Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
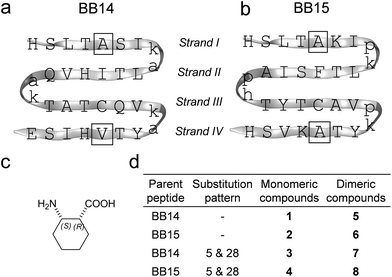
Peripheral cyclic β-amino acids balance the stability and edge-protection of β-sandwiches†
Gábor
Olajos
ac,
Anasztázia
Hetényi
b,
Edit
Wéber
a,
Titanilla
Szögi
 b,
Lívia
Fülöp
b and
Tamás A.
Martinek
b,
Lívia
Fülöp
b and
Tamás A.
Martinek
 *ac
*ac
aInstitute of Pharmaceutical Analysis, SZTE-MTA Lendület Foldamer Research Group, University of Szeged, Somogyi u. 4., H-6720 Szeged, Hungary. E-mail: martinek@pharm.u-szeged.hu
bDepartment of Medical Chemistry, University of Szeged, Dóm ter 8., H-6720 Szeged, Hungary
cMTA-SZTE Biomimetic Systems Research Group, University of Szeged, Dóm ter 8., H-6720 Szeged, Hungary
First published on 16th July 2018
Abstract
Engineering water-soluble stand-alone β-sandwich mimetics is a current challenge because of the difficulties associated with tailoring long-range interactions. In this work, single cis-(1R,2S)-2-aminocyclohexanecarboxylic acid mutations were introduced into the edge strands of the eight-stranded β-sandwich mimetic structures from the betabellin family. Temperature-dependent NMR and CD measurements, together with thermodynamic analyses, demonstrated that the modified peripheral strands exhibited an irregular and partially disordered structure but were able to exert sufficient shielding on the hydrophobic core to retain the predominantly β-sandwich structure. Although the frustrated interactions decreased the free energy of unfolding, the temperature of the maximum stabilities increased to or remained at physiologically relevant temperatures. We found that the irregular peripheral strands were able to prevent edge-to-edge association and fibril formation in the aggregation-prone model. These findings establish a β-sandwich stabilization and aggregation inhibition approach, which does not interfere with the pillars of the peptide bond or change the net charge of the peptide.
Introduction
The intricate programming rules of protein folding have been thoroughly investigated, and remarkable successes have been achieved in protein design by utilizing the natural α-amino acid building blocks.1–4 Protein mimetic structures containing unnatural monomers in the chain (foldamers) have opened new dimensions in constructing ordered hierarchical structures.5–7 In addition to the beauty of their structural diversity, foldamers have gained considerable interest for their ability to recognize macromolecular surfaces, which has led to numerous instances of protein–protein interaction inhibition.8–11 The design of foldamer helix mimetics is well established,12–17 including that of helix bundle quaternary structures.18–20 However, the construction of water-soluble β-sheet structures is still challenging.21–27 Using a top-down approach, the effects of α → β substitutions were systematically tested in foldameric β-hairpin systems. While α → β, αα → β2 and αα → β3 substitutions resulted in decreased structural stability,28 a reasonably high sheet content was achieved via the application of special β2,3-residues.29 A stable fold was attained by the incorporation of N-methyl-α-residues30,31 or cyclic γ-amino acids,32,33 which were also tested in a tertiary-folded system,32 but the position of the replacement proved to be an important factor to optimize. Our group investigated various β-amino side-chain topologies in the core of larger β-sandwich structures.34–37 The sequences exhibited dimerization- or membrane-induced β-sheet formation. Replacements with cis-(1R,2S)-2-aminocyclohexanecarboxylic acid (ACHC) were found to be able to maintain the interstrand H-bonding network and to fit sterically into the hydrophobic interior of the β-sandwich. However, the additional carbon atom in the backbone resulted in a backbone bulge and led to a reduced melting temperature, which prompted the question of whether ACHC substitution can be employed as an edge-protection approach.The general limitation of β-sheet design is the stability problem associated with a high aggregation propensity; this aggregation occurs at the open edges.38 Uncontrolled self-association not only prevents biological applications but also interferes with all phases of peptidic/protein drug development.39 The analysis of natural β-sheet proteins revealed that water solubility is improved by a negative design at the edge strands of β-sheets.40,41 An important natural edge protection strategy is the presence of irregularities (β-bulges) in the strand.42 These principles were used in the design of several edge-protection methods to prevent aggregation. A type of β-bulge was employed by replacing hydrophobic amino acids with lysine at the edges of the β-sheets.43
In this work, we utilized fibril-forming betabellin-1544 and non-aggregating betabellin-14 as templates and extended the ACHC substitutions to the edges of the β-sandwich. Here, we tested the effects of ACHC-generated peripheral bulges on the stability of the protein-mimetic models. We aimed to investigate the structural stability from multiple aspects: thermodynamic stability (free energy of reversible unfolding), thermal stability (thermal and cold denaturation) and stability against uncontrolled aggregation.
Materials and methods
Peptide synthesis and purification
All starting materials were commercially available. Tentagel R RAM resin was used as the solid support, and HATU was used as the coupling reagent. Couplings were performed under microwave irradiation in a 3-equivalent amino acid excess at 75 °C for 15 min for α-amino acids and for 30 min for β-residues. Histidine and cysteine were coupled at 50 °C. Peptides were cleaved with TFA/water/D,L-dithiothreitol/triisopropylsilane (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5) and precipitated in ice-cold diethyl ether. The resin was washed with acetic acid and water and was subsequently filtered and lyophilized. The peptides were purified with RP-HPLC on a C4 column (Phenomenex Jupiter, 4.6 × 250 mm). The HPLC eluents were (A) 0.1% TFA in water and (B) 0.1% TFA and 80% ACN in water, with a gradient from 25% to 55% B over 60 min, at a flow rate of 4 ml min−1. Dimeric peptides were obtained by air oxidation of the purified peptides (5 mg ml−1) in 20% DMSO/water at 37 °C for 24 h.45 The samples were diluted with water and purified analogously to the monomer peptides. Purity was confirmed by analytical RP-HPLC and ESI-MS measurements.
2.5) and precipitated in ice-cold diethyl ether. The resin was washed with acetic acid and water and was subsequently filtered and lyophilized. The peptides were purified with RP-HPLC on a C4 column (Phenomenex Jupiter, 4.6 × 250 mm). The HPLC eluents were (A) 0.1% TFA in water and (B) 0.1% TFA and 80% ACN in water, with a gradient from 25% to 55% B over 60 min, at a flow rate of 4 ml min−1. Dimeric peptides were obtained by air oxidation of the purified peptides (5 mg ml−1) in 20% DMSO/water at 37 °C for 24 h.45 The samples were diluted with water and purified analogously to the monomer peptides. Purity was confirmed by analytical RP-HPLC and ESI-MS measurements.
Circular dichroism measurements
CD measurements were performed with a Jasco J-815 CD spectrometer. The CD spectra were recorded using a 1 mm thermally jacketed quartz cell, from 250 to 195 nm, at a scan speed of 100 nm min−1 with 10 accumulations. The compounds were dissolved in Na-phosphate buffer (10 mM, pH 6.5), and the peptide concentration was 100 μM and 50 μM for monomeric and dimeric peptides, respectively. For thermal control, a Julabo water thermostat was used with a 10 min equilibration time for each temperature. The solvent baseline was subtracted. The stability of the peptide concentration was monitored via measuring the OD at 280 nM with a Shimadzu UV-1800 spectrophotomer (Fig. S1†).NMR experiments
The NMR spectra were recorded on a Bruker Avance III 600 MHz spectrometer equipped with a 5 mm CP-TCI triple-resonance cryoprobe at 298 and 308 K. The compounds were dissolved in d18-HEPES buffer (20 mM, pH 6.5) containing 10% D2O and 0.02% NaN3. The concentrations of the peptides were 500 μM and 250 μM for monomeric and dimeric peptides, respectively. For referencing, 4,4-dimethyl-4-silapentane-1-sulfonic acid was used as an external standard. Resonance assignment was performed according to the sequential assignment methodology based on 2D homonuclear TOCSY and NOESY. All spectra were acquired with the excitation sculpting solvent suppression pulse scheme with 2048 time domain points and 512 increments. TOCSY measurements were performed with homonuclear Hartman–Hahn transfer with a mixing time of 80 ms, using the DIPSI2 sequence for mixing. The number of scans was 16. The NOESY mixing time was 200 ms and the number of scans was 32. For the assignment of the 13C resonances, the 2D heteronuclear 13C-HSQC spectra were acquired under the same sample conditions as for the homonuclear experiments, but the buffer was prepared in D2O. Processing was performed using Topspin 3.5 (Bruker), a cosine-bell window function, single zero-filling, and automatic baseline correction. The spectra were analyzed using Sparky 3.114 (T. D. Goddard and D. G. Kneller, University of California, San Francisco).The SSP scores were calculated using the SSP software.46 The refDB random coil reference set based on the chemical shifts of properly referenced known protein structures was used. For the dimer structures, the built-in reference set for cysteine residues was replaced with the corresponding values of oxidized cysteine described in the literature.47 The resonances of the β- and D-α-residues were excluded from the calculations because of the lack of reference values. Although the neighborhood effects of the β- and D-α-residues on the adjacent α-residues cannot be ruled out, our earlier observations demonstrated no major systematic influence preventing the chemical shift analysis.35,37
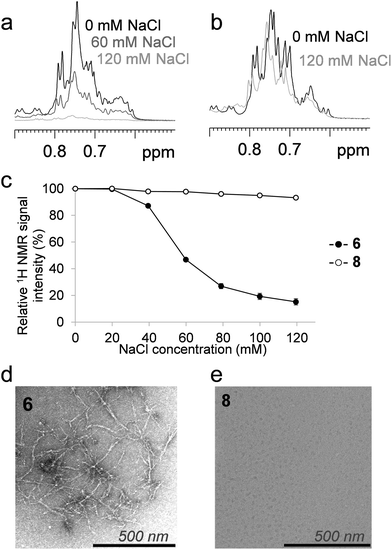
The salt-dependent aggregation of the compounds was tested by measuring the NMR-visible 1H signal intensity at various NaCl concentrations; 6 and 8 were dissolved in the same buffer at a concentration of 250 μM, and the 150 μl samples were titrated with 1 μl aliquots of a concentrated NaCl solution (3.0 M), resulting in a sample series with NaCl concentrations of 20, 40, 60, 80, 100 and 120 mM. The 1D 1H NMR spectra were acquired for all conditions, and the aliphatic regions were integrated. The integrals were corrected for sample dilution and sensitivity loss caused by high salt concentration with the signal attenuation of a reference sample (2 mM glucose).
Transmission electron microscopy
A 250 μM solution of 6 and 8 in 20 mM phosphate buffer (pH 6.5), containing 150 mM NaCl was placed on Formvar-carbon-coated 400-mesh copper grids (Electron Microscopy Sciences, Washington, PA) and stained negatively with uranyl acetate. The aggregates were characterized by TEM on a JEOL JEM-1400 transmission electron microscope (JEOL USA Inc., MA, USA) operating at 120 kV. The images were taken with an EM-15300SXV system and processed with the SightX Viewer software (also JEOL) routinely at a magnification of ×25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.
000.
Results and discussion
β-Sandwich design
Betabellins are 32-residue peptide sequences that are designed de novo to fold into four-stranded β-sheets by virtue of a palindromic pattern with alternating polar and nonpolar side chains. Their structure displays a hydrophobic and a polar face upon folding. The hydrophobic effects between the sheets can be enhanced by the covalent linkage of the betabellin monomers through a disulfide bridge involving the Cys21 residues, which results in 64-residue dimeric β-sandwiches.34 We chose and synthesized betabellin-14 (1) and -15 (2) as templates (Fig. 1a) because they display the best folding properties. Dimeric 2 was shown to form long fibrils via edge-to-edge aggregation and was proposed as a useful model for studying and inhibiting fibril formation.44 This offered a congener β-sandwich protein model for probing the effects of β-amino acid substitutions on stability and edge protection. According to our previous results, cis-(1R,2S)-ACHC (Fig. 1b) residues were adopted well by the β-sandwich, where the cis-backbone arrangement was crucial, according to the stereochemical patterning approach.13,48 Thus, we applied cis-(1R,2S)-ACHC mutations at the center of the two peripheral strands (strand I and IV), at positions 5 and 28. The 32-residue monomeric and 64-residue disulfide-linked dimeric betabellins and their analogs (Fig. 1c) were synthesized and studied.Dimerization-induced β-sheet formation
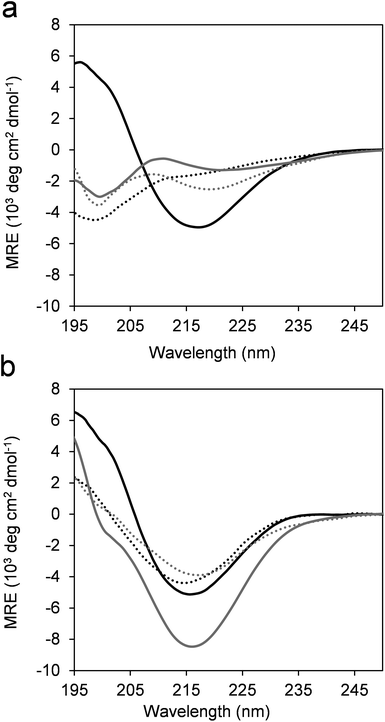
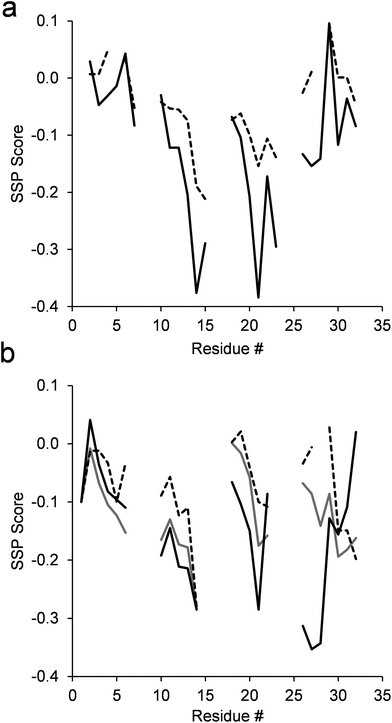
The overall folding propensity of the betabellins and their α/β-peptidic analogs was tested using CD spectroscopy. The β-sheet structure of the parent betabellin peptide 1 was already apparent from the CD curve in its monomeric form (Fig. 2a). In contrast, 2 and the ACHC-substituted analogs 3 and 4 exhibited U-shaped CD curves with a negative band at approximately 199 nm due to the presence of disordered conformations. In accordance with our previous results obtained for the core mutated betabellin-14 derivatives,36 the characteristic negative Cotton effect of the β-sheet appeared at 216 nm for the dimeric compounds 5–8 (Fig. 2b). This indicates dimerization-induced β-sheet folding for template 6 and the edge-substituted derivatives 7 and 8. The formation of 5via dimerization had little effect on the CD intensity, indicating an inherently stable β-sheet. Among the dimeric sequences, 6 displayed the highest intensity at 216 nm, which may indicate an increased β-sheet content and/or an altered overall folding pattern. To test the residue-level folding propensities of the dimeric β-sandwiches, NMR experiments were run.Chemical shift analysis was performed using the 1Hα and 13Cβ resonances (Table S1†). The 13Cα resonances are also sensitive to the secondary structure, but line broadening hindered the signal assignment for all sequences, especially for the flexible terminal residues. Therefore, the 13Cα signals were omitted from the comparative analysis. The chemical shifts were converted into residue-specific secondary structure propensity (SSP) scores for each residue, which estimate the folding tendency at the given positions.46 All sequences exhibited negative SSP scores along the sequence, reflecting the overall β-sheet propensity (Fig. 3). For the template sequence 5, the core strands II and III displayed order, while the terminal strands I and IV folded only partially into regular β-strand structures. Unexpectedly, the edge mutations in 7 had a marked effect on the folding propensity of the core strands. Despite the limited ability of the edge strands to fold into periodic secondary structures, there were long-range effects on the stability of the internal strands, supporting the view that the β-sandwich core made stabilizing shielding contacts with the more solvent-exposed edge strands without their regular folding. A similar pattern, but a less pronounced effect of the edge mutations was observed for 6 and 8. Notably, the core and edge strands displayed a more uniform folding tendency than did 5 and 7, which can explain the different CD fingerprints. For 6 and 8, NOESY contacts were clearly detected between the aromatic and aliphatic side-chains, supporting the formation of the hydrophobic core (Fig. S4†), but the spectral quality did not allow the detailed characterization of the backbone contacts. The temperature-dependent CD measurements (see later) suggested that the β-sandwiches of sequences 6–8 had higher thermodynamic stability at approximately 308 K; therefore, attempts were made to run NMR experiments at elevated temperatures. Successful assignment was only possible for 6 at 308 K (Fig. 3b). We found that the temperature stabilized the C-terminal edge strand and that this effect propagated to the β-sandwich core. This result is in accordance with the propensity of 6 for edge-to-edge aggregation and fibril formation.
The overlaps in the 2D homonuclear NMR spectra (Fig. S2 and 3†) and the residual flexibility of the peptides did not allow atomic-scale resolution in the structural characterization of the compounds.
Thermodynamic and thermal stability of the β-sandwich mimetics
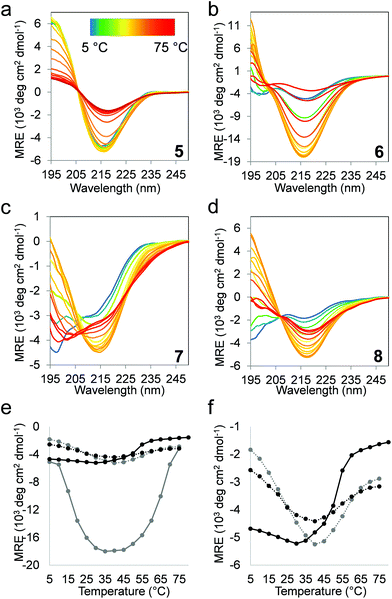
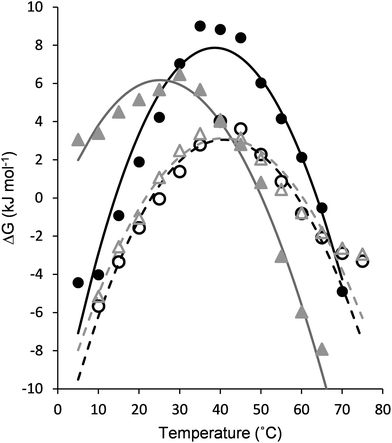
The folding behavior of proteins and protein-mimetic models can be assessed using the thermal denaturation profile. Temperature-dependent CD experiments were performed (Fig. 4a–d), and the β-sheet content (MRE216) was plotted against the temperature (Fig. 4e and f). Structural stability is often described on the basis of only the melting temperature (Tm). A more detailed picture can be obtained upon folding via the analysis of the stability curve, which is the free energy of unfolding as a function of temperature.49 To obtain a set of thermodynamic parameters, the CD-derived temperature-dependent stability data were fit against a theoretical model described by Privalov,50 and Becktel and Schellman.51 In this model, the temperature-independent heat capacity of unfolding (ΔCp), the temperature of the maximum stability (Ts), and the temperature-independent components of enthalpy (ΔHs) and entropy (ΔSs) of unfolding were obtained (Fig. 5 and Table 1). The maximum free energy of unfolding (ΔGs) and the temperatures for cold (Tc) and thermal (Tm) denaturation could also be calculated in the analysis.52| 5 | 6 | 7 | 8 | ||
|---|---|---|---|---|---|
| ΔHs | kJ mol−1 | 10.2 | 11.7 | 8.3 | 7.9 |
| ΔSs | J mol−1 K−1 | 13.6 | 12.2 | 16.3 | 15.5 |
| ΔCp | kJ mol−1 K−1 | 6.0 | 7.9 | 5.2 | 5.9 |
| ΔGs | kJ mol−1 | 6.2 | 7.9 | 3.1 | 3.1 |
| T s | °C | 25.8 | 39.1 | 41.8 | 41.7 |
| T c | °C | 0.7 | 14.1 | 21.6 | 23.0 |
| T m | °C | 50.3 | 63.8 | 60.4 | 59.1 |
The temperature-dependent CD experiments revealed a protein-like denaturation behavior for all β-sandwich mimetics, where both thermal and cold denaturations were observed. This curvature of the stability curve is a result of positive ΔCp, which is strongly connected to the solvent-mediated cooperative unfolding transition via the disruption of a manifold of weak interactions and the solvation of the polar and non-polar groups.53,54 The edge cis-ACHC replacements uniformly decreased the maximum stabilities (ΔGs) for the α/β sequences, but this loss of thermodynamic stability still affords maximum folded populations above 82% for both 7 and 8. On the other hand, the edge cis-ACHC replacements in 5 shifted Ts to a considerably higher temperature for 7, which led to a marked increase in thermal stability and the appearance of cold denaturation at room temperature. Sequences 6 and 8 displayed only a small difference in Ts, whereas room-temperature cold denaturation was also observed for 8. Cold denaturation occurs through the effective solvation of apolar side chains by water at low temperatures. This process can be promoted by the frustrated electrostatic interactions and the increased solvent accessibility of the hydrophobic core (trapped water). The H-bond network and back-bone curvature around the cis-ACHC residue are geometrically distorted,35–37 which certainly increases the solvent accessibility in the folded state. This is in line with the NMR findings of this work. The ΔCp per-residue value for the unfolding of native proteins is in the range 42–84 J mol−1 K−1,55 and it is increased by the percentage of the buried non-polar surface and the satisfied H-bonds in the folded state. Betabellins are designed to have a high ratio of buried non-polar side chains, and thus the parent sequences displayed ΔCp per-residue values of 93.8 and 124.0 J mol−1 for 5 and 6, respectively. The cyclic β-amino acid mutations changed these per-residue-based values to 81.7 and 92.0 J mol−1 for 7 and 8, respectively, which are still native protein-like figures despite their bulging geometry and frustrated H-bonds around the cis-ACHC residues. This finding indicated that the hydrophobic effect is an important stabilizing factor, which is facilitated by an irregular edge region that contacts the core region. Accordingly, the edge strands were able to convey stabilizing contacts to the core, as detected by NMR. A large weight of the hydrophobic effect in the overall stability of the α/β sequences can explain the good thermal stability of 7 and 8. The temperature-independent parts of the enthalpy and entropy values are determined by a complex interplay between the structural features and the enthalpy–entropy compensation that is encoded in the protein sequence itself, which makes the assignment of a single factor to the changes difficult. However, it is notable that ΔSs increased upon cis-ACHC replacements, pointing to an entropically favored unfolding of the α/β sequences, which again can be a sign of extra local rigidity induced by the cyclic β-residues with a possible contribution by the trapped water in the folded state. However, the less stable H-bonds in the folded states of 7 and 8 might be reflected in their less endothermic unfolding compared to 5 and 6. These β-residue-induced changes in the thermodynamics of β-sandwich folding are the opposite of those observed for protein helix segments.49
Stability against edge-to-edge aggregation
We tested the ability of the altered terminal strand geometry to inhibit salt-induced edge-to-edge aggregation and fibril formation, using 6 and 8 as models.44 Sequences 5 and 7 did not form fibrils under the conditions tested. The above-mentioned investigations were performed under NaCl-free conditions to maintain 6 in the monomeric form and to facilitate the detection of stability changes upon α → β substitutions. Diffusion-ordered NMR spectroscopy measurements under the low-salt conditions recorded for 6 and 8 did not indicate any significant differences in the sizes or shapes of the NMR-visible fraction of the peptides (Fig. S5†). The presence of NaCl scaled up the hydrophobic effects, which acted both as a β-sandwich stabilizing factor and a promoter of aggregation in the following experiments. At elevated salt concentrations (above 40 mM NaCl), a significant intensity loss was observed in the NMR spectra of 6 (Fig. 6a), likely due to the immediate formation of high molecular weight associates with extreme short transversal relaxation. In contrast, the NMR-visible fraction of the α/β-analog 8 did not display a marked decrease, even at physiological salt concentration (Fig. 6b). These results indicated that the aggregation of the α/β sequence is not induced at near-physiological salt concentrations. At a NaCl concentration of 120 mM, only 15% of the parent fibril-forming betabellin 6 remained in the NMR-visible form, in contrast with the 93% of 8 (Fig. 6c). In terms of the stability against aggregation at physiological NaCl concentrations, the ACHC-substituted analog 8 proved to be more stable than 6. To gain support for an edge-to-edge periodic association, TEM images were recorded for the high-salt content samples. In line with the literature results, NaCl triggered the edge-to-edge interactions of 6, and long thin fibrils were formed with an approximate size of 3.5 nm × 400 nm (Fig. 6d), while the electron microscopy investigation of the α/β-analog 8 revealed no aggregates in the sample (Fig. 6e).Conclusions
We successfully constructed peripheral cis-ACHC mutants of the de novo designed β-sandwich protein mimetics and their parent sequences. As revealed by the CD and NMR spectroscopy results, all 64-residue dimers exhibited high β-sheet content, facilitating the thermodynamic characterization of their structural stabilities. Residue-level investigations revealed that the β-sandwich templates studied exhibited different structural patterns. While 5 tended to stabilize through the formation of an ordered core strand region with shielding contacts from the irregular peripheral strands, 6 gained stability from the temperature-induced order of an edge strand. The cis-ACHC replacements promoted irregularity at the peripheral strands, which decreased the thermodynamic stability in terms of the free energy of unfolding, and cold denaturation was observed for both β-sandwich mimetic models. Importantly, the effects on thermal stabilities were not uniform. Depending on the parent sequence, the α → β mutations either increased or maintained the temperatures of the maximum stability, which were observed at approximately 40 °C, not far from physiological temperatures. In terms of sensitivity to aggregation, the irregular peripheral strands of 8 were effective as edge protection and successfully tuned down the unfavorable aggregation propensity of 6 at physiological salt concentration. Edge protection was achieved without interfering with the side-chain orientation, the electrostatic pattern of the H-bond pillars or the net charge. Overall, we conclude that cis-ACHC replacements at open edges can maintain sufficient thermodynamic stability to facilitate a predominant β-sandwich tertiary structure, provide favorable thermal stability profiles and offer an effective control over unwanted aggregation, with the potential benefit of contributing to protease resistance.56,57Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Hungarian Academy of Sciences, Lendület program (LP-2011-009) and GINOP-2.3.3-15- 2016-00010. E. W. thanks the Postdoctoral Fellowship Program 2014 of the Hungarian Academy of Sciences.References
- W. F. DeGrado, C. M. Summa, V. Pavone, F. Nastri and A. Lombardi, Annu. Rev. Biochem., 1999, 68, 779 CrossRef PubMed.
- M. G. Ryadnov and D. N. Woolfson, Nat. Mater., 2003, 2, 329 CrossRef PubMed.
- J. Kaplan and W. F. DeGrado, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 11566 CrossRef PubMed.
- A. L. Boyle and D. N. Woolfson, Chem. Soc. Rev., 2011, 40, 4295 RSC.
- D. J. Hill, M. J. Mio, R. B. Prince, T. S. Hughes and J. S. Moore, Chem. Rev., 2001, 101, 3893 CrossRef PubMed.
- C. M. Goodman, S. Choi, S. Shandler and W. F. DeGrado, Nat. Chem. Biol., 2007, 3, 252 CrossRef PubMed.
- D. Seebach, K. Beck Albert and J. Bierbaum Daniel, Chem. Biodiversity, 2004, 1, 1111 CrossRef PubMed.
- D. Seebach and J. Gardiner, Acc. Chem. Res., 2008, 41, 1366 CrossRef PubMed.
- J. A. Kritzer, J. D. Lear, M. E. Hodsdon and A. Schepartz, J. Am. Chem. Soc., 2004, 126, 9468 CrossRef PubMed.
- J. A. Kritzer, O. M. Stephens, D. A. Guarracino, S. K. Reznik and A. Schepartz, Bioorg. Med. Chem., 2005, 13, 11 CrossRef PubMed.
- M. Pelay-Gimeno, A. Glas, O. Koch and T. N. Grossmann, Angew. Chem., Int. Ed., 2015, 54, 8896 CrossRef PubMed.
- J. P. Plante, T. Burnley, B. Malkova, M. E. Webb, S. L. Warriner, T. A. Edwards and A. J. Wilson, Chem. Commun., 2009, 5091 RSC.
- I. M. Mandity, E. Weber, T. A. Martinek, G. Olajos, G. K. Toth, E. Vass and F. Fulop, Angew. Chem., Int. Ed., 2009, 48, 2171 CrossRef PubMed.
- T. A. Martinek and F. Fulop, Chem. Soc. Rev., 2012, 41, 687 RSC.
- L. M. Johnson and S. H. Gellman, Methods Enzymol., 2013, 523, 407 Search PubMed.
- B. B. Lao, K. Drew, D. A. Guarracino, T. F. Brewer, D. W. Heindel, R. Bonneau and P. S. Arora, J. Am. Chem. Soc., 2014, 136, 7877 CrossRef PubMed.
- C. M. Grison, J. A. Miles, S. Robin, A. J. Wilson and D. J. Aitken, Angew. Chem., Int. Ed., 2016, 55, 11096 CrossRef PubMed.
- D. S. Daniels, E. J. Petersson, J. X. Qiu and A. Schepartz, J. Am. Chem. Soc., 2007, 129, 1532 CrossRef PubMed.
- P. S. P. Wang and A. Schepartz, Chem. Commun., 2016, 52, 7420 RSC.
- C. M. Lombardo, G. W. Collie, K. Pulka-Ziach, F. Rosu, V. Gabelica, C. D. Mackereth and G. Guichard, J. Am. Chem. Soc., 2016, 138, 10522 CrossRef PubMed.
- S. H. Gellman, Curr. Opin. Chem. Biol., 1998, 2, 717 CrossRef PubMed.
- R. Guerois and M. López de la Paz, Protein design : methods and applications, Humana Press, Totowa, NJ, 2006 Search PubMed.
- M. Ramírez-Alvarado, T. Kortemme, F. J. Blanco and L. Serrano, Bioorg. Med. Chem., 1999, 7, 93 CrossRef.
- K. H. Mayo, E. Ilyina and H. Park, Protein Sci., 1996, 5, 1301 CrossRef PubMed.
- M. S. Searle, Pept. Sci., 2004, 76, 185 CrossRef PubMed.
- J. F. Espinosa, F. A. Syud and S. H. Gellman, Protein Sci., 2002, 11, 1492 CrossRef PubMed.
- H. E. Stanger and S. H. Gellman, J. Am. Chem. Soc., 1998, 120, 4236 CrossRef.
- G. A. Lengyel, R. C. Frank and W. S. Horne, J. Am. Chem. Soc., 2011, 133, 4246 CrossRef PubMed.
- G. A. Lengyel and W. S. Horne, J. Am. Chem. Soc., 2012, 134, 15906 CrossRef PubMed.
- J. Chatterjee, F. Rechenmacher and H. Kessler, Angew. Chem., Int. Ed., 2013, 52, 254 CrossRef PubMed.
- R. Spencer, K. H. Chen, G. Manuel and J. S. Nowick, Eur. J. Org. Chem., 2013, 2013, 3523 CrossRef.
- G. A. Lengyel, Z. E. Reinert, B. D. Griffith and W. S. Horne, Org. Biomol. Chem., 2014, 12, 5375 RSC.
- G. A. Lengyel, G. A. Eddinger and W. S. Horne, Org. Lett., 2013, 15, 944 CrossRef PubMed.
- Y. Yan and B. W. Erickson, Protein Sci., 1994, 3, 1069 CrossRef PubMed.
- Z. Hegedus, E. Weber, E. Kriston-Pal, I. Makra, A. Czibula, E. Monostori and T. A. Martinek, J. Am. Chem. Soc., 2013, 135, 16578 CrossRef PubMed.
- G. Olajos, A. Hetenyi, E. Weber, L. J. Nemeth, Z. Szakonyi, F. Fulop and T. A. Martinek, Chemistry, 2015, 21, 6173 CrossRef PubMed.
- Z. Hegedus, I. Makra, N. Imre, A. Hetenyi, I. M. Mandity, E. Monostori and T. A. Martinek, Chem. Commun., 2016, 52, 1891 RSC.
- J. M. Shifman, Structure, 2008, 16, 1751 CrossRef PubMed.
- W. Wang, Int. J. Pharm., 2005, 289, 1 CrossRef PubMed.
- J. S. Richardson and D. C. Richardson, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 2754 CrossRef PubMed.
- M. de Rosa, F. Bemporad, S. Pellegrino, F. Chiti, M. Bolognesi and S. Ricagno, FEBS J., 2014, 281, 4072 CrossRef PubMed.
- P. Craveur, A. P. Joseph, J. Rebehmed and A. G. de Brevern, Protein Sci., 2013, 22, 1366 Search PubMed.
- W. Wang and M. H. Hecht, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 2760 CrossRef PubMed.
- A. Lim, M. J. Saderholm, A. M. Makhov, M. Kroll, Y. Yan, L. Perera, J. D. Griffith and B. W. Erickson, Protein Sci., 1998, 7, 1545 CrossRef PubMed.
- J. P. W. Tam, C. R. Wu, W. Liu and J. W. Zhang, Peptides: Chemistry and biology, in A highly selective and effective reagent for disulfide bond formation in peptide synthesisa and protein folding, ESCOM Science Publishers, Leiden, The Netherlands, 1992 Search PubMed.
- J. A. Marsh, V. K. Singh, Z. Jia and J. D. Forman-Kay, Protein Sci., 2006, 15, 2795 CrossRef PubMed.
- H. Zhang, S. Neal and D. S. Wishart, J. Biomol. NMR, 2003, 25, 173 CrossRef PubMed.
- L. Berlicki, L. Pilsl, E. Weber, I. M. Mandity, C. Cabrele, T. A. Martinek, F. Fulop and O. Reiser, Angew. Chem., Int. Ed., 2012, 51, 2208 CrossRef PubMed.
- Z. E. Reinert and W. S. Horne, Chem. Sci., 2014, 5, 3325 RSC.
- P. L. Privalov, Thermochim. Acta, 1990, 163, 33 CrossRef.
- W. J. Becktel and J. A. Schellman, Biopolymers, 1987, 26, 1859 CrossRef PubMed.
- D. Sanfelice, E. Morandi, A. Pastore, N. Niccolai and P. A. Temussi, ChemPhysChem, 2015, 16, 3599 CrossRef PubMed.
- A. Cooper, Biophys. Chem., 2005, 115, 89 CrossRef PubMed.
- A. Cooper, Biophys. Chem., 2000, 85, 25 CrossRef PubMed.
- C. Ganesh, N. Eswar, S. Srivastava, C. Ramakrishnan and R. Varadarajan, FEBS Lett., 1999, 454, 31 CrossRef PubMed.
- J. Frackenpohl, P. I. Arvidsson, J. V. Schreiber and D. Seebach, ChemBioChem, 2001, 2, 445 CrossRef PubMed.
- D. F. Hook, P. Bindschadler, Y. R. Mahajan, R. Sebesta, P. Kast and D. Seebach, Chem. Biodiversity, 2005, 2, 591 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/C8OB01322E |
| This journal is © The Royal Society of Chemistry 2018 |