens and to the consequent high activity of non-exoelectrogenic microorganisms, which consumed part of the electrons through pathways competitive to electricity generation. In fact, the methanogenic archaeal family of Methanobacteriaceae, belonging to the order of Methanobacteriales, accounted for 38% of the active anode-attached community in the thermophilic MFC. Methanobacteriales lack cytochromes and methanophenazine, and are thus able to perform hydrogenotrophic, but not acetoclastic, methanogenesis.58 Therefore, Methanobacteriaceae cannot compete for the substrate with exoelectrogenic microorganisms, but their metabolism decreases the availability of electrons for electricity production. Methanobacteriaceae have been previously found in a glucose-fed (1.8 g L−1) MFC operated at room temperature, and indicated as one of the causes for low bioelectricity production, as about 16% of the electrons were directed to methane production.14 Rismani-Yazdi et al.59 reported methane production by Methanobacteriaceae in a mesophilic (39 °C) cellulose-fed MFC only at the beginning of the operation, whereas Hussain et al.60 reported Methanobacteriaceae in a thermophilic (50 °C) syngas-fed MFC. Such microorganisms likely decreased the efficiency of their MFC by performing hydrogenotrophic methanogenesis.
The family of Hydrogenophilaceae, which accounted for 46% of the active planktonic community in the thermophilic MFC, includes the thermophilic Hydrogenophylus sp., which could have consumed a share of electrons by H2 oxidation,61 lowering power production in the thermophilic MFC. Thermodesulfobiaceae, found among the anode-attached families, were also found among the planktonic community (Fig. 5). Coprothermobacter is able to perform extracellular electron transfer,52 but further studies are required to understand its possible involvement in long-range electron transfer to the anode.
In the thermophilic MFC, the family of Comamonadaceae was the most abundant membrane-attached family and, similarly to the mesophilic MFC, it was likely related to aerobic metabolism and thus, oxygen consumption. Armatimonadetes, which accounted for 17% of the active membrane-attached community, is also an order of aerobic microorganisms.62
4.3 Xylose degradation pathways
In the mesophilic MFC, the xylose consumption and metabolite production profiles (Fig. 2) suggest that xylose was firstly converted to volatile fatty acids, which were subsequently oxidized to CO2 and H2O likely mainly by Geobacteraceae, which dominated the anode-attached active community. Interestingly, the power density remained stable for about 30 hours after the depletion of acetate and butyrate. A possible explanation is that acetate and butyrate were accumulated and oxidized intracellularly, thus not detectable in the anolyte and resulting in a flow of electrons directed outside the cell to the anode.4 In fact, after substrate depletion, the soluble COD remained stable (Fig. 2), suggesting that the electron donor was not in the anolyte but likely inside the cells. Also Marshall and May20 observed the same phenomenon and decided to starve a pure culture of Thermincola for two cycles before electrochemical measurements to avoid interferences from the intracellularly accumulated acetate, and its associated storage products.In the thermophilic MFC, xylose was consumed relatively fast, but acetate, the only metabolite found in the anolyte, was not fully consumed even after 144 hours. The power density peak obtained just after the xylose depletion suggests that exoelectrogenic thermophiles were growing on xylose, but the microbial community was lacking effective acetate-utilizing microorganisms. However, it should be noted that the profiles in Fig. 2 were obtained in the feeding cycle “IX”, whereas the samples for microbial community analysis were collected at the end of cycle “XI”. The anodic potential, which increased from cycle IX to cycle XI in the thermophilic MFC (Fig. 1b), suggests a possible shift in the microbial community.
4.4 Performance of the MFCs
In the mesophilic MFC, the shape of the polarization curve (the stable slope in the last part of the curve) suggests low mass transfer limitation, as expected in MFCs using soluble sugars as the substrate. The low CE (12 and 3% for the mesophilic and thermophilic MFC, respectively) was attributed to the MFC design, which was not optimized for power production. The slow rate of oxygen reduction in the cathodic surface and the low proton conduction through the membrane are often the main causes of low power production in air-cathode MFCs.63 In fact, a CE up to 82% was obtained in a xylose-fed, two-chamber MFC (75 mL anodic chamber volume) using 50 mM ferricyanide for the cathodic reaction and a cation exchange membrane.64 Haavisto et al.,28 with a similar inoculum and substrate, obtained an 18% higher CE than the one obtained in this study operating a mesophilic (37 °C) upflow microbial fuel cell in continuous mode using ferricyanide at the cathode. Huang and Logan65 obtained a power production of 13 W m−3 (61% CE) using a xylose-fed air cathode MFC, against the 1.1 W m−3 (12% CE) obtained in this study. However, the anodic chamber of their MFC was equipped with four carbon brushes (6 cm diameter and 7 cm length each), against the single carbon brush (1.5 cm diameter and 5 cm length) used in this study, and their xylose load was three times higher.The structure of the active microbial community in the thermophilic MFC, lacking a known effective exoelectrogen such as Geobacter and including competitors such as methanogenic archaea, was likely the main cause for the lower power produced from the thermophilic MFC in comparison to the mesophilic MFC (Fig. 3a). In fact, the non-exoelectrogenic anode-attached microbial community in the thermophilic MFC likely caused a high internal resistance (560 Ω). Temperature also affects oxygen solubility in water, resulting in a decreased availability of oxygen at high temperature. In fact, the oxygen concentration at the cathode was about 7.0 and 5.6 g L−1 in the mesophilic and thermophilic MFC, respectively. In the thermophilic MFC, the power overshoot curve (Fig. 3b), previously reported in MFCs,66,67 prevented the detection of possible mass transfer limitations. A multiple-cycle method, consisting in running the MFC at a fixed resistance for one entire batch cycle, can be applied to avoid overshoot.68
5. Conclusions
The composition of the anode-attached, planktonic and membrane-attached microbial community, and the active subpopulation, was evaluated in a mesophilic (37 °C) and a thermophilic (55 °C) xylose-fed MFC. This study contributes in understanding of the microbial communities directly and indirectly involved in mesophilic and thermophilic electricity generation. An active microbial community dominated by Geobacteraceae was enriched and shown to sustain power production in mesophilic (37 °C) MFCs, whereas thermophilic (55 °C) power production was hampered by the development of competitors such as hydrogenotrophic methanogens and hydrogen oxidizers. A RNA-based analysis is required to understand the role of the microorganisms in MFCs, as a DNA-based analysis may lead to overestimation or underestimation of the contribution of certain species on power production.A different inoculum source, possibly from thermophilic anaerobic processes, and a different start-up strategy, for example by using a poised anode potential or by suppressing the methanogenic archaea e.g. by addition of bromoethanesulphonic acid (BESA), could be viable alternatives to facilitate the establishment of an efficient thermophilic exoelectrogenic biofilm in future studies. The power production from pure cultures of potentially exoelectrogenic thermophilic microorganisms, for example species of the Thermodesulfobiaceae family detected from the thermophilic anodes in this study, must also be evaluated to confirm their role in electricity production.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Marie Skłodowska-Curie European Joint Doctorate (EJD) in Advanced Biological Waste-To-Energy Technologies (ABWET) funded from Horizon 2020 under grant agreement no. 643071. JH and AML would also like to acknowledge the Academy of Finland (New Indigo ERA-Net Energy 2014; Project no. 283013) for financial support. The authors gratefully thank Mira Sulonen (TUT, Finland) for helping with the performance analysis, Antti Nuottajärvi (TUT, Finland) for helping with the experimental set-up, and the Viinikanlahti municipal wastewater treatment plant (Tampere, Finland) for providing the activated sludge used as inoculum.References
- S. K. Butti, G. Velvizhi, M. L. K. Sulonen, J. M. Haavisto, E. Oguz Koroglu, A. Yusuf Cetinkaya, S. Singh, D. Arya, J. Annie Modestra, K. Vamsi Krishna, A. Verma, B. Ozkaya, A.-M. Lakaniemi, J. A. Puhakka and S. Venkata Mohan, Renewable Sustainable Energy Rev., 2016, 53, 462–476 CrossRef CAS.
- R. Kumar, L. Singh, Z. A. Wahid and M. F. Din, Int. J. Energy Res., 2015, 39, 1048–1067 CrossRef CAS.
- S. Kalathil and D. Pant, RSC Adv., 2016, 6, 30582–30597 RSC.
- R. M. Snider, S. M. Strycharz-glaven, S. D. Tsoi, J. S. Erickson and L. M. Tender, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 15467–15472 CrossRef CAS PubMed.
- R. Kumar, L. Singh and A. W. Zularisam, Renewable Sustainable Energy Rev., 2016, 56, 1322–1336 CrossRef CAS.
- S. P. Bonanni, G. D. Schrott and J. P. Busalmen, Biochem. Soc. Trans., 2012, 40, 1274–1279 CrossRef PubMed.
- K. Rabaey, N. Boon, M. Höfte and W. Verstraete, Environ. Sci. Technol., 2005, 39, 3401–3408 CrossRef CAS PubMed.
- T. H. Pham, N. Boon, P. Aelterman, P. Clauwaert, L. De Schamphelaire, L. Vanhaecke, K. De Mayer, M. Höfte, W. Verstraete and K. Rabaey, Appl. Microbiol. Biotechnol., 2008, 77, 1119–1129 CrossRef CAS PubMed.
- K. P. Nevin, H. Richter, S. F. Covalla, J. P. Johnson, T. L. Woodard, A. L. Orloff, H. Jia, M. Zhang and D. R. Lovley, Environ. Microbiol., 2008, 10, 2505–2514 CrossRef CAS PubMed.
- M.-S. Kim and Y.-J. Lee, Int. J. Hydrogen Energy, 2010, 35, 13028–13034 CrossRef CAS.
- D. Sun, J. Chen, H. Huang, W. Liu, Y. Ye and S. Cheng, Int. J. Hydrogen Energy, 2016, 41, 16523–16528 CrossRef CAS.
- M. A. Rosenbaum, H. Y. Bar, Q. K. Beg, D. Segrè, J. Booth, M. A. Cotta and L. T. Angenent, Bioresour. Technol., 2011, 102, 2623–2628 CrossRef CAS PubMed.
- D. Wu, D. Xing, X. Mei, B. Liu, C. Guo and N. Ren, Int. J. Hydrogen Energy, 2013, 38, 15568–15573 CrossRef CAS.
- K. Chung and S. Okabe, Appl. Microbiol. Biotechnol., 2009, 83, 965–977 CrossRef CAS PubMed.
- T. Catal, K. Li, H. Bermek and H. Liu, J. Power Sources, 2008, 175, 196–200 CrossRef CAS.
- Z. Du, H. Li and T. Gu, Biotechnol. Adv., 2007, 25, 464–482 CrossRef CAS PubMed.
- Q. Fu, N. Fukushima, H. Maeda, K. Sato and H. Kobayashi, Biosci., Biotechnol., Biochem., 2015, 79, 1200–1206 CrossRef CAS PubMed.
- Q. Fu, H. Kobayashi, H. Kawaguchi, J. Vilcaez, T. Wakayama, H. Maeda and K. Sato, J. Biosci. Bioeng., 2013, 115, 268–271 CrossRef CAS PubMed.
- K. C. Wrighton, P. Agbo, F. Warnecke, K. A. Weber, E. L. Brodie, T. Z. DeSantis, P. Hugenholtz, G. L. Andersen and J. D. Coates, ISME J., 2008, 2, 1146–1156 CrossRef CAS PubMed.
- C. W. Marshall and H. D. May, Energy Environ. Sci., 2009, 2, 699–705 CAS.
- B. G. Lusk, Q. F. Khan, P. Parameswaran, A. Hameed, N. Ali, B. E. Rittmann and C. I. Torres, Environ. Sci. Technol., 2015, 49, 14725–14731 CrossRef CAS PubMed.
- Q. Fu, H. Kobayashi, H. Kawaguchi, T. Wakayama, H. Maeda and K. Sato, Environ. Sci. Technol., 2013, 47, 12583–12590 CrossRef CAS PubMed.
- J. De Vrieze, L. Regueiro, R. Props, R. Vilchez-Vargas, R. Jáuregui, D. H. Pieper, J. M. Lema and M. Carballa, Biotechnol. Biofuels, 2016, 9, 244 CrossRef PubMed.
- D. K. Newman and R. Kolter, Nature, 2000, 405, 94–97 CrossRef CAS PubMed.
- M. Miskan, M. Ismail, M. Ghasemi, J. Md Jahim, D. Nordin and M. H. Abu Bakar, Int. J. Hydrogen Energy, 2016, 41, 543–552 CrossRef CAS.
- M. Lu, S. Chen, S. Babanova, S. Phadke, M. Salvacion, A. Mirhosseini, S. Chan, K. Carpenter, R. Cortese and O. Bretschger, J. Power Sources, 2017, 356, 274–287 CrossRef CAS.
- P. Dessì, A.-M. Lakaniemi and P. N. L. Lens, Water Res., 2017, 115, 120–129 CrossRef PubMed.
- J. M. Haavisto, M. E. Kokko, C.-H. Lay and J. A. Puhakka, Int. J. Hydrogen Energy, 2017, 42, 27494–27501 CrossRef CAS.
- A. E. Mäkinen, M. E. Nissilä and J. A. Puhakka, Int. J. Hydrogen Energy, 2012, 37, 12234–12240 CrossRef.
- S.-E. Oh, B. Min and B. E. Logan, Environ. Sci. Technol., 2004, 38, 4900–4904 CrossRef CAS PubMed.
- R. I. Griffiths, A. S. Whiteley, A. G. O'Donnell and M. J. Bailey, Appl. Environ. Microbiol., 2000, 66, 5488–5491 CrossRef CAS PubMed.
- J. G. Caporaso, C. L. Lauber, W. A. Walters, D. Berg-Lyons, C. A. Lozupone, P. J. Turnbaugh, N. Fierer and R. Knight, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4516–4522 CrossRef CAS PubMed.
- P. D. Schloss, S. L. Westcott, T. Ryabin, J. R. Hall, M. Hartmann, E. B. Hollister, R. A. Lesniewski, B. B. Oakley, D. H. Parks, C. J. Robinson, J. W. Sahl, B. Stres, G. G. Thallinger, D. J. Van Horn and C. F. Weber, Appl. Environ. Microbiol., 2009, 75, 7537–7541 CrossRef CAS PubMed.
- J. J. Kozich, S. L. Westcott, N. T. Baxter, S. K. Highlander and P. D. Schloss, Appl. Environ. Microbiol., 2013, 79, 5112–5120 CrossRef CAS PubMed.
- R Development Core Team, R Found. Stat. Comput. Vienna, Austria, ISBN 3-900051-07-0, http//www.R-project.org Search PubMed.
- W. Zhi, Z. Ge, Z. He and H. Zhang, Bioresour. Technol., 2014, 171, 461–468 CrossRef CAS PubMed.
- C. Gao, A. Wang, W.-M. Wu, Y. Yin and Y.-G. Zhao, Bioresour. Technol., 2014, 167, 124–132 CrossRef CAS PubMed.
- K. L. Lesnik and H. Liu, Appl. Microbiol. Biotechnol., 2014, 98, 4187–4196 CrossRef CAS PubMed.
- Q. Jiang, D. Xing, R. Sun, L. Zhang, Y. Feng and N. Ren, RSC Adv., 2016, 6, 85149–85155 RSC.
- X. Mei, C. Guo, B. Liu, Y. Tang and D. Xing, RSC Adv., 2015, 5, 78136–78141 RSC.
- S. Saheb Alam, F. Persson, B. Wilén, M. Hermansson and O. Modin, Sci. Rep., 2015, 5, 18433 CrossRef CAS PubMed.
- A. Kumar, L. H.-H. Hsu, P. Kavanagh, F. Barrière, P. N. L. Lens, L. Lapinsonnière, J. H. Lienhard V, U. Schröder, X. Jiang and D. Leech, Nat. Rev. Chem., 2017, 1, 24 CrossRef CAS.
- T. P. Sciarria, A. Tenca, A. D'Epifanio, B. Mecheri, G. Merlino, M. Barbato, S. Borin, S. Licoccia, V. Garavaglia and F. Adani, Bioresour. Technol., 2013, 147, 246–253 CrossRef CAS PubMed.
- C. S. Kang, N. Eaktasang, D.-Y. Kwon and H. S. Kim, Bioresour. Technol., 2014, 165, 27–30 CrossRef CAS PubMed.
- M. Grattieri, K. Hasan, R. D. Milton, S. Abdellaoui, M. Suvira, B. Alkotaini and S. D. Minteer, Sustainable Energy Fuels, 2017, 1, 1568–1572 CAS.
- G. Zhang, Q. Zhao, Y. Jiao, K. Wang, D.-J. Lee and N. Ren, Water Res., 2012, 46, 43–52 CrossRef CAS PubMed.
- Y. Song, L. Xiao, I. Jayamani, Z. He and A. M. Cupples, J. Microbiol. Methods, 2015, 108, 4–11 CrossRef CAS PubMed.
- T. Shimoyama, A. Yamazawa, Y. Ueno and K. Watanabe, Microbes Environ., 2009, 24, 188–192 CrossRef PubMed.
- R. A. Timmers, M. Rothballer, D. P. B. T. B. Strik, M. Engel, S. Schulz, M. Schloter, A. Hartmann, B. Hamelers and C. Buisman, Appl. Microbiol. Biotechnol., 2012, 94, 537–548 CrossRef CAS PubMed.
- D. Xing, S. Cheng, B. E. Logan and J. M. Regan, Appl. Microbiol. Biotechnol., 2010, 85, 1575–1587 CrossRef CAS PubMed.
- J. R. Kim, S. Cheng, S.-E. Oh and B. E. Logan, Environ. Sci. Technol., 2007, 41, 1004–1009 CrossRef CAS PubMed.
- M. C. Gagliano, C. M. Braguglia, M. Petruccioli and S. Rossetti, FEMS Microbiol. Ecol., 2015, 91, 1–12 CrossRef PubMed.
- K. Sasaki, M. Morita, D. Sasaki, J. Nagaoka, N. Matsumoto, N. Ohmura and H. Shinozaki, J. Biosci. Bioeng., 2011, 112, 469–472 CrossRef CAS PubMed.
- B. C. Jong, B. H. Kim, I. S. Chang, P. W. Y. Liew, Y. F. Choo and G. S. Kang, Environ. Sci. Technol., 2006, 40, 6449–6454 CrossRef CAS PubMed.
- M. Wei, F. Harnisch, C. Vogt, J. Ahlheim, T. R. Neu and H. H. Richnow, RSC Adv., 2015, 5, 5321–5330 RSC.
- T. Yamada, T. Yamauchi, K. Shiraishi, P. Hugenholtz, A. Ohashi, H. Harada, Y. Kamagata, K. Nakamura and Y. Sekiguchi, ISME J., 2007, 1, 246–255 CrossRef CAS PubMed.
- P. Liu, Q. Qiu and Y. Lu, Appl. Environ. Microbiol., 2011, 77, 3884–3887 CrossRef CAS PubMed.
- R. K. Thauer, A.-K. Kaster, H. Seedorf, W. Buckel and R. Hedderich, Nat. Rev. Microbiol., 2008, 6, 579–591 CrossRef CAS PubMed.
- H. Rismani-Yazdi, S. M. Carver, A. D. Christy, Z. Yu, K. Bibby, J. Peccia and O. H. Tuovinen, Bioresour. Technol., 2013, 129, 281–288 CrossRef CAS PubMed.
- A. Hussain, P. Mehta, V. Raghavan, H. Wang, S. R. Guiot and B. Tartakovsky, Enzyme Microb. Technol., 2012, 51, 163–170 CrossRef CAS PubMed.
- N. R. Hayashi, T. Ishida, A. Yokota, T. Kodama and Y. Igarashi, Int. J. Syst. Bacteriol., 1999, 49, 783–786 CrossRef PubMed.
- K. C. Y. Lee, P. F. Dunfield and M. B. Stott, in The Prokaryotes: Other Major Lineages of Bacteria and The Archaea, ed. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt and F. Thompson, Springer Berlin Heidelberg, Berlin, Heidelberg, 2014, pp. 447–458 Search PubMed.
- H. Rismani-Yazdi, S. M. Carver, A. D. Christy and O. H. Tuovinen, J. Power Sources, 2008, 180, 683–694 CrossRef CAS.
- A. E. Mäkinen, C. H. Lay, M. E. Nissilä and J. a. Puhakka, Int. J. Hydrogen Energy, 2013, 38, 15606–15612 CrossRef.
- L. Huang and B. E. Logan, Appl. Microbiol. Biotechnol., 2008, 80, 655–664 CrossRef CAS PubMed.
- S. M. Carver, P. Vuoriranta and O. H. Tuovinen, J. Power Sources, 2011, 196, 3757–3760 CrossRef CAS.
- P.-C. Nien, C.-Y. Lee, K.-C. Ho, S. S. Adav, L. Liu, A. Wang, N. Ren and D.-J. Lee, Bioresour. Technol., 2011, 102, 4742–4746 CrossRef CAS PubMed.
- V. J. Watson and B. E. Logan, Electrochem. Commun., 2011, 13, 54–56 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra12316g |
| This journal is © The Royal Society of Chemistry 2018 |