A simple, modular synthesis of bifunctional peptide-polynorbornenes for apoptosis induction and fluorescence imaging of cancer cells†
Nan
Xie
 a,
Ke
Feng
a,
Ke
Feng
 *b,
Jianqun
Shao
a,
Bin
Chen
b,
Chen-Ho
Tung
b and
Li-Zhu
Wu
*b,
Jianqun
Shao
a,
Bin
Chen
b,
Chen-Ho
Tung
b and
Li-Zhu
Wu
 *b
*b
aSchool of Pharmaceutical Sciences, Capital Medical University, Beijing 100069, P. R. China
bKey Laboratory of Photochemical Conversion and Optoelectronic Materials, Technical Institute of Physics and Chemistry & School of Future Technology, University of CAS, the Chinese Academy of Sciences, Beijing 100190, P. R. China. E-mail: kefeng@mail.ipc.ac.cn; lzwu@mail.ipc.ac.cn
First published on 28th November 2017
Abstract
Bifunctional peptide-polynorbornenes, comprised of four modules, D-enantiomeric, protease-resistant sequence D[KLAKLAK]2 peptide as the anticancer motif, cell-penetrating peptide TAT sequence as the “transmission adapter”, red fluorescence RhB as the imaging reporter, and biocompatible PEG as the hydrophilic solubilizer, were designed and fabricated via a simple ROMP (ring-opening metathesis polymerization) strategy. Attributed to the integration of both KLA and TAT peptides, these bioactive copolymers exhibit the synergistic induction of tumor cell apoptosis. Confocal laser scanning microscopy and flow cytometric analyses reveal that these bifunctional peptide KLA–TAT copolymers target or act on mitochondria and disrupt the mitochondrial membrane potential (MMP) after efficient internalization with the aid of the cell-penetrating TAT motif, thus inducing mitochondrial-dependent apoptosis of cancer cells with enhanced activity. More importantly, this modularly designed ROMP strategy could integrate the multi-functionalities of targeting, imaging and cancer therapeutics into a single polymeric scaffold, and endow synthetic polymers with tremendous potential to serve as a new type of agent for cancer diagnostics and treatment.
Introduction
The deregulation of programmed cell death, or apoptosis, has been regarded as one of the most essential alterations in cell physiology that dictates malignant growth and is supposed to be a hallmark of most and perhaps all types of cancer.1,2 Many anticancer agents have been explored to improve therapeutic efficacy by inducing apoptosis in cancer cells, and overcome the limitations of traditional antineoplastic drugs, such as lack of selectivity, undesirable side effects, multi-drug resistance (MDR) as well as poor solubility in aqueous solution.3,4 The α-helical amphiphilic [KLAKLAK]2 (KLA) peptide has been recognized as one of the typically apoptotic stimuli, which intrinsically disrupts the mitochondrial membrane and initiates apoptotic processes in almost all kinds of cancer cells.5–8 Extensive attempts have been made to utilize apoptotic peptide as an anticancer agent; however, its therapeutic application is still thwarted by fast proteolytic degradation, poor cell membrane permeability, limited cellular accumulation, and complicated preparation procedures even when combined with tumor homing ligands, antibodies or nanocomplexes.9–14Ring-opening metathesis polymerization (ROMP) had been testified as a simple and effective toolbox to fabricate random, block, and even alternating functionalized copolymers.15–17 Utilizing Grubbs’ third-generation catalyst as the initiator, cyclic olefins, especially polynorbornenes, can be facilely fabricated from diverse monomeric modules that are capable of containing chromophores,18–21 hydrophilic and hydrophobic building blocks,22–29 hydrogen-bond interfaces,30–33 monosaccharide or oligosaccharide,34–37 DNA or protein analogues,38–42 as well as designable bioactive peptides43–50 with well-controlled molecular weight and narrow polydispersity indices. Applying the modular ROMP strategy, we had introduced functional peptide sequences, including receptor-targeted peptides,51,52 nuclear localization signaling peptides and cell-penetrating peptides53 into the polynorbornenyl scaffold to facilitate targeting capability towards cancer cells, organelles, and even certain tissues. Attributed to the repeating peptide motifs on a polymeric chain, the multivalent effect and the EPR (enhanced permeation and retention) effect could increase the signal-to-noise ratio for bioimaging, enhance target-binding specificity, and improve the bioavailability or theranostic efficacy.54–56
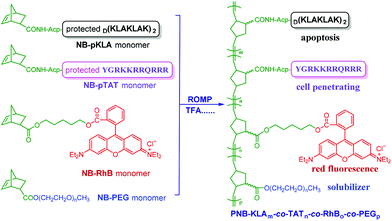
In this contribution, we intend to incorporate both the apoptotic peptide KLA and the cell-penetrating peptide TAT into one polynorbornenyl chain to fabricate anticancer copolymers through modular ROMP. On the one hand, the integration of TAT, a well-known CPP derived from human immunodeficiency virus (HIV) transactivator protein,57,58 could facilitate the cellular transport and accumulation of anticancer copolymers in a hypotonic environment; on the other hand, the multivalent tethering of the bioactive KLA motifs on polynorbornenes would enhance mitochondria-targeting specificity and sluggish proteolytic degradation, thus improving the bioavailability and anticancer activity of the copolymers. As shown in Scheme 1, the bifunctional peptide-polynorbornenes were designed and architected with four modules: D-enantiomeric, protease-resistant sequence D[KLAKLAK]2 peptide as the anticancer motif, cell-penetrating peptide TAT sequence as the “transmission adapter” for enhanced membrane permeability, strong red fluorescence rhodamine B as the imaging reporter, and biocompatible poly(ethylene glycol) as the hydrophilic solubilizer. Based on the intrinsic properties of each functional module, we expected that these bifunctional peptide-polynorbornenes would disrupt the mitochondrial membrane after efficient internalization with the aid of a cell-penetrating TAT motif, and induce mitochondrial-dependent apoptosis of cancer cells with enhanced activity. More importantly, this modularly designed ROMP strategy could integrate the multi-functionalities of targeting, imaging and cancer therapeutics into a single polymeric scaffold, and endow synthetic polymers with tremendous potential to serve as a new type of agent for cancer diagnostics and treatment.
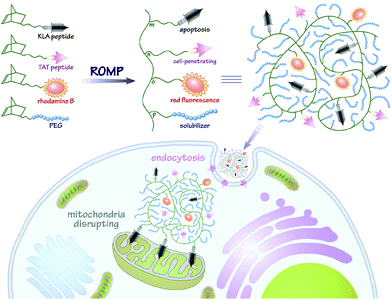 | ||
| Scheme 1 Fabrication of bifunctional peptide-polynorbornenes and possible mechanism for apoptosis induction and fluorescence imaging in cancer cells. | ||
Experimental
General
All experiments with air- and moisture-sensitive intermediates and compounds were conducted under an inert atmosphere using standard Schlenk techniques. Mass spectra were recorded on a Bruker APEX IV FTMS spectrometer. NMR spectra were recorded on a Bruker Advance DPX 400 MHz spectrometer and referenced using the residual proton signal of the solvent. Elemental analyses were performed on a Carlo Erba 1106 elemental analyzer. UV-vis spectra were recorded using a Hitachi U-3900 spectrophotometer. Fluorescence measurements and the corresponding lifetime were determined using the time-correlated single-photon counting method on an F-900 Edinburgh Analytical Instruments system. Gel-permeation chromatography analyses were carried out on a multi-angle, digital signal processing light scattering system (Wyatt, USA) using a Shimadzu LC-10A pump coupled to a Dawn Heleos II light scattering detector (equipped with a 100 mW GaAs laser) and a Optilab rEX interferometric refractometer with THF or DMF (with 25 mM LiBr at 313 K) as eluents on a 10 μm linear MZ-Gel SDplus column; the flow rate used for all measurements was 0.5 mL min−1. No calibration standards were used, and dn/dc values were obtained for each injection by assuming 100% mass elution from the columns. Mw, Mn and PDIs were treated using an Astra 5 software package. Dynamic light scattering (DLS) measurements were performed on a DynaPro NanoStar system (Wyatt, USA) with the Dynamics 7.0 software package. Zeta potential measurements were performed in 10 mM PBS buffer with a pH value of 7.4 at 298 K using a Nano-ZS90 Zetasizer system (Malvern, UK). The zeta potential (ζ) was calculated from the electrophoretic mobility using the Smoluchowski equation as the average values of three runs.Materials and synthesis
exo-Bicyclo[2.2.1]hept-5-ene-2-carboxylic acid was synthesized according to published procedures.25 5-Rhodamine-B-formyloxypentyl-exo-bicyclo[2.2.1]-hept-5-ene-2-carboxylate (NB-RhB monomer), methoxypolyethylene-glycol-550-exo-bicyclo[2.2.1]-hept-5-ene-2-carboxylate (NB-PEG monomer) and the cell-penetrating peptide monomer (NB-pTAT monomer) were synthesized according to the procedure of our previous report.51,53 2-Chlorotrityl chloride resin (100–200 mesh) and Fmoc protected amino acids were purchased from GL Biochem Ltd (Shanghai, China). Other reagents and solvents were obtained from commercial suppliers and used as received without further purification.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) provided the NB-pKLA monomer (1.27 g) as an off-white powder. HR-ESI MS: m/z = cacld. 1177.7567, found 1177.7533 for [M − 2H]2−, C116H203N21O292−. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 12.11 (br, 1H), 7.86 (m, 15H), 6.68 (m, 5H), 6.38 (br, 1H), 6.11 (m, 2H), 4.11 (m, 14H), 3.02 (m, 2H), 2.87 (m, 13H), 2.75 (s, 1H), 2.18 (m, 2H), 2.00 (m, 1H), 1.36 (m, 124H), 0.82 (m, 24H). Anal. cacld for C116H205N21O29: C, 59.09; H, 8.76; N, 12.47. Found: C, 59.77; H, 9.10; N, 12.02.
1) provided the NB-pKLA monomer (1.27 g) as an off-white powder. HR-ESI MS: m/z = cacld. 1177.7567, found 1177.7533 for [M − 2H]2−, C116H203N21O292−. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 12.11 (br, 1H), 7.86 (m, 15H), 6.68 (m, 5H), 6.38 (br, 1H), 6.11 (m, 2H), 4.11 (m, 14H), 3.02 (m, 2H), 2.87 (m, 13H), 2.75 (s, 1H), 2.18 (m, 2H), 2.00 (m, 1H), 1.36 (m, 124H), 0.82 (m, 24H). Anal. cacld for C116H205N21O29: C, 59.09; H, 8.76; N, 12.47. Found: C, 59.77; H, 9.10; N, 12.02.
Cell culture
Human cervical carcinoma HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM). The medium contains 10% fetal bovine serum (FBS), 100 mg L−1 streptomycin and 100 U mL−1 penicillin. Cells were maintained under a humidified atmosphere of 5% CO2 at 37 °C.Cell viability assay
Cellular viability was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells growing in the log phase were seeded into a 96-well cell-culture plate at a density of 5 × 103 cells per well, and incubated overnight prior to adding the peptide polymers and KLA monomer at various concentrations. After 48 h incubation, the medium in the wells was discarded and the cells were washed with a PBS buffer and then incubated in 100 μL fresh medium and 25 μL of 5 mg mL−1 MTT solution in PBS. After incubation for an additional 4 h, the formazan crystals generated by dehydrogenases in live cells were dissolved in DMSO and the absorbance was measured at 570 nm by using a microplate reader (Multiskan MK3, Thermo Electron Corporation). The viability of the cells can be calculated as the ratio of the mean absorbance of the treatment group to the mean absorbance of the control group. The MTT assays of cells incubated in the medium without the polymers were set as the control.Apoptosis assay
Cell apoptosis was measured by dual staining with an annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) kit (Beyotime Institute of Biotechnology, China) according to the manufacturer's instructions. Briefly, cells were seeded at a density of 5 × 105 cells per well on each 35 mm glass bottom culture dish (NEST Biotechnology Co., Ltd). After overnight incubation, 10 μM PNB-KLA15-co-PEG85 or PNB-KLA15-co-TAT10-co-PEG75 was added to each dish and the cells were allowed to incubate for another 4, 8, 24, 36 and 48 h, respectively. After washing with PBS 3 times, the cells were stained with annexin V-FITC and PI, and observed by using a confocal laser scanning microscope (CLSM, Leica TCS-SP8) equipped with a 63× oil-immersion objective lens at an excitation wavelength of 488 nm and 530 nm. Light emissions were collected using a green (520/30 nm) and red (610/30 nm) filter set. For flow cytometry analysis, the cells were seeded into 6-well plates at a density of 5 × 105 cells per well and incubated overnight. Thereafter, the cells were treated with the peptide polymers and digested with trypsin prior to staining, and then analyzed by using a BD LSRFortessa flow cytometer (BD Biosciences).Measurement of the mitochondrial membrane potential
The mitochondrial membrane potential was determined using a 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolyl-carbocyanine iodide (JC-1) kit (KeyGEN bioTECH, China) according to the manufacturer's instructions. Cells were seeded into 6-well plates at a density of 5 × 105 cells per well. After overnight incubation, 10 μM PNB-KLA15-co-TAT10-co-PEG75 was added and the cells were allowed to incubate for another 12, 24 and 48 h. After washing with PBS, the cells were trypsinized, stained with JC-1, and analyzed with a flow cytometer. Parallelly, a fresh batch of treated cells was stained with JC-1 and visualized under a CLSM at an excitation wavelength of 488 nm and 530 nm. Light emissions were collected using green (520/30 nm) and red (610/30 nm) filters for the detection of the monomer and aggregation form of JC-1.Cellular internalization of polymers
Cells were plated in a 6-well plate at a density of 5 × 105 cells per well and incubated overnight. Then the polymers were added from a concentrated stock solution in PBS at a final concentration of 10 μM and incubated for 4 h. After discarding the media and rinsing three times with PBS, the cells were harvested by trypsinization. After centrifugation, the cell pellets were washed carefully with PBS, resuspended in PBS and analyzed immediately using a flow cytometer operating at an excitation wavelength of 561 nm and with a 610/30 nm emission filter set.Colocalization imaging
Cells were seeded at a density of 1 × 105 cells per well on each 35 mm glass bottom culture dish and incubated overnight. Then the polymers were added from a concentrated stock solution in PBS at a final concentration of 10 μM and incubated for 4 h. After discarding the media and rinsing three times with PBS, the cells were stained with MitoTracker Green for 30 min and imaged directly by CLSM at an excitation wavelength of 560 nm and with a 610/30 nm emission filter set.Statistical analysis
Statistical analysis was performed by using ANOVA test. The results were expressed as mean ± SD% and considered to be statistically significant when p < 0.05.Results and discussion
Synthesis and characterization of copolymers
Applying the strategy discussed in our recent report,53 the KLA and TAT peptide monomers were facilely prepared by a standard Fmoc (fluorenylmethyloxycarbonyl) solid phase peptide synthesis (SPPS). Maynard and Grubbs firstly applied this method to the construction of monomeric norbornenes.43 The synthetic procedure involved the step-by-step chain extension of individual protected amino acid units via active hydroxyl groups on the 2-chlorotrityl chloride resin. As the chain termination was modified by employing exo-norbornenyl carboxylic monoacid as the monomeric synthon, fully protected peptide monomers could be obtained after cleavage from the resin beads. During this process, 6-aminohexanoic acid was inserted as a soft spacer to enhance the molecular flexibility of the peptide monomers and improve their reactivity during polymerization. The protected peptide monomers NB-pKLA and NB-pTAT were well characterized by NMR, MS and EA, and could be subjected to subsequent copolymerization without any need for further purification.As shown as Fig. 1, utilizing the one-pot ROMP strategy by initiation with the Grubbs’ third-generation catalyst, the norbornenyl copolymers could be conveniently derived from varied monomers to integrate multiple functionalities for a diversity of biomedical applications. In order to optimize these polymers and evaluate the synergistic effect of the KLA and TAT motifs, the tri-component copolymers PNB-KLAm-co-TATn-co-PEGp were synthesized from adjustable feeding proportions of peptide NB-pKLA, NB-pTAT and hydrophilic NB-PEG monomers. A fixed DP (degree of polymerization) value of 100 was applied to all polynorbornenes to guarantee the molar ratio of the total feeding amounts of monomeric norbornenes to the Grubbs’ initiator. The NB-pKLA proportion m varied from 5 to 20, the NB-pTAT proportion n was empirically chosen as 10, and hydrophilic NB-PEG was incorporated as needed to keep the DP constant. For additional imaging purpose, 5% of red fluorescence NB-RhB monomer was introduced and endowed the copolymer PNB-KLA15-co-TAT10-co-RhB5-co-PEG70 with fluorescent probing characteristics. To investigate the localization and confirm the enhanced internalization in cancer cells, PNB-KLA15-co-RhB5-co-PEG80 and PNB-TAT10-co-RhB5-co-PEG85 were analogously synthesized in the absence of either KLA or TAT peptide for control experiments.
The bioactivation of the peptide copolymers could be completed by the removal of side-chain protecting groups via TFA-mediated cleavage. Consistent with our previous studies,51 the copolymers showed well-controlled molecular weights as well as comparable polydispersity indices (PDIs) after deprotection; these results could be verified by GPC measurements (Table 1). The molecular weights and corresponding polymeric particle diameters determined by Rayleigh scattering with DLS are in good accordance with the results from GPC (Table S1†). By bearing a high proportion of the hydrophilic PEG solubilizer, the deprotected PNB-KLAm-co-TATn-co-RhBo-co-PEGp copolymers exhibit excellent water solubility and are likely to stay as single-chain coils in aqueous media. In order to minimize the ionic effect, the deprotected peptide-polynorbornenes were adequately dialyzed against physiologic saline or sterile distilled water at 4 °C and prepared as a stock solution for further use. By examining the photophysical properties of PNB-KLAm-co-TATn-co-RhBo-co-PEGp, we demonstrated that the incorporation of the red fluorescence module rhodamine B is critical in achieving the imaging capability for the copolymers. As expected, the polymeric analogues exhibited intense absorption with λabs at 565 nm and strong emission with λem at 592 nm, almost identical to that of molecular rhodamine B (Fig. S1†). Additionally, the luminescence quantum yield (Φ, 0.50) and lifetime (τ, 1.66 ns) of PNB-KLA15-co-TAT10-co-RhB5-co-PEG70 are also very similar to those of its polymeric analogues bearing different peptide branches.53
| Polymera | M n/kDa | M w/kDa | PDI | ζ /mV |
|---|---|---|---|---|
| a Measured in DMF with 25 mM LiBr at 313 K. b Measured in 10 mM PBS buffer (pH 7.4) with a concentration of 1.0 mg mL−1 at 298 K. | ||||
| PNB-pKLA5-co-pTAT10-co-PEG85 | 113.1 | 145.2 | 1.28 | — |
| PNB-KLA5-co-TAT10-co-PEG85 | 90.5 | 117.0 | 1.29 | +10.3 ± 1.9 |
| PNB-pKLA10-co-pTAT10-co-PEG80 | 125.4 | 156.3 | 1.25 | — |
| PNB-KLA10-co-TAT10-co-PEG80 | 97.7 | 123.9 | 1.27 | +12.7 ± 2.1 |
| PNB-pKLA15-co-pTAT10-co-PEG75 | 120.2 | 148.3 | 1.23 | — |
| PNB-KLA15-co-TAT10-co-PEG75 | 98.1 | 120.6 | 1.23 | +12.2 ± 1.7 |
| PNB-pKLA20-co-pTAT10-co-PEG70 | 114.7 | 152.3 | 1.33 | — |
| PNB-KLA20-co-TAT10-co-PEG70 | 101.8 | 133.2 | 1.31 | +14.0 ± 2.7 |
| PNB-pKLA15-co-PEG85 | 84.5 | 100.1 | 1.18 | — |
| PNB-KLA15-co-PEG85 | 75.6 | 89.9 | 1.19 | +11.5 ± 1.1 |
| PNB-pKLA15-co-pTAT10-co-RhB5-co-PEG70 | 112.6 | 141.7 | 1.26 | — |
| PNB-KLA15-co-TAT10-co-RhB5-co-PEG70 | 92.4 | 115.8 | 1.25 | +16.8 ± 1.5 |
| PNB-pTAT10-co-RhB5-co-PEG85 | 95.3 | 118.9 | 1.25 | — |
| PNB-TAT10-co-RhB5-co-PEG85 | 80.8 | 102.1 | 1.26 | +14.7 ± 1.8 |
| PNB-pKLA15-co-RhB5-co-PEG80 | 81.6 | 99.3 | 1.22 | — |
| PNB-KLA15-co-RhB5-co-PEG80 | 74.1 | 90.2 | 1.22 | +13.9 ± 2.0 |
| PNB-pTAT10-co-PEG90 | 85.1 | 105.4 | 1.24 | — |
| PNB-TAT10-co-PEG90 | 74.5 | 93.4 | 1.25 | +11.6 ± 2.3 |
Synergistic inhibition of cell proliferation
Our recent research illustrated that the cell-penetrating peptide (CPP) TAT sequence would greatly enhance the cellular internalization of polynorbornenes, which also made the promising application of functional peptide to cellular pro-apoptosis. On the basis of this result, we integrated both KLA and TAT peptides into the same polymeric scaffold, and expected to endow the polynorbornenes with the synergistic inhibition of cell proliferation. The cell proliferation profiles of the MTT assay demonstrated the great difference between the bifunctional KLA–TAT and solo-type polymers. As shown in Fig. 2, the solo-type PNB-KLA15-co-mPEG85 copolymer or monomeric NB-KLA exhibited no clear cytotoxicity against HeLa cells even when their concentration was higher than 50 μM. This could be reasonably explained by the poor cell-penetrating capability of the polymeric scaffold in the absence of the TAT motifs. A control experiment from the solo-type PNB-TAT10-co-mPEG90 copolymer also indicated a comparably low cytotoxicity. In contrast, all bifunctional KLA–TAT polymers showed a clearly synergistic effect and presented a significant cytotoxicity towards HeLa cells on typical dose–response curves even when the KLA peptide concentrations were lower than 10 μM. The toxic potency was proportional to the doping rate of the KLA peptide within the PNB-KLAx-co-TAT10-co-PEG90−x (x = 5, 10, and 15) copolymers, which had half inhibitory concentration (IC50) values of 7.86 ± 0.39, 5.79 ± 0.47 and 3.43 ± 0.21 μM, respectively (Table 2, based on the effective concentration of the KLA peptide). The highest KLA doping rate of 20 in PNB-KLA20-co-TAT10-co-PEG70 did not give the lowest IC50 value (5.20 ± 0.35 μM), which might be attributed to the mismatching of the TAT peptide in the copolymer thereafter reducing synergism. The corresponding fluorescent copolymer PNB-KLA15-co-TAT10-co-RhB5-co-PEG70 was held at the same KLA–TAT doping ratio as optimization, and a comparable IC50 value was observed at 4.63 ± 0.27 μM in HeLa cells.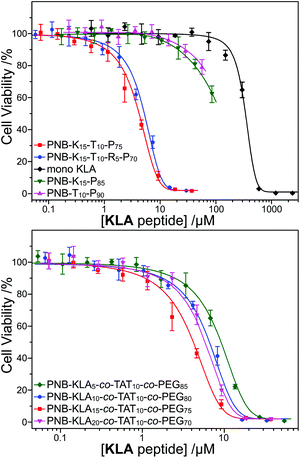 | ||
| Fig. 2 Cell proliferation profiles of the polymer and monomer at various concentrations in HeLa cells after 48 h incubation (n = 3). | ||
Analysis of localization and cellular uptake
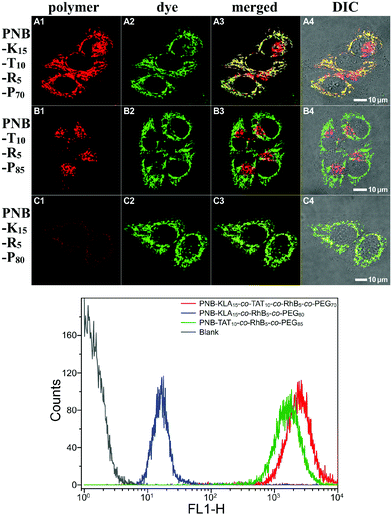
In order to assess the intracellular localization as well as the cellular internalization of these bifunctional peptide KLA–TAT polymers, fluorescence imaging studies were carried out with the red emissive RhB-labelled analogues in HeLa cells. Representative images revealing the colocalization of the polymers with MitoTracker Green are shown in Fig. 3. PNB-KLA15-co-TAT10-co-RhB5-co-PEG70 or PNB-KLA15-co-RhB5-co-PEG80 displayed a relatively high level of mitochondrial localization with a Pearson's correlation coefficient of 0.90 ± 0.04, while PNB-TAT10-co-RhB5-co-PEG85 was just found in the cytosol around the nucleus and showed essentially no mitochondrial (Pearson's r = 0.07 ± 0.02) but lysosomal localization.53 These distinct localization profiles should be closely associated with the different physicochemical properties originated from either the TAT or KLA motif. The former weakly locates lysosomes due to its guanidinium-rich structure, while the latter dominates the translocations through mitochondrial membranes by the helical lipophilic and cationic [KLAKLAK]2 residues.6 Besides, PNB-KLA15-co-TAT10-co-RhB5-co-PEG70 exhibited much stronger red fluorescence in HeLa cells than that of PNB-KLA15-co-RhB5-co-PEG80, indicating that efficient transport through the plasma membrane of cancer cells could be achieved by the incorporation of the TAT motif on the polymeric scaffold. This observation was also consistent with quantitative analysis by flow cytometry. Without the aid of the cell-penetrating TAT module, the PNB-KLA15-co-RhB5-co-PEG80 polymer was almost negligibly taken up by HeLa cells, while the treatment with PNB-KLA15-co-TAT10-co-RhB5-co-PEG70 led to a notable increase in red fluorescence from rhodamine B by nearly 2 orders of magnitude (Fig. 3). All these results implied that the combination of the KLA and TAT modules potentiates the bioactivity of polynorbornenes by both cell membrane penetration and mitochondria targeting.Evaluation of cell apoptosis
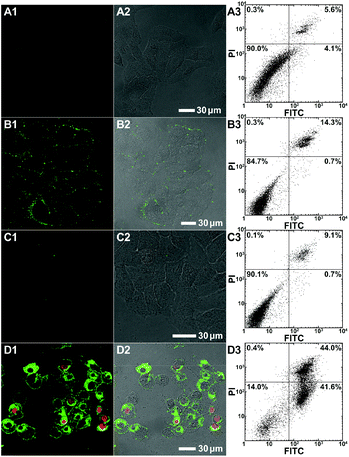
To evaluate the polymer-induced cell death through apoptosis, HeLa cells co-incubated with either the bifunctional KLA–TAT or solo-type polymer were stained with annexin V-FITC/propidium iodide (PI). As a specific marker for apoptosis, annexin V binds specifically to the exposed phosphatidylserine (PS) on the cell surface in early-stage apoptosis, and then PI results in a rapid staining of nuclear chromatin in membrane-compromised cells during late apoptosis or even necrosis. As is evident from Fig. 4, the cells treated with 10 μM of PNB-KLA15-co-PEG85 only had a slight enhancement in green fluorescence compared to that in blank cells, indicating the negligible cytotoxicity of the KLA polymer in the absence of the cell-penetrating TAT module. A similar result was also obtained in the case of PNB-TAT10-co-PEG90. In comparison, the cells treated with the same concentration of PNB-KLA15-co-TAT10-co-PEG75 showed bright green staining in the cytoplasm as well as red staining in the nucleus, indicating the presence of both early- and late-stage apoptosis in HeLa cells. These results were further substantiated by flow cytometry (Fig. 4, A3–D3); more than 85% of the tumor cells survived after the treatment with the solo-type PNB-KLA15-co-PEG85 polymer or PNB-TAT10-co-PEG90 polymers; on the contrary, PNB-KLA15-co-TAT10-co-PEG75 exerted a significant apoptosis in up to 86% of HeLa cells because of the synergistic effect in the presence of both the TAT and KLA motifs. We also made a time course observation on apoptotic cells within 48 h. As shown in Fig. 5, the counting number of dead cells increased with the prolonged incubation time after the treatment with the bifunctional KLA–TAT polymer; the time-dependent apoptosis was also in good agreement with the MTT and flow cytometry results.Cell apoptotic mechanism
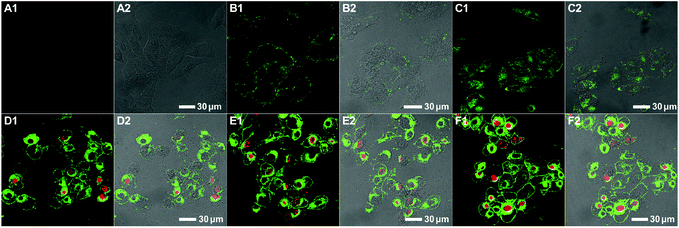
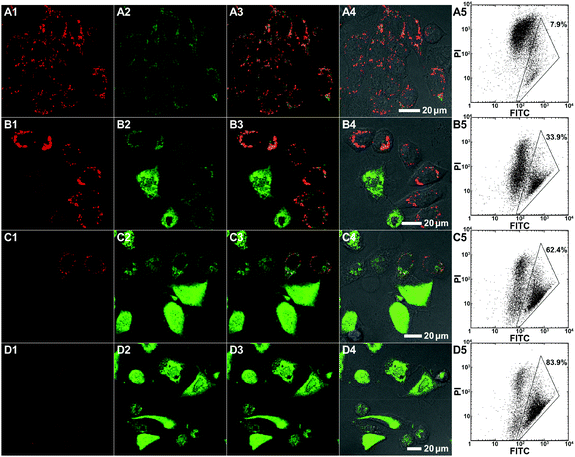
Both colocalization and apoptosis experiments demonstrated the cell apoptosis induced by the bifunctional KLA–TAT polymer via a mitochondrial-dependent pathway that is usually associated with the disruption in the mitochondrial membrane potential (MMP). Thus, the lipophilic cationic probe JC-1 was employed in our case to monitor the potential-dependent accumulation in the mitochondria. The normal MMP of healthy cells stained with the JC-1 probe allows for the formation and sequestration in mitochondria from JC aggregates with red fluorescence. Once the MMP was lost in apoptotic cells, the JC aggregates would dissipate into the cytoplasm as a monomeric form, indicating green fluorescence. Thus, the status of the mitochondria as well as apoptotic cells could be distinguished by the ratio of green-to-red fluorescence. As shown as the CLSM images in Fig. 6, a clear decrease in red fluorescence after the treatment with PNB-KLA15-co-TAT10-co-PEG75 revealed the collapse of the mitochondrial membrane potential in HeLa cells in a time-dependent manner. Within the prolonged incubation period of 48 h, the apoptotic process could be evidenced by the increasing ratio of green-to-red fluorescence in HeLa cells. The flow cytometric analyses (Fig. 6, A5–D5) quantitatively illustrated these gradual changes in green fluorescence as the labelled triangle area with a percentage of 7.9% at initiation, 33.9% for 12 h, 62.4% for 24 h, and 83.9% for 48 h after incubation with the PNB-KLA15-co-TAT10-co-PEG75 polymer. Apparently, the potentiation in the cell apoptosis of the bifunctional peptide KLA–TAT polymer should benefit from a TAT-mediated cell-penetrating and KLA-targeted mitochondria disruption pathway.Conclusions
In summary, we constructed the bifunctional peptide-polynorbornenes via modular ROMP with a polymeric architecture comprised of four modules, bifunctional peptides with D-enantiomeric, protease-resistant sequence D[KLAKLAK]2 peptide as the anticancer motif, cell-penetrating peptide TAT sequence as the “transmission adapter”, red fluorescence RhB as the imaging reporter, and biocompatible PEG as the hydrophilic solubilizer. Confocal laser scanning microscopy and flow cytometric analyses demonstrated that these bifunctional peptide-polynorbornenes would disrupt the mitochondrial membrane after efficient internalization with the aid of the cell-penetrating TAT motif, and induce mitochondrial-dependent apoptosis of cancer cells with enhanced activity. This modularly designed ROMP strategy integrated both bioimaging and cell apoptosis into a single polymeric scaffold, and endowed synthetic polymers with tremendous potential to serve as a new type of agent for cancer diagnostics and treatment.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Key R&D Program of China (2017YFA0206900), the Ministry of Science and Technology of China (2014CB239402), the National Natural Science Foundation of China (21204052, 21372232 and 91427303), the Beijing Natural Science Foundation (2172019), and the Strategic Priority Research Program of CAS (XDB17000000). The authors also acknowledge the technical support provided by the Core Facility Center at Capital Medical University.Notes and references
- D. Hanahan and R. A. Weinberg, Cell, 2000, 100, 57–70 CrossRef CAS PubMed.
- S. W. Fesik, Nat. Rev. Cancer, 2005, 5, 876–885 CrossRef CAS PubMed.
- M. E. Davis, Z. Chen and D. M. Shin, Nat. Rev. Drug Discovery, 2008, 7, 771–782 CrossRef CAS PubMed.
- T. K. Kelly, D. D. De Carvalho and P. A. Jones, Nat. Biotechnol., 2010, 28, 1069–1078 CrossRef CAS PubMed.
- M. M. Javadpour, M. M. Juban, W.-C. J. Lo, S. M. Bishop, J. B. Alberty, S. M. Cowell, C. L. Becker and M. L. McLaughlin, J. Med. Chem., 1996, 39, 3107–3113 CrossRef CAS PubMed.
- H. M. Ellerby, W. Arap, L. M. Ellerby, R. Kain, R. Andrusiak, G. D. Rio, S. Krajewski, C. R. Lombardo, R. Rao, E. Ruoslahti, D. E. Bredesen and R. Pasqualini, Nat. Med., 1999, 5, 1032–1038 CrossRef CAS PubMed.
- A. J. Marks, M. S. Cooper, R. J. Anderson, K. H. Orchard, G. Hale, J. M. North, K. Ganeshaguru, A. J. Steele, A. B. Mehta, M. W. Lowdell and R. G. Wickremasinghe, Cancer Res., 2005, 65, 2373–2377 CrossRef CAS PubMed.
- J. Li, K. Hu, H. Chen, Y. Wu, L. Chen, F. Yin, Y. Tian and Z. Li, Chem. Commun., 2017, 53, 10452–10455 RSC.
- L. Agemy, D. Friedmann-Morvinski, V. R. Kotamraju, L. Roth, K. N. Sugahara, O. M. Girard, R. F. Mattrey, I. M. Verma and E. Ruoslahti, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 17450–17455 CrossRef CAS PubMed.
- W.-H. Chen, G.-F. Luo, W.-X. Qiu, Q. Lei, L.-H. Liu, D.-W. Zheng, S. Hong, S.-X. Cheng and X.-Z. Zhang, Chem. Mater., 2016, 28, 6742–6752 CrossRef CAS.
- Z.-Y. Qiao, W.-J. Zhao, Y. Cong, D. Zhang, Z. Hu, Z.-Y. Duan and H. Wang, Biomacromolecules, 2016, 17, 1643–1652 CrossRef CAS PubMed.
- T. Suma, J. Cui, M. Müllner, S. Fu, J. Tran, K. F. Noi, Y. Ju and F. Caruso, J. Am. Chem. Soc., 2017, 139, 4009–4018 CrossRef CAS PubMed.
- X. Yan, Q. Yu, L. Guo, W. Guo, S. Guan, H. Tang, S. Lin and Z. Gan, ACS Appl. Mater. Interfaces, 2017, 9, 6804–6815 CAS.
- J. Zong, S. L. Cobb and N. R. Cameron, Biomater. Sci., 2017, 5, 872–886 RSC.
- A. Fürstner, Angew. Chem., Int. Ed., 2000, 39, 3012–3043 CrossRef.
- T. M. Trnka and R. H. Grubbs, Acc. Chem. Res., 2000, 34, 18–29 CrossRef.
- J. A. Love, J. P. Morgan, T. M. Trnka and R. H. Grubbs, Angew. Chem., Int. Ed., 2002, 41, 4035–4037 CrossRef CAS PubMed.
- A. R. Morales, C. O. Yanez, Y. Zhang, X. Wang, S. Biswas, T. Urakami, M. Komatsu and K. D. Belfield, Biomaterials, 2012, 33, 8477–8485 CrossRef CAS PubMed.
- N. Xie, K. Feng, B. Chen, M. Zhao, S. Peng, L.-P. Zhang, C.-H. Tung and L.-Z. Wu, J. Mater. Chem. B, 2014, 2, 502–510 RSC.
- Y. Li, Y. Bai, N. Zheng, Y. Liu, G. A. Vincil, B. J. Pedretti, J. Cheng and S. C. Zimmerman, Chem. Commun., 2016, 52, 3781–3784 RSC.
- M. Hollauf, P. W. Zach, S. M. Borisov, B. J. Muller, D. Beichel, M. Tscherner, S. Kostler, P. Hartmann, A. C. Knall and G. Trimmel, J. Mater. Chem. C, 2017, 5, 7535–7545 RSC.
- K. Lienkamp, A. E. Madkour, A. Musante, C. F. Nelson, K. Nüsslein and G. N. Tew, J. Am. Chem. Soc., 2008, 130, 9836–9843 CrossRef CAS PubMed.
- E. M. Kolonko, J. K. Pontrello, S. L. Mangold and L. L. Kiessling, J. Am. Chem. Soc., 2009, 131, 7327–7333 CrossRef CAS PubMed.
- K. Miki, A. Kimura, K. Oride, Y. Kuramochi, H. Matsuoka, H. Harada, M. Hiraoka and K. Ohe, Angew. Chem., 2011, 123, 6697–6700 CrossRef.
- K. Feng, N. Xie, B. Chen, L.-P. Zhang, C.-H. Tung and L.-Z. Wu, Macromolecules, 2012, 45, 5596–5603 CrossRef CAS.
- S. A. Barnhill, N. C. Bell, J. P. Patterson, D. P. Olds and N. C. Gianneschi, Macromolecules, 2015, 48, 1152–1161 CrossRef CAS.
- J.-F. Lutz, J.-M. Lehn, E. W. Meijer and K. Matyjaszewski, Nat. Rev. Mater., 2016, 1, 16024 CrossRef CAS.
- J.-X. Yang, Y.-Y. Long, L. Pan, Y.-F. Men and Y.-S. Li, ACS Appl. Mater. Interfaces, 2016, 8, 12445–12455 CAS.
- D.-Y. Kim, S. Shin, W.-J. Yoon, Y.-J. Choi, J.-K. Hwang, J.-S. Kim, C.-R. Lee, T.-L. Choi and K.-U. Jeong, Adv. Funct. Mater., 2017, 27, 1606294 CrossRef.
- S. K. Yang, A. V. Ambade and M. Weck, Chem. Soc. Rev., 2011, 40, 129–137 RSC.
- E. Elacqua, A. Croom, K. B. Manning, S. K. Pomarico, D. Lye, L. Young and M. Weck, Angew. Chem., Int. Ed., 2016, 55, 15873–15878 CrossRef CAS PubMed.
- U. Drechsler, R. J. Thibault and V. M. Rotello, Macromolecules, 2002, 35, 9621–9623 CrossRef CAS.
- S. R. Mane, A. Sathyan and R. Shunmugam, Sci. Rep., 2017, 7, 44857 CrossRef CAS PubMed.
- S.-G. Lee, J. M. Brown, C. J. Rogers, J. B. Matson, C. Krishnamurthy, M. Rawat and L. C. Hsieh-Wilson, Chem. Sci., 2010, 1, 322–325 RSC.
- Y. I. Oh, G. J. Sheng, S.-K. Chang and L. C. Hsieh-Wilson, Angew. Chem., Int. Ed., 2013, 125, 12012–12015 CrossRef.
- R. S. Loka, F. Yu, E. T. Sletten and H. M. Nguyen, Chem. Commun., 2017, 53, 9163–9166 RSC.
- F. Fan, C. Cai, L. Gao, J. Li, P. Zhang, G. Li, C. Li and G. Yu, Polym. Chem., 2017, 8, 6709–6719 RSC.
- A. M. Rush, D. A. Nelles, A. P. Blum, S. A. Barnhill, E. T. Tatro, G. W. Yeo and N. C. Gianneschi, J. Am. Chem. Soc., 2014, 136, 7615–7618 CrossRef CAS PubMed.
- X. Lu, F. Jia, X. Tan, D. Wang, X. Cao, J. Zheng and K. Zhang, J. Am. Chem. Soc., 2016, 138, 9097–9100 CrossRef CAS PubMed.
- H. Xing, Y. Bai, Y. Bai, L. H. Tan, J. Tao, B. Pedretti, G. A. Vincil, Y. Lu and S. C. Zimmerman, J. Am. Chem. Soc., 2017, 139, 3623–3626 CrossRef CAS PubMed.
- P. W. Lee, S. A. Isarov, J. D. Wallat, S. K. Molugu, S. Shukla, J. E. P. Sun, J. Zhang, Y. Zheng, M. Lucius Dougherty, D. Konkolewicz, P. L. Stewart, N. F. Steinmetz, M. J. A. Hore and J. K. Pokorski, J. Am. Chem. Soc., 2017, 139, 3312–3315 CrossRef CAS PubMed.
- R. J. Banga, S. A. Krovi, S. P. Narayan, A. J. Sprangers, G. Liu, C. A. Mirkin and S. T. Nguyen, Biomacromolecules, 2017, 18, 483–489 CrossRef CAS PubMed.
- H. D. Maynard, S. Y. Okada and R. H. Grubbs, Macromolecules, 2000, 33, 6239–6248 CrossRef CAS.
- K. S. Roberts and N. S. Sampson, J. Org. Chem., 2003, 68, 2020–2023 CrossRef CAS PubMed.
- R. M. Conrad and R. H. Grubbs, Angew. Chem., Int. Ed., 2009, 48, 8328–8330 CrossRef CAS PubMed.
- A. L. Parry, P. H. H. Bomans, S. J. Holder, N. A. J. M. Sommerdijk and S. C. G. Biagini, Angew. Chem., Int. Ed., 2008, 47, 8859–8862 CrossRef CAS PubMed.
- P. R. Patel, R. C. Kiser, Y. Y. Lu, E. Fong, W. C. Ho, D. A. Tirrell and R. H. Grubbs, Biomacromolecules, 2012, 13, 2546–2553 CrossRef CAS PubMed.
- A. P. Blum, J. K. Kammeyer, J. Yin, D. T. Crystal, A. M. Rush, M. K. Gilson and N. C. Gianneschi, J. Am. Chem. Soc., 2014, 136, 15422–15437 CrossRef CAS PubMed.
- A. P. Blum, J. K. Kammeyer and N. C. Gianneschi, Chem. Sci., 2016, 7, 989–994 RSC.
- H. Xia, H. Fu, Y. Zhang, K.-C. Shih, Y. Ren, M. Anuganti, M.-P. Nieh, J. Cheng and Y. Lin, J. Am. Chem. Soc., 2017, 139, 11106–11116 CrossRef CAS PubMed.
- N. Xie, K. Feng, B. Chen, M. Zhao, L.-P. Zhang, C.-H. Tung, L.-Z. Wu and S. Peng, Chem. Commun., 2014, 50, 9539–9542 RSC.
- N. Xie, K. Feng, B. Chen, C.-H. Tung and L.-Z. Wu, New J. Chem., 2016, 40, 3252–3260 RSC.
- K. Feng, N. Xie, B. Chen, C.-H. Tung and L.-Z. Wu, Biomacromolecules, 2016, 17, 538–545 CrossRef CAS PubMed.
- D. F. Baban and L. W. Seymour, Adv. Drug Delivery Rev., 1998, 34, 109–119 CrossRef CAS PubMed.
- H. Maeda, J. Wu, T. Sawa, Y. Matsumura and K. Hori, J. Controlled Release, 2000, 65, 271–284 CrossRef CAS PubMed.
- A. K. Iyer, G. Khaled, J. Fang and H. Maeda, Drug Discovery Today, 2006, 11, 812–818 CrossRef CAS PubMed.
- S. Fawell, J. Seery, Y. Daikh, C. Moore, L. L. Chen, B. Pepinsky and J. Barsoum, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 664–668 CrossRef CAS.
- S. B. Fonseca, M. P. Pereira and S. O. Kelley, Adv. Drug Delivery Rev., 2009, 61, 953–964 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: UV-Vis absorption and fluorescence spectra, NMR and mass spectra for monomers and polymers. See DOI: 10.1039/c7py01730h |
| This journal is © The Royal Society of Chemistry 2018 |