Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ugi multicomponent reaction to prepare peptide–peptoid hybrid structures with diverse chemical functionalities†
Manuel
Hartweg
a,
Charlotte J. C.
Edwards-Gayle
bc,
Elham
Radvar
a,
Dominic
Collis
a,
Mehedi
Reza
d,
Michael
Kaupp
efg,
Jan
Steinkoenig
 efg,
Janne
Ruokolainen
d,
Robert
Rambo
c,
Christopher
Barner-Kowollik
efg,
Janne
Ruokolainen
d,
Robert
Rambo
c,
Christopher
Barner-Kowollik
 efg,
Ian W.
Hamley
efg,
Ian W.
Hamley
 b,
Helena S.
Azevedo
b,
Helena S.
Azevedo
 a and
C. Remzi
Becer
a and
C. Remzi
Becer
 *a
*a
aSchool of Engineering and Materials Science, Queen Mary University, London, E1 4NS, UK. E-mail: r.becer@qmul.ac.uk
bDepartment of Chemistry, University of Reading, Whiteknights, Reading, RG6 6AD, UK
cDiamond Light Soruce Ltd., Harwell Science and Innovation Campus, Didcot, OX11 0DE, UK
dDepartment of Applied Physics, Aalto University, Finland
eMacromolecular Architectures, Institut für Technische Chemie und Polymerchemie, Karlsruhe Institute of Technology (KIT), Engesserstraße 18, 76128 Karlsruhe, Germany
fInstitute of Biological Interfaces, Karlsruhe Institute of Technology (KIT), Hermann-von-Helmholtz-Platz 1, 76344 Eggenstein-Leopoldshaven, Germany
gSchool for Chemistry, Physics and Mechanical Engineering, Queensland University of Technology (QUT), 2 George Street, QLD 4000, Brisbane, Australia
First published on 21st December 2017
Abstract
Monodisperse sequenced peptides and peptoids present unique nano-structures based on their self-assembled secondary and tertiary structures. However, the generation of peptide and peptoid hybrid oligomers in a sequence-defined manner via Ugi multicomponent reaction has not yet been studied. Herein, we report a synthetic strategy that enables both the modification of peptides as well as the generation of sequence-defined peptide–peptoid hybrid structures. Our synthetic methodology rests on the fusion of solid phase peptide synthesis with Ugi multicomponent reactions. We evidence that a diversity of chemical functionalities can be inserted into peptides or used in the design of peptide–peptoid hybrids exploiting a wide functional array including amines, carboxylic acids, hydrocarbons, carbohydrates as well as polymers, introducing a sequence-defined synthetic platform technology for precision peptoid hybrids.
Introduction
Nature has created sequence defined peptides to carry out specific functions, such as molecular recognition and biocatalysis, in the course of evolution. Today, even though synthetic macromolecules are widely used for a large number of applications, achieving an absolute control over their primary structure still remains as a key challenge for chemists.1–3 Therefore, our fundamental understanding of the role of monomer sequence and its influence on the secondary and tertiary structures of peptides and proteins has to be enhanced dramatically.Amongst a range of methods for the synthesis of such sequence-defined macromolecules, Merrifield solid phase peptide synthesis (SPPS) method has been the most successful.4–6,7–17 Solid-phase protocols typically use excess of reagents and require two steps, which are the coupling and deprotection steps, for incorporation of each repeat unit. Besides, extremely efficient reactions are required to ensure monodisperse formation of peptide chains in high yields.18,19
Complementary to the synthesis of peptides, Zuckermann et al. reported the synthesis of peptoids via solid-phase submonomer synthesis. Polypeptoids are accepted as peptidomimetic polymers20–22 with a substituent on the amide nitrogen, which significantly influences the formation of higher order structures due to suppression of a hydrogen bonding.23,24 Moreover, the N-substitution eliminates the chirality of the more flexible peptoid backbone,25 which transforms peptoids to a generally side-chain dominated system, dissimilar to peptides in which conformation and packing are dictated primarily by the chemical structure of the backbone. However, compared to peptides, engineered peptoids display a range of interesting properties, such as protein-mimetic secondary and tertiary structures.26–29 They have been demonstrated to form stable nanosheets in vivo,30–32 as well as to improve tissue accumulation for reduced excretion rates.33,34 Further successful examples for the peptoid synthesis can be listed as solid-phase synthesis via Fmoc-strategy,35 bromoacetyl-bromide method in solution-phase,36 N-heterocyclic carbene mediated ring-opening polymerization of N-substituted N–carboxy-anhydrides,37,38 and Ugi 4-component reaction (Ugi-4CR).39 However, up to date, only very little research was carried out on peptide–peptoid hybrids. For example, a few studies have reported the synthesis of sequence defined hybrids with various biological applications.40–44 Also the synthesis of peptide–peptoid block conjugates,42,45,46 using native chemical ligation (NCL),10 fragment condensation,47 and copper-catalyzed azide–alkyne [3 + 2] cycloaddition48 have been reported. In the present study, Ugi–4CR was utilized in order to generate highly functional and sterically demanding peptoid units. Recently, the Ugi-4CR was already applied for the sequence defined polymers, the functionalization of peptide arrays49 synthesis of polypeptoids from γ, δ, ε-amino acids,50 and alternating peptide–peptoid copolymers from51 Ugi-4CR chemistry generally displays the for solid-phase synthesis crucial high efficiency,52–55 compatibility with amino acid chemistry,56 while allowing incorporation of a range of chemical diversity into peptoid side chains.57 The method benefits from its simplicity and remarkably high diversity for modification of peptides or synthesis of novel peptoid and hybrid structures.
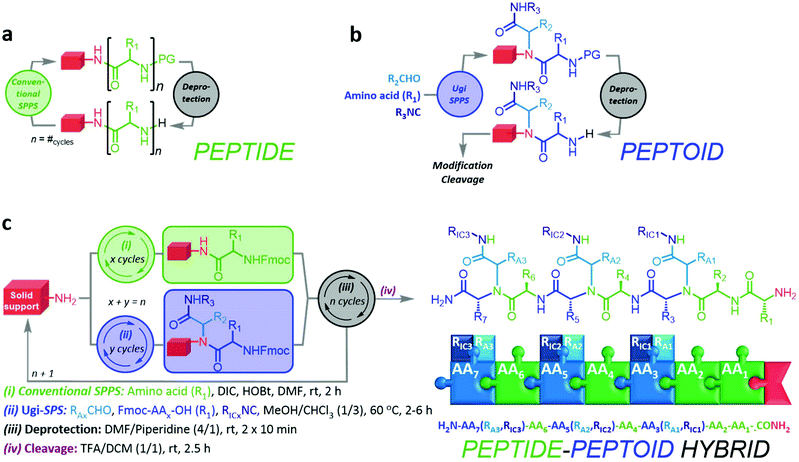
Herein, we report an iterative synthesis method towards highly functional, sequence-defined, and monodisperse peptides, peptoids, and peptide–peptoid hybrids (Fig. 1). In an extended solid-phase protocol, the methodology fuses SPPS and Ugi-4CR chemistry for the incorporation of peptide and peptoid units, respectively. Furthermore, we investigate the self-assembly behavior and defragmentation pattern of selected compounds to demonstrate the diversity of these hybrid structures. Moreover, we propose a nomenclature for petpoid structures generated by the means of Ugi-4CR, as well as for peptide Ugi-peptoid hybrids.
Results and discussion
The preliminary considerations indicated that the distinct reaction protocols of conventional SPPS and Ugi-SPPS are suitable for iterative, random, or programmed sequence-defined solid phase synthesis. Therefore, a nomenclature was suggested (ESI 1†), indicating the whether a unit is of peptide or peptoid nature, and which R-group the peptide (RAA) or two independent R-groups the Ugi-peptoids (RA and RIC) are carrying. However, for the Ugi-SPS process, the combinations of several amino acids (i.e. glycine, phenylalanine and lysine), aldehydes (i.e. butyraldehyde, undecanal, undecenal, glucose aldehyde, and PEG-aldehyde), and isocyanides (cyclohexyl, and 2-morpholinoethyl isocyanide) were investigated. An Fmoc-Rink-amide coupling strategy was employed throughout the process for both coupling methodologies (Fig. 1). The process typically breaks down into six steps: (i) automated deprotection of the resin, (ii) automated Fmoc-solid phase peptide synthesis (Fmoc SPPS) of the anchors, (iii) manual Fmoc-SPPS (iv) manual Ugi-Fmoc-solid phase peptoid synthesis (Ugi-Fmoc-SPPS), (v) cleavage and (vi) purification. In order to validate the successful combination of SPPS and Ugi-SPPS, compounds 1–6 (Fig. 2) were prepared. Thus, peptide anchors (H2N–FGF-CONH2 in 1, 2, 7) and (H2N-KKKK-CONH2 in 3–6, 8–12) were prepared via automated SPPS on Rink-amide resin. Anchors were then deprotected to generate the free amines for incorporation of subsequent peptide or peptoid units. Afterwards, in contrast to conventional SPPS, where the amine moieties are reacted with coupling reagents and an amino acid, amines were reacted with an excess of aldehyde (4.00 eq.) in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 MeOH/CHCl3. It is important to note that the use of both MeOH and CHCl3 was crucial to achieve full and selective conversion of the terminal amine into a peptoid sequence. CHCl3 is essential to ensure sufficient swelling of the resin, whereas MeOH is required to promote Ugi reaction. After complete imine formation, the respective Fmoc-protected amino acid (5.00 eq.) and isocyanide (6.00 eq.) were added to react for a further 2–6 hours. Terminal free amine has been formed as a result of the subsequent deprotection step in 1
3 MeOH/CHCl3. It is important to note that the use of both MeOH and CHCl3 was crucial to achieve full and selective conversion of the terminal amine into a peptoid sequence. CHCl3 is essential to ensure sufficient swelling of the resin, whereas MeOH is required to promote Ugi reaction. After complete imine formation, the respective Fmoc-protected amino acid (5.00 eq.) and isocyanide (6.00 eq.) were added to react for a further 2–6 hours. Terminal free amine has been formed as a result of the subsequent deprotection step in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 pyridine/DMF. Successive coupling steps were performed accordingly to obtain the desired sequence of the final peptoid or peptide/peptoid hybrids. Following the last coupling reaction or the final deprotection step, oligomers were cleaved off the solid support with 1
4 pyridine/DMF. Successive coupling steps were performed accordingly to obtain the desired sequence of the final peptoid or peptide/peptoid hybrids. Following the last coupling reaction or the final deprotection step, oligomers were cleaved off the solid support with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
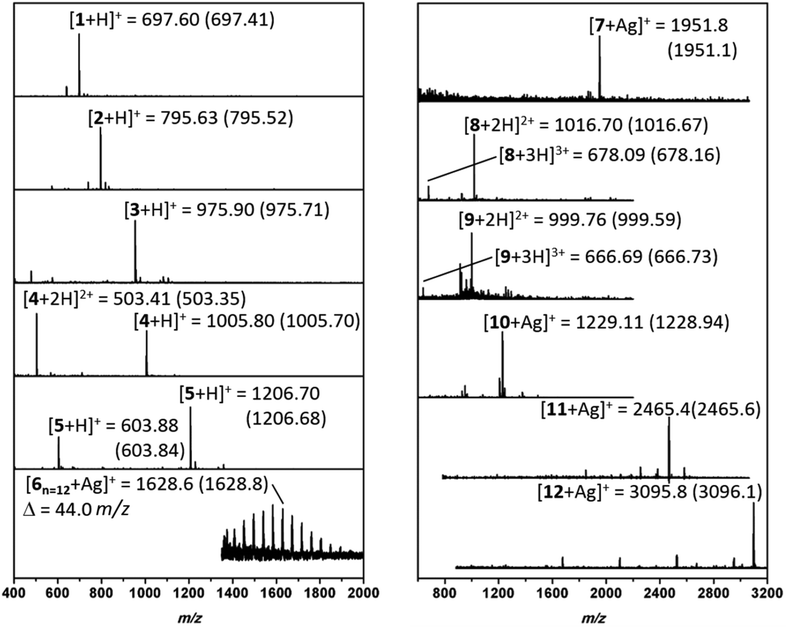
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 TFA/DCM mixture and crude mixtures were analyzed. Electrospray ionization mass spectrometer (ESI-MS) or matrix assisted laser desorption ionization time of flight mass spectrometer (MALDI-ToF MS) were used to investigate the conversion and chemoselectivity of oligomers (Fig. 3). Furthermore, reverse phase high performance liquid chromatography (RP-HPLC) or size exclusion chromatography (SEC) was conducted to confirm the purity of monodisperse oligomers. SEC analysis of 1 and 2 have displayed narrow peaks and indicate the formation of the desired sequences (Fig. S6 and S7†).
1 TFA/DCM mixture and crude mixtures were analyzed. Electrospray ionization mass spectrometer (ESI-MS) or matrix assisted laser desorption ionization time of flight mass spectrometer (MALDI-ToF MS) were used to investigate the conversion and chemoselectivity of oligomers (Fig. 3). Furthermore, reverse phase high performance liquid chromatography (RP-HPLC) or size exclusion chromatography (SEC) was conducted to confirm the purity of monodisperse oligomers. SEC analysis of 1 and 2 have displayed narrow peaks and indicate the formation of the desired sequences (Fig. S6 and S7†).
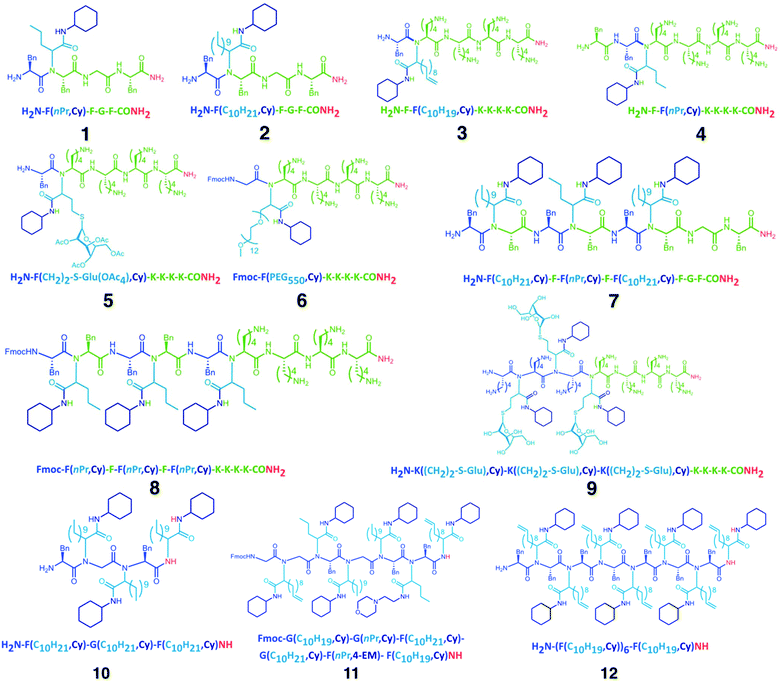 | ||
| Fig. 2 Chemical structures (1–12) of the peptide/peptoid hybrid structures synthesized in this study. Green and blue units represent peptide and peptoid units, respectively. The letter codes for the structures are displayed according to ESI 1.† | ||
Subsequently, to obtain more polar hybrids, H2N-KKKK-CONH2 was used as an anchor. The reactions with undecenal or butyraldehyde, Fmoc-Phe-OH, and cyclohexyl isocyanide, as well as successive SPPS with Fmoc-Phe-OH, yielded oligomeric 3 and 4 at high purity based on RP-HPLC traces (Fig. 3). Compound 3 has two main isomers, one from cis and one from the trans, of the bis-amide formed as a result of Ugi-4CR. Furthermore, ESI-MS analysis was carried out for 3 and 4, where mainly singly and doubly charged cations were observed (Fig. 3). Subsequently, the addition of a glucose aldehyde resulted in a chemoselective formation of glycopeptoid–peptide hybrid 5 that was confirmed by RP-HPLC and ESI-MS (Fig. S14† and Fig. 3). An acetyl-protected sugar-derived aldehyde was synthesized via base catalyzed thiol-Michael addition using tetra-acetylated thioglucose (ESI†). Additionally, Ugi-4CR was used as a ligation tool in the synthesis of the hybrid 6. Therefore, PEG-aldehyde was synthesized by Swern oxidation reaction (ESI†).58 Thus, PEG500 was used as a starting material and ligated to the anchor compound 6. The success of the reaction was determined by MALDI-ToF, whereby the PEG signals shift by Δm/z 917.2 (Fig. S15†), which corresponds exactly to the mass of peptoid/aldehyde.
To further expand the versatility of this synthetic process, several hybrids with different backbone architectures, such as alternating 7, and 8 and block 9 co-oligomers, were prepared. Starting from the same H2N-FGF-CONH2 peptide anchor, three peptoid sequences were attached with a phenylalanine peptide sequence in between the peptoids. The MS analysis of compound 7 (Fig. S17†) demonstrated high purity even in the absence of any purification steps. Also, H2N–KKKK–CONH2 was designed as a more polar anchor and used in the synthesis of 8 and 9. ESI-MS (Fig. S19†) shows the formation of the 9-mer displaying the singly, doubly and triply charged cations. Compound 9 is designed as a glycopeptoid containing three glycan units 9. Acetyl-protected glucose aldehyde was used in the synthesis and following to the deacetylation of glucose units, ESI-MS revealed signals for the doubly and triply charged cations (Fig. 3). RP-HPLC analysis of compound 9 also indicated the formation of an oligomer in high purity.
In the final set of hybrid structures, homopeptoids were studied to obtain highly functional oligopeptoids. Initially, 10 was prepared and analyzed as a trimer. MS analysis (Fig. 3) indicated a small amount of a side product, which was assigned as the trimer incorporating only two peptoid and one peptide sequence instead of the third peptoid unit. The main reason for this effect is ascribed as the steric hindrance as the reaction rate significantly suffers when bulky reagents are used in the Ugi reaction. The isotopic pattern of the main peak (Fig. S24†) is in good agreement with the theoretical.
Moreover, hexamer compound 11 using different starting compounds and heptamer compound 12 with identical units and molecular weights of approximately 3 kDa were synthesized. In the case of 11, the MALDI-ToF MS analysis indicated that when 2-morpholinoethyl isocyanide was used alternatively, the reaction was not chemoselective and thus only one single peptide sequence was incorporated in the backbone (Fig. 3). MALDI-ToF MS spectrum of 12 shows a series of peaks with a low intensity and Δm/z of one single Ugi-repetition unit was detected (Fig. 3). Nevertheless, the major peak was assigned to be the desired compound 12 and the isotopic pattern of 8 and 9 are found to be in good agreement.
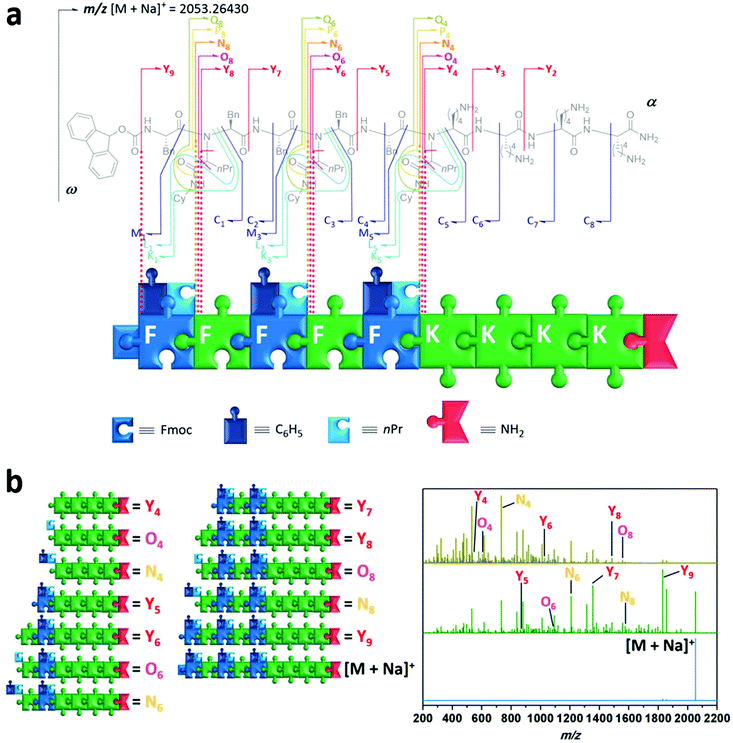
Having demonstrated the versatility of the synthetic process to prepare sequence-defined peptide–peptoid hybrid structures, we then investigated their properties, namely their ability for information storage and self-assembly into nanostructures. Sequential deciphering of hybrid oligomers can be complicated. Due to the use of a multicomponent reaction, both peptide as well as peptoid units may be present within the hybrid. The selected analysis technique must be able to determine whether a sequence has a peptidic or peptoidic nature, and in case of a peptoid unit it should provide information regarding the composition of two incorporated R-groups derived from the Ugi-4CR. Although various analytical tools for the characterisation and sequencing of macromolecules have been developed in the past decades, biopolymers are usually decoded fast and reliably via tandem mass spectroscopy (MS/MS).59 However, no universal rules apply for the MS/MS dissociation patterns of non-natural polymers. For the specific fragmentation pathways, to decipher structural information of the peptide/peptoid hybrid macromolecules one has to refer to established polymer and peptide fragmentation assignments.60 As illustrated (Fig. 4a), the composition of amino acids of peptide units can be assigned using their one letter code (i.e. K = lysine, F = phenylalanine). The nature of Ugi-peptoid units and their R–groups derived from the aldehyde and isocyanide can be assigned by the one letter code for the amino acid, as well as the abbreviations for the respective side chain units (i.e. nPr, Cy). In order to demonstrate the power of data storage on designed peptide–peptoid hybrid structures, compound 8 was analysed in detail via ESI-MS/MS. The precursor ion ([M + Na]+ (m/z 2053.26)) was isolated and activated with increasing collisional energy (Fig. 4b blue trace, ESI section 4†). As expected, even at lower energies, a fragment (Y9) was detected initially, which displays the cleavage of the urethane bond of the Fmoc-group (Δm/z 222). When the collisional energy was increased (Fig. 4b green and gold spectrum), further fragmentation patterns were observed. To our delight, we have not only observed fragmentation patterns from the main chain (C, M, N and Z), but also the specific side chain patterns could be identified. These outcomes provided structural information about both R–groups of the N–substitution of the peptoid units and thus revealing the location of every starting compound used in the respective Ugi-reaction. This was enabled by the bis-amide nature of the formed Ugi-scaffold, whereby the side chain amide moiety was fragmented on both sides of the amide bond (P, Q, O, as well as K and L). For instance, the fragmentation of the peptoid sequence situated at the eight sequence can be envisioned (N8vs. O8vs. Y8). The fragment of the oligomer including sequence eight, N8 (m/z 1685.09) displays Δm/z 127.12 compared to O8 (m/z 1557.97), which corresponds to CyNHC(H)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. Additionally, Y8 (m/z 1500.95) provides information about the aldehyde derived R-group. A Δm/z 57.02 value compared to Y8 belongs to CH3(CH2)2CH2 and displays the use of butyraldehyde (resulting in nPr as R-group) as a monomer in the Ugi-reaction. Similar observations were made for the sequence at location six and four. These observations were evidenced from α → ω (N, O, P, Q and Y patterns) and ω → α (C, K, L, M and C patterns) for each peptoid sequence, whereas the peptide unit based fragments only revealed conventional peptide signals (C and Y patterns). For peptide and peptoid sequences, also conventional X and Z as well as A and B patterns were observed, but neglected for clarity.
O. Additionally, Y8 (m/z 1500.95) provides information about the aldehyde derived R-group. A Δm/z 57.02 value compared to Y8 belongs to CH3(CH2)2CH2 and displays the use of butyraldehyde (resulting in nPr as R-group) as a monomer in the Ugi-reaction. Similar observations were made for the sequence at location six and four. These observations were evidenced from α → ω (N, O, P, Q and Y patterns) and ω → α (C, K, L, M and C patterns) for each peptoid sequence, whereas the peptide unit based fragments only revealed conventional peptide signals (C and Y patterns). For peptide and peptoid sequences, also conventional X and Z as well as A and B patterns were observed, but neglected for clarity.
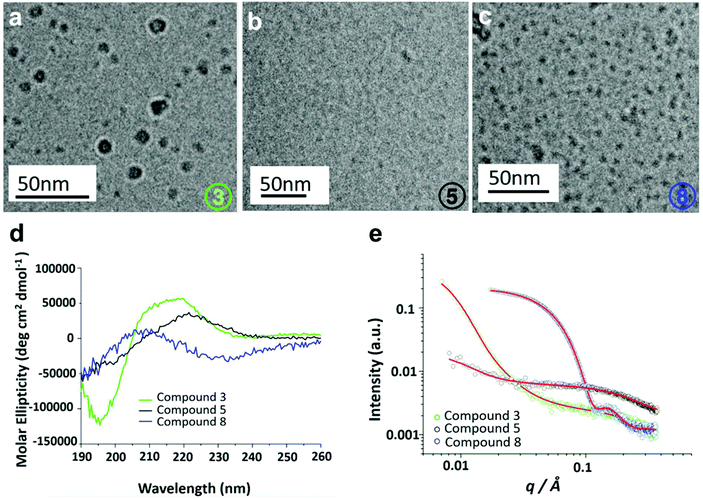
Secondary structures of 3, 5 and 8 were determined using circular dichroism (CD) spectroscopy. It was found that the structures were intrinsically disordered, identified through a negative peak around 200 nm (Fig. S29†). The self-assembly properties of these three compounds were then investigated using cryogenic transmission electron microscopy (Cryo-TEM) and small-angle X-ray scattering (SAXS). Cryo-TEM images of 3 (Fig. 5a) revealed the presence of slightly larger globular structures with an average diameter of 14.95 nm. Compound 5 showed a small population of irregular sized aggregates or clusters (Fig. 5b). Finally, for compound 8 mixture of larger globular like structures and smaller micelle like assemblies with an average diameter of 3.125 nm are observed (Fig. 5c). SAXS was used to further probe the self-assembled structures of the hybrid oligomers, providing information on the structure and dimensions of the assemblies. Scattering intensities and form factor models are shown (Fig. S30†). Scattering intensities of 3 and 5 were fitted based on a coexistence of clusters (described by a mass fractal form factor) and monomers (described by generalized Gaussian coil form factor). The models are in good agreement with cryo-TEM results, which showed a population of irregular aggregated structures. SAXS data for compound 8 fitted to a ‘spherical shell’ form factor model, corresponding to a core–shell micelle structure, also consistent with Cryo-TEM results. Parameters of fits are listed in Tables S2 and S3.†
Conclusions
In summary, two chemical synthesis techniques, i.e. solid-phase peptide synthesis (SPPS) and Ugi multicomponent reaction were combined to obtain peptide/peptoid hybrid structures with defined sequences and functionalities. Thus, a series of model peptide/peptoid hybrids were prepared. As evident from the various analysis techniques applied on the hybrids, both reactions proceeded at very high conversions and no further purification was required, demonstrating the power of the combined iterative approach and the diversity of the chemical groups that can be inserted in peptide sequences via Ugi reactions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was partially supported by the European Commission Horizon2020 programme (EU-ITN EuroSequences Proposal No: 642083). C. R. B. also acknowledges EPSRC (EP/P009018/1) for the financial support. D. C. and H. S. A. thank the support from the EU-funded project “SuprHApolymers” (PCIG14-GA-2013-631871). I. W. H. thanks EPSRC (EP/L020599/1) for support and Diamond Light Source and the University of Reading for co-funding the PhD studentship of C. E. G. Diamond Light Source is thanked for the award of beamtime on beamline B21. C. B.-K. acknowledges the Sonderforschungsbereich 1176 (project A3) funded by the German Research Council for continued support. J. S.'s project is funded by a Landesgraduierten Scholarship of the State of Baden-Wuerttemberg. D. W. P. C. and H. S. A. thank the support from the EU-funded project “SuprHApolymers” (PCIG14-GA-2013-631871).Notes and references
- Y. Brudno and D. R. Liu, Chem. Biol., 2009, 16, 265 CrossRef CAS PubMed.
- G. Gody, T. Maschmeyer, P. B. Zetterlund and S. Perrier, Nat. Commun., 2013, 4, 2505 Search PubMed.
- N. G. Engelis, A. Anastasaki, G. Nurumbetov, N. P. Truong, V. Nikolaou, A. Shegiwal, M. R. Whittaker, T. P. Davis and D. M. Haddleton, Nat. Chem., 2017, 9, 171 CrossRef CAS PubMed.
- R. B. Merrifield, J. Am. Chem. Soc., 1963, 85, 2149 CrossRef CAS.
- R. B. Merrifield, Angew. Chem., 1985, 97, 801 CrossRef CAS.
- R. B. Merrifield, Angew. Chem., Int. Ed. Engl., 1985, 24, 799 CrossRef.
- R. N. Zuckermann, J. M. Kerr, S. B. H. Kent and W. H. Moos, J. Am. Chem. Soc., 1992, 114, 10646 CrossRef CAS.
- S. J. Danishefsky, K. F. McClure, J. T. Randolph and R. B. Ruggeri, Science, 1993, 260, 1307 CAS.
- M. Schuster, P. Wang, J. C. Paulson and C.-H. Wong, J. Am. Chem. Soc., 1994, 116, 1135 CrossRef CAS.
- P. E. Dawson, T. W. Muir, I. Clark-Lewis and S. B. Kent, Science, 1994, 266, 776 CAS.
- O. J. Plante, E. R. Palmacci and P. H. Seeberger, Science, 2001, 291, 1523 CrossRef CAS PubMed.
- P. Espeel, L. L. G. Carrette, K. Bury, S. Capenberghs, J. C. Martins, F. E. Du Prez and A. Madder, Angew. Chem., Int. Ed., 2013, 52, 13261 CrossRef CAS PubMed.
- T. G. W. Edwardson, K. M. M. Carneiro, C. J. Serpell and H. F. Sleiman, Angew. Chem., Int. Ed., 2014, 53, 4567 CrossRef CAS PubMed.
- D. Ponader, S. Igde, M. Wehle, K. Märker, M. Santer, D. Bléger and L. Hartmann, Beilstein J. Org. Chem., 2014, 10, 1603 CrossRef PubMed.
- R. K. Roy, A. Meszynska, C. Laure, L. Charles, C. Verchin and J.-F. Lutz, Nat. Commun., 2015, 6, 7237 CrossRef PubMed.
- A. A. Ouahabi, M. Kotera, L. Charles and J.-F. Lutz, ACS Macro Lett., 2015, 4, 1077 CrossRef.
- M. R. Silk, J. Newman, J. C. Ratcliffe, J. F. White, T. Caradoc-Davies, J. R. Price, S. Perrier, P. E. Thompson and D. K. Chalmers, Chem. Commun., 2017, 53, 6613 RSC.
- T. Freichel, S. Eierhoff, N. L. Snyder and L. Hartmann, J. Org. Chem., 2017, 82, 9400 CrossRef CAS PubMed.
- S. Billiet, K. De Bruycker, F. Driessen, H. Goossens, V. Van Speybroeck, J. M. Winne and F. E. Du Prez, Nat. Chem., 2014, 6, 815 CrossRef CAS PubMed.
- J. Sun and R. N. Zuckermann, ACS Nano, 2013, 7, 4715 CrossRef CAS PubMed.
- C. Lavilla, G. Yilmaz, V. Uzunova, R. Napier, C. R. Becer and A. Heise, Biomacromolecules, 2017, 18, 1928 CrossRef CAS PubMed.
- C. Bonduelle, H. Oliveira, C. Gauche, J. Huang, A. Heise and S. Lecommandoux, Chem. Commun., 2016, 52, 11251 RSC.
- B. Laufer, J. Chatterjee, A. O. Frank and H. Kessler, J. Pept. Sci., 2009, 15, 141 CrossRef CAS PubMed.
- N. Gangloff, J. Ulbricht, T. Lorson, H. Schlaad and R. Luxenhofer, Chem. Rev., 2016, 116, 1753 CrossRef CAS PubMed.
- A. M. Rosales, H. K. Murnen, S. R. Kline, R. N. Zuckermann and R. A. Segalman, Soft Matter, 2012, 8, 3673 RSC.
- B.-C. Lee, R. N. Zuckermann and K. A. Dill, J. Am. Chem. Soc., 2005, 127, 10999 CrossRef CAS PubMed.
- B.-C. Lee, T. K. Chu, K. A. Dill and R. N. Zuckermann, J. Am. Chem. Soc., 2008, 130, 8847 CrossRef CAS PubMed.
- C. Bonduelle, J. Huang, E. Ibarboure, A. Heise and S. Lecommandoux, Chem. Commun., 2012, 48, 8353 RSC.
- U. I. M. Gerling-Driessen, N. Mujkic-Ninnemann, D. Ponader, D. Schöne, L. Hartmann, B. Koksch, U. I. M. Gerling-Driessen, D. Schöne, B. Koksch, D. Ponader, N. Mujkic-Ninnemann and L. Hartmann, Biomacromolecules, 2015, 16, 2394 CrossRef CAS PubMed.
- B. Sanii, R. Kudirka, A. Cho, N. Venkateswaran, G. K. Olivier, A. M. Olson, H. Tran, R. M. Harada, L. Tan and R. N. Zuckermann, J. Am. Chem. Soc., 2011, 133, 20808 CrossRef CAS PubMed.
- K. T. Nam, S. A. Shelby, P. H. Choi, A. B. Marciel, R. Chen, L. Tan, T. K. Chu, R. A. Mesch, B.-C. Lee, M. D. Connolly, C. Kisielowski and R. N. Zuckermann, Nat. Mater., 2010, 9, 454 CrossRef CAS PubMed.
- R. V. Mannige, T. K. Haxton, C. Proulx, E. J. Robertson, A. Battigelli, G. L. Butterfoss, R. N. Zuckermann and S. Whitelam, Nature, 2015, 526, 415 CrossRef CAS PubMed.
- J. Seo, G. Ren, H. Liu, Z. Miao, M. Park, Y. Wang, T. M. Miller, A. E. Barron and Z. Cheng, Bioconjugate Chem., 2012, 23, 1069 CrossRef CAS PubMed.
- J. Seo, B. C. Lee and R. N. Zuckermann, in Comprehensive Biomaterials, Elsevier, 2011, vol. 2, p. 53 Search PubMed.
- R. J. Simon, R. S. Kania, R. N. Zuckermann, V. D. Huebner, D. A. Jewell, S. Banville, S. Ng, L. Wang, S. Rosenberg and C. K. Marlowe, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 9367 CrossRef CAS.
- T. Hjelmgaard, S. Faure, C. Caumes, E. De Santis, A. A. Edwards and C. Taillefumier, Org. Lett., 2009, 11, 4100 CrossRef CAS PubMed.
- L. Guo and D. Zhang, J. Am. Chem. Soc., 2009, 131, 18072 CrossRef CAS PubMed.
- J. Huang and A. Heise, Chem. Soc. Rev., 2013, 42, 7373 RSC.
- L. A. Wessjohann, D. G. Rivera and O. E. Vercillo, Chem. Rev., 2009, 109, 796 CrossRef CAS PubMed.
- S. P. Shukla, J. C. Manarang and D. G. Udugamasooriya, Eur. J. Med. Chem., 2017, 137, 1 CrossRef CAS PubMed.
- T. J. Desai and D. G. Udugamasooriya, Biochem. Biophys. Res. Commun., 2017, 486, 545 CrossRef CAS PubMed.
- J. Singh, S. P. Shukla, T. J. Desai and D. G. Udugamasooriya, Bioorg. Med. Chem., 2016, 24, 4470 CrossRef CAS PubMed.
- T. J. Desai, J. E. Toombs, J. D. Minna, R. A. Brekken and D. G. Udugamasooriya, Oncotarget., 2016, 7, 30678 Search PubMed.
- J. M. Matharage, J. D. Minna, R. A. Brekken and D. G. Udugamasooriya, ACS Chem. Biol., 2015, 10, 2891 CrossRef CAS PubMed.
- K. Pels and T. Kodadek, ACS Comb. Sci., 2015, 17, 152 CrossRef CAS PubMed.
- Y. Gao and T. Kodadek, Chem. Biol., 2013, 20, 360 CrossRef CAS PubMed.
- P. M. Levine, T. W. Craven, R. Bonneau and K. Kirshenbaum, Org. Biomol. Chem., 2013, 11, 4142 CAS.
- D. Pasini, Molecules, 2013, 18, 9512 CrossRef CAS PubMed.
- B. Ridder, D. S. Mattes, A. Nesterov-Mueller, F. Breitling and M. A. R. Meier, Chem. Commun., 2017, 53, 5553 RSC.
- X. Zhang, S. Wang, J. Liu, Z. Xie, S. Luan, C. Xiao, Y. Tao and X. Wang, ACS Macro Lett., 2016, 5, 1049 CrossRef CAS.
- A. Al Samad, J. De Winter, P. Gerbaux, C. Jerome and A. Debuigne, Chem. Commun., 2017, 53, 12240 RSC.
- I. Ugi, A. Dömling and W. Hörl, Endeavour, 1994, 18, 115 CrossRef CAS.
- A. Dömling, W. Wang and K. Wang, Chem. Rev., 2012, 112, 3083 CrossRef PubMed.
- E. Ruijter, R. Scheffelaar and R. V. A. Orru, Angew. Chem., Int. Ed., 2011, 50, 6234 CrossRef CAS PubMed.
- M. Hartweg and C. R. Becer, Green Chem., 2016, 18, 3272 RSC.
- A. F. S. Barreto, O. E. Vercillo, M. A. Birkett, J. C. Caulfield, L. A. Wessjohann and C. K. Z. Andrade, Org. Biomol. Chem., 2011, 9, 5024 CAS.
- S. C. Solleder, K. S. Wetzel and M. A. R. Meier, Polym. Chem., 2015, 6, 3201 RSC.
- K. Omura and D. Swern, Tetrahedron, 1978, 34, 1651 CrossRef CAS.
- K. Biemann, Int. J. Mass Spectrom., 2007, 259, 1 CrossRef CAS.
- C. Wesdemiotis, N. Solak, M. J. Polce, D. E. Dabney, K. Chaicharoen and B. C. Katzenmeyer, Mass Spectrom. Rev., 2011, 30, 523 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Polymerization kinetics and analysis, competition binding assays. See DOI: 10.1039/c7py01953j |
| This journal is © The Royal Society of Chemistry 2018 |