Cyclic azole-homologated peptides from Marine sponges
Tadeusz F.
Molinski

Department of Chemistry and Biochemistry, and Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, 9500 Gilman Dr 0358, La Jolla, California 92093, USA. E-mail: tmolinski@ucsd.edu
First published on 28th November 2017
Abstract
This review discusses the chemistry of cyclic azole-homologated peptides (AHPs) from the marine sponges, Theonella swinhoei, other Theonella species, Calyx spp. and Plakina jamaicensis. The origin, distribution of AHPs and molecular structure elucidations of AHPs are described followed by their biosynthesis, bioactivity, and synthetic efforts towards their total synthesis. Reports of partial and total synthesis of AHPs extend beyond peptide coupling reactions and include creative construction of the non-proteinogenic amino acid components, mainly the homologated heteroaromatic and α-keto-β-amino acids. A useful conclusion is drawn regarding AHPs: despite their rarity, exotic structures and the potent protease inhibitory properties of some members, their synthesis is under-developed and beckons solutions for outstanding problems towards their efficient assembly.
Introduction
During the 1970s and 1980s – two decades that embraced the ‘log phase’ of discovery of marine natural products – attention was mainly given to non-polar, lipophilic compounds including terpenoids, polyketides, lipids – particularly polyhalogenated compounds from red algae. Relatively few peptides were described from marine organisms in this period. For example, although in 1985 the tally of marine natural products, listed by the late D. John Faulkner in the first three of his comprehensive reviews on the subject, was about 1700,1 fewer than 200 of these were peptides and amino acid (aa) acylates.2 By the end of 2015, the RSC database MarinLit showed the inventory had grown to 1450 peptides among a total of 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 255 marine natural products.3†
255 marine natural products.3†
Structural diversity of Marine-derived peptides
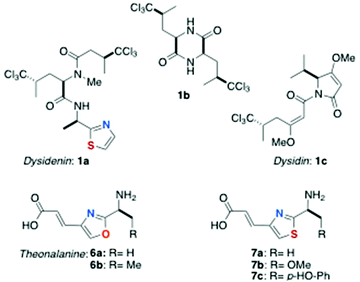
The structures of most peptides from marine organisms are cyclic; consequently they lack exposed N- and C-termini and are relatively non-polar; a property that allows their facile recovery and purification beginning with solvent partitioning.‡ A notable characteristic of marine-derived peptides is the frequent presence of highly modified aa residues, many of them in the non-proteinogenic D-configuration. For example, two unusual metabolites from Dysidea herbacea, (−)-dysidenin (1a)4a and diketopiperazine 1b (Fig. 1) reported in the late 1970s by Kazlauskas and coworkers from an Australian specimen, are the first examples of L-5,5,5-trichloroleucine (TCL) containing peptide natural products. The structure of (+)-dysidin (1c), elucidated by Hofheinz and Oberhänsli, revealed a trichloro-metabolite suggestive of biosynthetic catabolism of TCL to (S)-4,4,4-trichloroisovaleric acid and its homologation by one malonate unit.4c Peptide 1a also displays the cysteine-derived D-2-(2-thiazolyl)alanine. Subsequent reports have expanded the list of oxazole- and thiazole-containing peptides from marine organisms, including a special subclass: the azole-homologated peptides (AHPs) containing azole-homologated amino acid residues (AHAs). The chemistry of AHPs is the subject of this review.Origin and classification of azole-homologated peptides
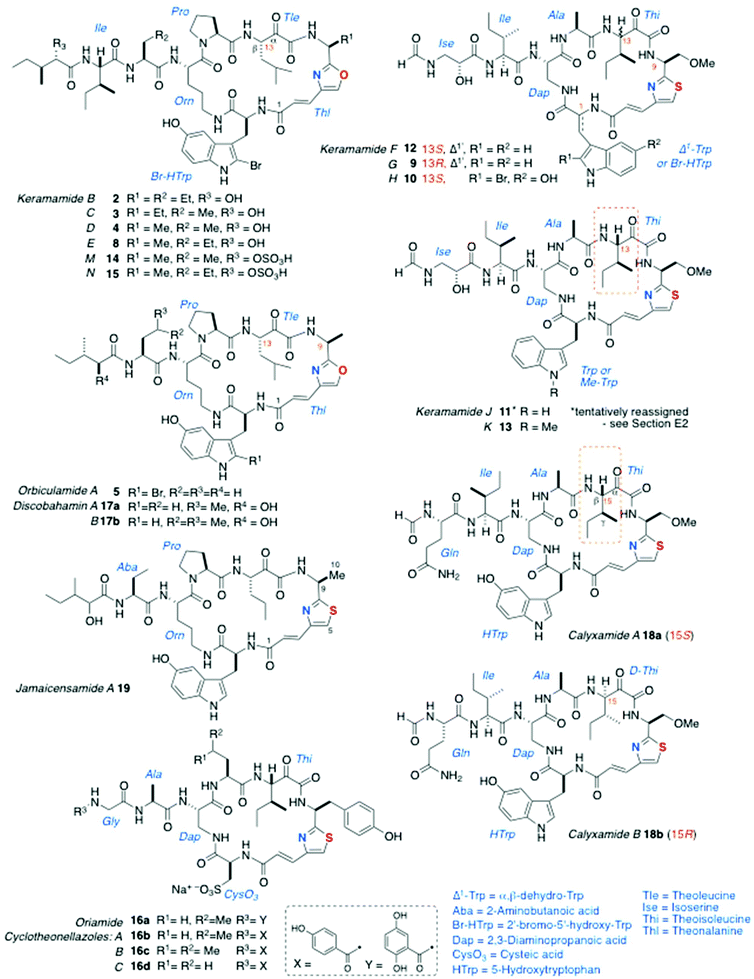
Cyclic AHPs are biosynthesised by non-ribosomal synthase/polyketide synthase (NRPS-PKS) and appear to be limited to a few genera of marine sponges. Structurally, they can be classified as Type I or Type 2, based on the presence of conserved tetrads of aa residues within the macrocyclic ring. Peptides of Type 1 contain an oxazole-homologated Ala, Aba, or O-Me-Ser (for the single exception, vide infra), ring-closed by a tetrad of conserved aa residues: in sequence, an α-keto-β-amino-alkanoic acid (mostly originating from Leu or Ile), Pro, Orn, and Trp (or 5-HO-Trp or a variant, thereof). In Type 2 peptides, the first two residues are retained, but Pro and Orn are replaced consistently by Ala and 2,3-diaminopropanoic acid (Dap), respectively. In all AHPs, the α-amino group of the Orn or Dap residue is extended through an amide bond to one or two additional aa residues, and the terminal aa is N-acylated by a short-chain carboxylic acid.The structures of keramamides B–D (2–4)5 and orbiculamide A (5)6 (Fig. 2) from the marine sponge Theonella swinhoei, representing the first Type 1 AHPs, were published ‘back-to-back’ in 1991 by the groups of Kobayashi and Fusetani, respectively. In the structures of 2–5, the heteroaromatic unit is oxazole-homologated L-Ala – so-called ‘theonalanine’ (6a)6 or its homolog 6b – and an α-keto-β-amino acid derived from isoleucine: so-called ‘theoleucine’.6 All other AHPs display one of the thiazole-containing residues 7a–c. Most AHPs described to date were isolated from T. swinhoei (which is also a prolific producer of polyketides, e.g. swinholide A7), but a few were found in Calyx spp. and one from Discodermia jamaicensis.
Subsequent to the reports of 2–5, additional AHPs in the keramamide series from T. swinhoei were disclosed: (in approximate chronological order) keramamides E (8), G (9), H (10) and J (11),8 F (12),9 (F, G, H and J contain the rare D-isoserine residue), K (13),10 and M (14) and N (15), the O-sulphate esters of 4 and 8, respectively.11§
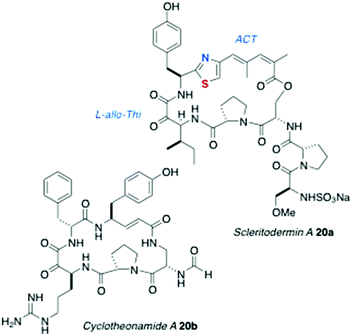
The structure of oriamide (16a), from a South African Theonella aff. swinhoei, is closest to a Type 2 AHP with exceptions to the rule: the Trp-derived residue is replaced by cysteic acid, the Ala-derived AHA is replaced by one derived from Tyr, and the N-terminal group is a 2,5-dihydroxybenzamide.13 The homologs discobahamins A and B (17a,b)14 and diastereomeric calyxamides A and B (18a,b)15 were isolated from Atlantic and Pacific Discodermia species, respectively. Most recently, jamaicensamide A (19) – so far, the only thiazole-homologated Type 1 AHP – was discovered in the rare Bahamian sponge Plakina jamaicensis.16 Cycloneothellazoles A–C (16b–d) are p-hydroxybenzamide analogs of 16a from the same sponge that delivered the latter.17 Finally, of related interest, the depsipeptide scleritodermin A (20a, Fig. 3) from the sponge Scleritoderma nodosum,12 mostly resembles AHPs 16a–d: 20a contains a thiazole-double homologated Tyr (‘ACT’) linked to a diastereomeric α-keto-β-amino aa (L-keto-allo-isoleucine, or L-allo-Thi).
Biological activity
Compounds 2, 5, 8–18 have been reported along with biological activity, mostly modest levels of cytotoxicity against cultured murine cancer cells (e.g. P388, L1210). Keramamides B–D (2–4) inhibit superoxide generation (∼30–50 nM) by human neutrophils stimulated with a chemotactic peptide.5 Of outstanding interest is the potent protease activities of cyclotheonellazoles A–C (16b–d): all were shown to be nanomolar inhibitors of chymotrypsin and subnanomolar inhibitors of elastase.17 The thrombin inhibitory activity of another peptide, cyclotheonamide A (20b, IC50 ∼100 nM) from T. swinhoei has been more extensively studied,20 and suggests a common link between the four compounds and protease activity: the α-keto-β-amino acid residue appears to be a transition state mimic within the hydrolytic active site of the enzymes. The protease activity of other AHPs – all of which bear the same or homologous aa residue – is under-investigated, but a worthy subject and opportunity for future investigations.Biosynthesis
Comprehensive studies of molecular genetics and biochemistry of the prolific sponge T. swinhoei by Piel and coworkers using late-generation genetic tools,18 revealed the origin of many of its peptide metabolites.19 A single symbiotic γ-proteobacterium, ‘Entotheonella sp.’ that lives within the tissue of T. swinhoei is the ‘talented producer’ responsible for biosynthesis of virtually all peptides associated with this sponge, including the keramamides5,8–12 and other unrelated peptides.19¶ For example, genetic evidence shows the CthA2 and KerA5 genes of Entotheonella sp. shows high specificity for adenylation of diaminopropanoic acid (Dap) in cyclotheonamide (and also 16a–d, 18a,b) and Leu, respectively, which is transformed to theoleucine in about half the keramamides (9–13).19 The 13R configuration of some AHPs (e.g.9 and 18b) is unexpected as the gene cluster lacks an epimerase domain,19 however, it is known this centre is prone to spontaneous epimerisation (vide infra).Synthetic studies
Remarkably, few synthetic studies on AHPs have been reported and only one total synthesis, that of a stereoisomer of the keramamide J (11) that questions the configuration of the natural product. The lion's share of the work has been design and synthesis of the non-proteinogenic aa residues; particularly the α-keto-β-amino acids and the AHA residues. Amide bond couplings follow standard protocols but strategic choices of which amide bond to close during macrolactamization is critical to any macrocyclic peptide synthesis.Approach to keramamide B (2): Shioiri and Hughes
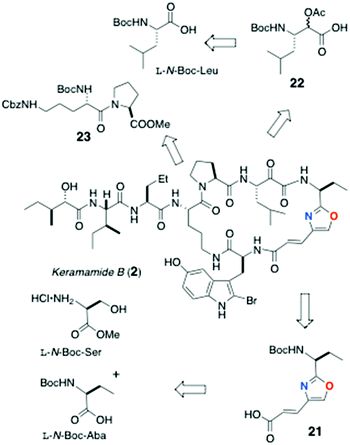
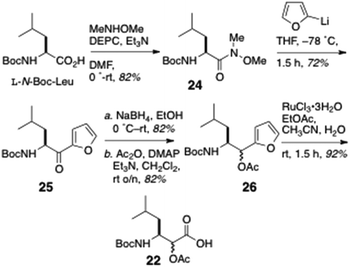
The partial synthesis of keramamide B comprising the preparation of the oligopeptide fragments 21, 22 and 23 was described by Shioiri and Hughes (Scheme 1).22 The protected oxazole-homologated aa residue 21 was derived from L-N-Boc-Ser and N-Boc-2-aminobutanoic acid (N-Boc-Aba), while the protected α-keto-β-amino acid residue (carried as its reduced α-acetoxy acid analog, 22) was elaborated from a 2-substituted furan derived from L-N-Boc-Leu. The last fragment prepared was the differentially N-protected L-Orn-Pro-OMe dipeptide 23.In the forward direction (Scheme 2), L-N-Boc-Leu was converted to the Weinreb amide 24 under standard conditions, then transformed to the 2-furylketone 25 (2-lithiofuran, −78 °C). Reduction of 25 (NaBH4) followed by O-acetylation gave an inconsequential mixture of α-acetoxy esters (26; the stereocentre would be removed after hydrolysis-oxidation). Perruthenate oxidation of 26 gave the free carboxylic acid 22.
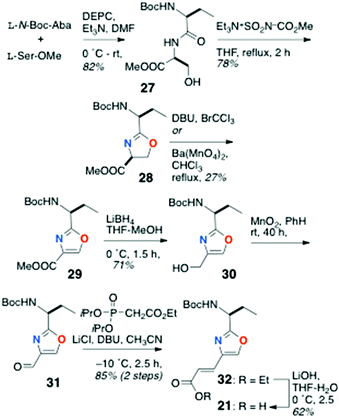
Preparation of the oxazole-homologated aa residue 21 (Scheme 3) started with amide bond formation between L-N-Boc-Aba and L-Ser methyl ester followed by cyclodehydration of the dipeptide product 27 with the Burgess reagent to oxazoline 28 which was subsequently oxidized to afford oxazole 29 in low yield (27%).
Reduction of the ester group of 29 (LiBH4) to primary alcohol 30, followed by oxidation with activated MnO2, delivered aldehyde 31 which was subjected to Horner–Wadsworth–Emmons olefination under Masamune-Roush conditions to provide the protected ethyl ester 32 in good yield: saponification of the latter delivered 21.
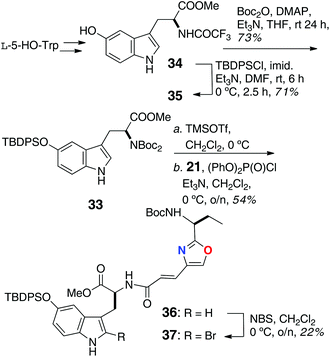
The remaining peptide fragments were assembled from common L-amino acids and condensed, mostly, using the coupling reagent diethylcyanophosphoridate (DEPC; Schemes 4 and 5). The N,N,O-protected 2-bromo-5-hydroxytryptophan (33) was elaborated as shown in Scheme 4 from L-HO-Trp, through intermediates 34 and 35, was coupled [(PhO)2P(O)Cl] with oxazole-containing aa 21 to dipeptide 36. Bromination of the latter at C-2 of Trp (NBS) gave 37 in low yield (22%).
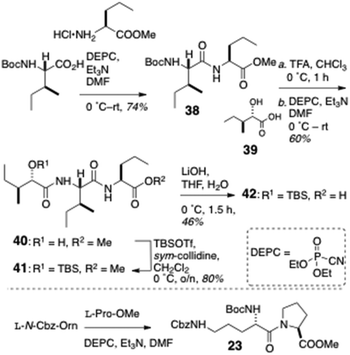
Condensation of L-N-Boc-Ile with L-methyl 2-aminopentanoate hydrochloride (Scheme 5) gave dipeptide 38 which – after removal of the Boc group – was coupled to L-isoleucic acid (39) to deliver dipeptide acylate 40. O-Protection of 40 (TBSOTf, sym-collidine) gave methyl ester 41: saponification of the latter delivered protected dipeptide 42. Finally, dipeptide L-(Nα-Boc-Nε-Cbz)-Orn-L-Pro-OMe (23) was accessed through DEPC coupling of the respective Orn and Pro precursors.
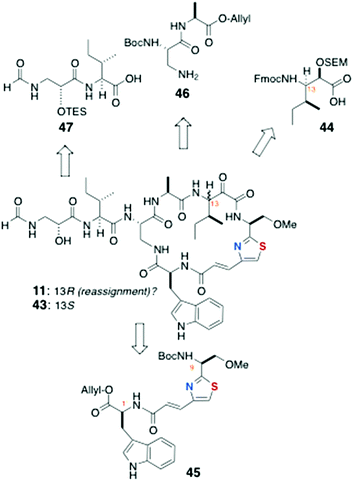
13-epi-Keramamide J (43): Sowinski and Toogood
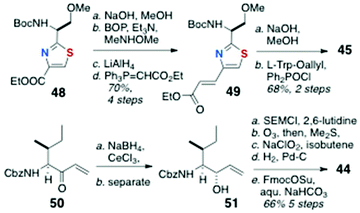
The total synthesis of 43, the C-13 epimer of keramamide J (11), was achieved by Toogood and coworkers using a different approach to both the AHP and the α-keto-β-amino aa residues and order of assembly of the cyclic peptide (Scheme 6) from the defined fragments 44–47.23 In the event, it was found the product, although bearing the constitution and stereochemistry assigned by Kobayashi to keramamide J,8 was not identical with the natural product raising questions about the configuration of C-13 and drawing to the conclusion that reassignment of the latter is necessary (vide infra).Preparation of the thiazole-homologated aa (Scheme 7) began with the known thiazole-2-carboxylate ethyl ester 48, derived earlier by the authors in high enantiomeric purity from N-Tr-O-methylserine thioamide via a modified Hantsch synthesis.24 After saponification of 48, coupling of the liberated carboxylic acid with MeNHOMe (BOP, Et3N) gave Weinreb amide which was reduced to the corresponding aldehyde (LiAlH4) and extended by Wittig olefination to conjugated thiazole 49. The latter was saponified and coupled to L-Trp-OAllyl [Ph2P(O)Cl] to provide dipeptide 45 (68%, 2 steps).
 | ||
| Scheme 7 13-epi-Keramamide J: Preparation of protected α-keto-β-amino acid 44 and thiazole-homologated aa 45. | ||
The key strategy for carrying forward the sensitive α-keto-β-amino aa residue of 11 was masking the latter as an SEM protected α-hydroxyester 44 (Scheme 7), and unmasking, at a late stage, by hydrolysis-oxidation. Vinyl ketone 50, prepared from N-Cbz-L-Ile by vinylation of the corresponding Weinreb amide, was reduced under Luche conditions (NaBH4, CeCl3) to a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of allylic alcohols, one of which, 51, was separated and carried forward through five steps: SEM protection, ozonolysis to the aldehyde, Pinnick oxidation, and N-protecting group interchange (NHCbz to NHFmoc) to deliver 44.
1 mixture of allylic alcohols, one of which, 51, was separated and carried forward through five steps: SEM protection, ozonolysis to the aldehyde, Pinnick oxidation, and N-protecting group interchange (NHCbz to NHFmoc) to deliver 44.
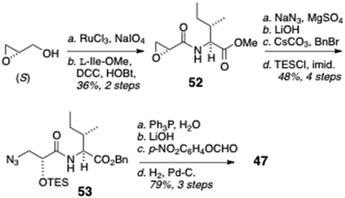
The N-formyl peptide side chain (Scheme 8) was assembled in a straightforward manner starting with commercially available (S)-glycidol (Scheme 8). Perruthenate oxidation of the primary alcohol, followed by amide bond coupling with L-Ile methyl ester (DCC, HOBt), gave the acylated amino ester 52 that underwent regiospecific epoxide opening by azide in the presence of a mild Lewis acid (NaN3, MgSO4) to provide an azide intermediate that was transformed by ester interchange (OMe to OBn) and protection of the secondary OH group (TESCl, imidazole) to give azido benzyl ester 53. Staudinger reduction of 53 (Ph3P, H2O) followed by saponification of the COOBn group and formylation of the liberated primary amine (p-NO2C6H4OCHO) gave 47.
Assembly of the peptide bonds and macrocyclisation (Scheme 9) began with removal of the Boc group from 45 (HCl, Et2O) and peptide coupling with 44 (DCC, HOBt, Hünig's base), followed by removal of the allyl group (Pd(Ph3P)4, dimedone) and coupling with dipeptide 46 to deliver the cyclisation precursor 54 (56% over four steps).
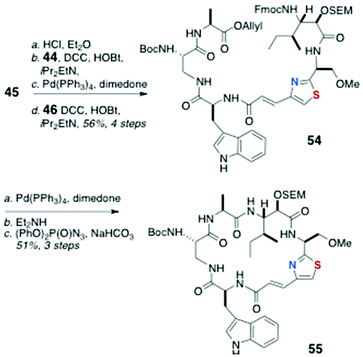 | ||
| Scheme 9 13-epi-Keramamide J: Fragment assembly by amide bond coupling-macrocyclisation to cyclic peptide 55. | ||
After removal of both N and C terminal protecting groups of 54 in separate steps, a dilute solution of the product was cyclised with diphenylphosphoryl azide [(PhO)2P(O)N3, NaHCO3] to afford the cyclic peptide 55 in 51% yield over three steps.
Completion of the synthesis required attachment of the N-formyl dipeptidyl chain 47 to the core macrocycle (Scheme 10). Treatment of 55 with mineral acid (HCl, MeOH) simultaneously removed both the Boc and SEM protecting groups, which allowed coupling of the amine product with fragment 47 to give 56, the completed carbon framework of the target molecule.
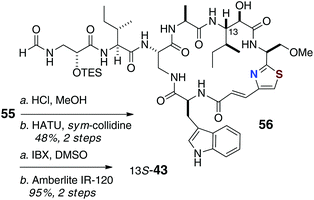 | ||
| Scheme 10 13-epi-Keramamide J: Final assembly (deprotection, oxidation) of 13-epi-keramamide B (43). | ||
Oxidation of the C-13 hydroxyl group in 56 (IBX, DMSO) followed by removal of the TES group in the presence of strong cation-exchange resin (Amberlite IR-120) gave a product 43 that, although of the same molecular mass as isolated keramamide J (11), was different by NMR.
From a thorough comparison of the NMR and chiroptical properties of the synthetic product, 13-epi-keramamide J (43), and the highly similar 13S keramamide F (12), the authors made a strong case that their end product was the targeted 13S epimer, the structure assigned by Kobayashi,8 but the natural product appears to be 13R (13-epi-keramamide J). It's possible that free 13S-Ile arose during oxidative degradation of natural 13R-keramamide J by epimerisation of the labile α-centre. The authors also noted, as did Fusetani with orbiculamide A (5),6 that partial C-13 epimerisation of synthetic 43 occurs under the alkaline hydrogen peroxide conditions, or even spontaneously upon standing in solution.23
Other synthetic studies
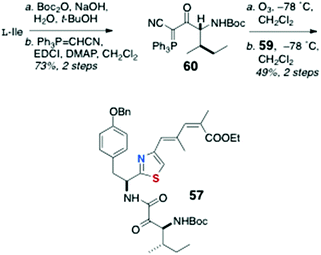
As mentioned earlier, little has been published on the total synthesis of AHPs, but key pilot studies show the way forward. Synthesis of peptide components for assembly of keramamide F (12)9 were reported by Sowinski and Toogood,25 based largely on their successful synthesis of 48 (see Scheme 7) and a new method for preparation of Δ1-Trp.26Scleritodermin A (20a, Fig. 3),12 which resembles 16d,13,17 has been the subject of one report by Serra and coworkers who described the synthesis of the component aa units and assembly of the constituent dipeptide 57 (Scheme 11).27 Efficient preparation of thiazole-homologated Tyr, 58, starting with L-p-benzyloxy-Phe, was achieved using Wipf's oxazoline-thiazoline interchange (H2S, Et3N, MeOH),28 followed by oxidation-elimination (DAST, DBU, BrCCl3). Conversion of the COOMe group of 58 to conjugated thiazole 59 was effected via the corresponding aldehydes through iterative Wittig-type olefination reactions. Finally, L-Ile was converted to the α-cyano phosphorane 60 (Scheme 12) which, after ozonolysis, was condensed with 59 using the Wasserman protocol29 to give ‘theoisoleucine’ dipeptide 57 in 49% yield.
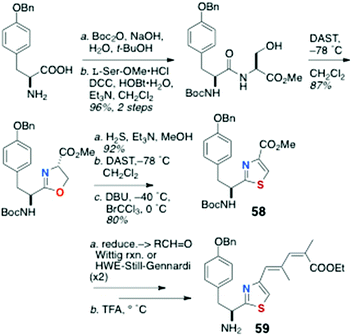 | ||
| Scheme 11 Preparation of thiazole-homologated ‘theoisoleucine’ dipeptide 59 of scleritodermin A (20a). | ||
This approach cleverly avoids the masked α-hydroxycarboxamide derivatives of the Shioiri and Toogood approaches and attendant problems of oxidation-epimerisation and gives the dipeptide in fewer steps.
Conclusions
The marine sponge-derived azole-homologated peptides (AHPs) – based on a macrocyclic core of five aa residues, four of them non-proteinogenic – are remarkable in terms of structural complexity and potential for discovery of inhibitors of serine proteases [e.g. elastase inhibitors, cyclotheonellazoles (16b–c).17Recent advances in metagenomic analysis have identified some of the genes responsible for keramamide biosynthesis in sponges: biosynthesis of these and other unrelated peptides are attributed to a single symbiotic γ-proteobacterium, ‘Entotheonella’, which lives interstitially within the sponge tissue.19
Published approaches to syntheses of AHPs have been few, and only one total synthesis – that of 13-epi-keramamide J (43) – have appeared. Nevertheless, the AHPs have inspired significant advances and creative endeavours towards the total syntheses of keramamide B (2), keramamide J (11) and the ‘hybrid’ peptide, scleritodermin A (20a). No doubt, design and execution of efficient targeted total synthetic schemes towards procurement of useful amounts of sample will evolve hand-in-hand with exploration of their biological properties.
Finally, a comment on stereo-assignment of AHPs is timely. To date, the aa residues in the macrocyclic cores of AHPs have been found to be L-configured with the exception of the α-keto-β-amino acid which appears to exhibit stereochemical plasticity. While mostly L-configured, this residue occasionally occurs in the D-configuration. Given its propensity for epimerisation (vide supra), the relatively acidic β-CH may be responsible for the 13R configuration in ‘keto-L-Ile’ assigned to keramamide G (9) and possibly keramamide J (11),8 but this alone does not account for an outstanding anomaly. Inversion of C-13 from L- to D- would give a ‘keto-allo-D-Ile’ residue that, upon oxidative-cleavage and acid hydrolysis, would yield the diastereomeric allo-D-Ile.|| Equivalent residue in calyxamides A and B (18a,b)15 is a more surprising finding: the two peptides have antipodal ‘keto-Ile’ residues corresponding to L-Ile and D-Ile, which implies both α- and β-stereocentres are inverted in 18b. Finally, the corresponding residue in scleritodermin A (20a)12 is replaced by ‘keto-L-allo-Ile’ that is inverted only at the β-stereocentre with respect to proteinogenic L-Ile; a rare, but not unprecedented finding. Findings of new AHPs should go hand-in-hand with critical stereochemical evaluation of α-keto-β-amino acid residue.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The author is grateful for funding support from the National Institutes of Health (AI100776).References
-
(a) D. J. Faulkner, Nat. Prod. Rep., 1984, 1, 251 RSC
; D. J. Faulkner, Nat. Prod. Rep., 1984, 1, 551 Search PubMed
; D. J. Faulkner, Nat. Prod. Rep., 1986, 3, 1 Search PubMed
.
-
C. M. Ireland, T. F. Molinski, T. M. Zabriskie, T. C. McKee, J. C. Swersey and M. P. Foster, in Bioorganic and Marine Chemistry, ed. P. J. Scheuer, Springer-Verlag, Berlin, 1989, vol. 3 Search PubMed
.
- http: //pubs.rsc.org/marinlit/ .
-
(a) R. Kazlauskas, R. O. Lidgard, R. J. Wells and W. Vetter, Tetrahedron Lett., 1977, 18, 3183 CrossRef
; (b) R. Kazlauskas, P. T. Murphy and R. J. Wells, Tetrahedron Lett., 1978, 19, 4945–4948 CrossRef
; (c) W. Hofheinz and W. E. Oberhänsli, Helv. Chim. Acta, 1977, 60, 660–669 CrossRef CAS
.
- J. Kobayashi, F. Itagaki, H. Shigemori, M. Ishibashi, K. Takahashi, M. Ogura, S. Nagasawa, T. Nakamura and H. Hirota, J. Am. Chem. Soc., 1991, 113, 7812–7813 CrossRef CAS
.
- N. Fusetani, T. Sugawara, S. Matsunaga and H. Hirota, J. Am. Chem. Soc., 1991, 113, 7811–7813 CrossRef CAS
.
- S. Carmeli and Y. Kashman, Tetrahedron Lett., 1985, 26, 511 CrossRef
.
- J. Kobayashi, F. Itagaki, I. Shigemori, T. Takao and Y. Shimonishi, Tetrahedron, 1995, 51, 2525–2532 CrossRef CAS
.
- F. Itagaki, H. Shigemori, M. Ishibashi, T. Nakamura, T. Sasaki and J. Kobayashi, J. Org. Chem., 1992, 57, 5540–5542 CrossRef CAS
.
- H. Uemoto, Y. Yahiro, H. Shigemori, M. Tsuda, T. Takao, Y. Shimonishi and J. Kobayashi, Tetrahedron, 1998, 54, 6719–6724 CrossRef CAS
.
- M. Tsuda, H. Ishiyama, K. Masuko, T. Takao, Y. Shimonishi and J. Kobayashi, Tetrahedron, 1999, 55, 12543–12548 CrossRef CAS
.
- E. W. Schmidt, C. Ravento-Suarez, M. Bifano, A. T. Menen-dez, C. R. Fairchild and D. J. Faulkner, J. Nat. Prod., 2004, 67, 475–478 CrossRef CAS PubMed
.
- L. Chill, Y. Kashman and M. Schleyer, Tetrahedron, 1997, 53, 16147–16152 CrossRef CAS
.
- S. P. Gunasekera, S. A. Pomponi and P. J. McCarthy, J. Nat. Prod., 1994, 57, 79–83 CrossRef CAS
.
- M. Kimura, T. Wakimoto, Y. Egami, K. C. Tan, Y. Ise and I. Abe, J. Nat. Prod., 2012, 75, 290–294 CrossRef CAS PubMed
.
- M. T. Jamison and T. F. Molinski, J. Nat. Prod., 2016, 79, 2243 CrossRef CAS PubMed
.
- M. Issac, M. Aknin, A. Gauvin-Bialecki, N. De Voogd, A. Ledoux, M. Frederich, Y. Kashman and S. Carmeli, J. Nat. Prod., 2017, 80, 1110–1116 CrossRef CAS PubMed
.
- K. M. Fisch, C. Gurgui, N. Heycke, S. A. van der Sar, S. A. Anderson, V. L. Webb, S. Taudien, M. Platzer, B. K. Rubio, S. J. Robinson, P. Crews and J. Piel, Nat. Chem. Biol., 2009, 5, 494–501 CrossRef CAS PubMed
.
- M. C. Wilson, T. Mori, C. Ruckert, A. R. Uria, M. J. Helf, K. Takada, C. Gernert, U. A. E. Steffens, N. Heycke, S. Schmitt, C. Rinke, E. J. N. Helfrich, A. O. Brachmann, C. Gurgui, T. Wakimoto, M. Kracht, M. Crusemann, U. Hentschel, I. Abe, S. Matsunaga, J. Kalinowski, H. Takeyama and J. Piel, Nature, 2014, 506, 58 CrossRef CAS PubMed
.
-
(a) N. Fusetani, S. Matsunaga, H. Matsumoto and Y. Takebayashi, J. Am. Chem. Soc., 1990, 112, 7053–7054 CrossRef CAS
; (b) S. D. Lewis, A. S. Ng and J. J. Baldwin, Thromb. Res., 1993, 70, 173–190 CrossRef CAS PubMed
; (c) B. E. Maryanoff, X. Qiu, K. P. Padmanabhan, A. Tulinsky, H. Almond Jr., P. Andrade-Gordon, M. N. Greco, J. A. Kauffman, K. C. Nicolaou, A. Liu, P. H. Brungs and N. Fusetani, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 8048–8052 CrossRef CAS PubMed
.
-
(a) T. Hamada, S. Matsunaga, G. Yano and N. Fusetani, J. Am. Chem. Soc., 2005, 127, 110–118 CrossRef CAS PubMed
; (b) T. Hamada, T. Sugawara, S. Matsunaga and N. Fusetani, Tetrahedron Lett., 1994, 35, 719–720 CrossRef CAS
.
- T. Shioiri and R. J. Hughes, Heterocycles, 2003, 61, 23 CrossRef CAS
.
- J. A. Sowinski and P. L. Toogood, Chem. Commun., 1999, 981–982 RSC
.
-
J. A. Sowinski, Thesis, Ph.D., University of Michigan, 1997
.
- J. A. Sowinski and P. L. Toogood, J. Org. Chem., 1996, 61, 7671 CrossRef CAS PubMed
.
- J. A. Sowinski and P. L. Toogood, Tetrahedron Lett., 1995, 36, 67–70 CrossRef CAS
.
- D. Sellanes, E. Manta and G. Serra, Tetrahedron Lett., 2007, 48, 1827–1830 CrossRef CAS PubMed
.
- P. Wipf, C. P. Miller, S. Venkatraman and P. C. Fritch, Tetrahedron Lett., 1995, 36, 6395–6398 CrossRef CAS
.
- H. H. Wasserman and R. Zhang, Tetrahedron Lett., 2002, 43, 3743–3746 CrossRef CAS
.
- J. Kobayashi, J. M. Sato, M. Ishibashi, H. Shigemori, T. Nakamura and Y. Ohizumi, J. Chem. Soc., Perkin Trans. 1, 1991, 2609–2611 RSC
.
- E. W. Schmidt, M. K. Harper and D. J. Faulkner, J. Nat. Prod., 1997, 60, 779–782 CrossRef CAS
.
Footnotes |
| † I am happy to thank John Blunt for extracting this data from MarinLit. |
| ‡ An alternative interpretation reads, “marine natural product peptides are peptides successfully isolated by chemists through solvent extraction.”! |
| § Keramamides A30 and L,10 also from T. swinhoei,10 are not discussed in this review and their structures are absent from Fig. 2: both are cyclic peptides of a different class – a Type 3, if you will – the first members of which, mozamides A and B, were reported by Faulkner and coworkers.31 See ESI of ref. 16. ‘Keramamide I’ seems to be an ordinal omission: a compound of this name is absent from the literature. |
| ¶ Interestingly, sequencing of the Entotheonella spp. genomes has revealed the gene clusters responsible for a range of other Theonella NRPS-PKS natural products, such as konbamides, onnamides, polytheonamides [ref. 21], nazuamides and psymberin [ref. 18 and 19]. |
| || Fusetani alludes to ‘partially racemized Leu’ obtained from strongly alkaline oxidative degradation of orbiculamide A (5); strictly, this is C-2 epimerization and the end product would be D-allo-Leu.6 Other papers are also less clear on this point. |
| This journal is © The Royal Society of Chemistry 2018 |