Open Access Article
Open Access ArticleA novel thermal detection method based on molecularly imprinted nanoparticles as recognition elements†
Francesco
Canfarotta
 *a,
J.
Czulak
*a,
J.
Czulak
 a,
K.
Betlem
a,
K.
Betlem
 b,
A.
Sachdeva
b,
A.
Sachdeva
 c,
K.
Eersels
c,
K.
Eersels
 d,
B.
van Grinsven
d,
B.
van Grinsven
 d,
T. J.
Cleij
d and
M.
Peeters
d,
T. J.
Cleij
d and
M.
Peeters
 *b
*b
aMIP Diagnostics Ltd., Fielding Johnson Building, University of Leicester, LE1 7RH, UK. E-mail: fc114@leicester.ac.uk
bManchester Metropolitan University, Faculty of Science & Engineering, Div. of Chemistry & Environmental Science, John Dalton Building, Chester Street, M15GD, Manchester, UK. E-mail: m.peeters@mmu.ac.uk; Tel: +44 (0)1612471450
cWellcome Centre for Mitochondrial Research & Northern Institute for Cancer Research, Newcastle University, Framlington Place, Newcastle-upon-Tyne NE2 4HH, UK
dMaastricht University Maastricht Science Programme, P.O. Box 616, 6200 MD Maastricht, The Netherlands
First published on 4th January 2018
Abstract
Molecularly Imprinted Polymers (MIPs) are synthetic receptors that are able to selectively bind their target molecule and, for this reason, they are currently employed as recognition elements in sensors. In this work, MIP nanoparticles (nanoMIPs) are produced by solid-phase synthesis for a range of templates with different sizes, including a small molecule (biotin), two peptides (one derived from the epithelial growth factor receptor and vancomycin) and a protein (trypsin). NanoMIPs are then dipcoated on the surface of thermocouples that measure the temperature inside a liquid flow cell. Binding of the template to the MIP layer on the sensitive area of the thermocouple tip blocks the heat-flow from the sensor to the liquid, thereby lowering the overall temperature measured by the thermocouple. This is subsequently correlated to the concentration of the template, enabling measurement of target molecules in the low nanomolar regime. The significant improvement in the limit of detection (a magnitude of three orders compared to previously used MIP microparticles) can be attributed to their high affinity, enhanced conductivity and increased surface-to-volume ratio. It is the first time that these nanosized recognition elements are used in combination with thermal detection, and it is the first report on MIP-based thermal sensors for determining protein levels. The developed thermal sensors have a high selectivity, fast measurement time (<5 min), and data analysis is straightforward, which makes it possible to monitor biomolecules in real-time. The set of biomolecules discussed in this manuscript show that it is possible to cover a range of template molecules regardless of their size, demonstrating the general applicability of the biosensor platform. In addition, with its high commercial potential and biocompatibility of the MIP receptor layer, this is an important step towards sensing assays for diagnostic applications that can be used in vivo.
1. Introduction
Molecularly Imprinted Polymers (MIPs) are synthetic receptors with recognition sites that are complementary to the template in terms of size, shape, and orientation of functional groups.1–3 Bulk imprinting, the traditional approach to synthesize MIPs, leads to the formation of microparticles that are not straightforward to incorporate into sensing devices.4 The preparation of nanoparticles from a solid-phase support (hereafter referred to as nanoMIPs) holds great potential both in the diagnostic and therapeutic sector, also in light of their good biocompatibility.5,6 Currently, nanoMIPs produced by solid-phase synthesis are applied as recognition elements in both optical and electrochemical sensors and assays.7,8 In addition, they can be considered as replacement for antibodies and have been used for the detection of several targets in ELISA-type assays.9–13 The use of synthetic antibodies has a number of distinct advantages over their natural counterparts, including their cost-effectiveness, simple and scalable synthesis, and that no animals are required for the production process.14,15 Due to their robust nature, nanoMIPs can be employed in harsh chemical environments such as extremes of pH, temperature and also in organic solvents.16,17 Since nanoMIPs are not proteins, they are not susceptible to degradation or proteolysis.12 This makes them re-usable and viable for use in more remote geographical areas where healthcare infrastructure is limited. It is also beneficial for the in situ monitoring of biomolecules, since field sensors may need to withstand relatively harsh conditions depending on their designated use.18Whilst MIPs are extremely suitable for extraction and quantification of target molecules in complex matrices, their use in biosensors remains limited. This can be explained by two factors; first, difficulties to incorporate MIPs into sensors platforms and, second, lack of detection methods that allow simple interpretation and are suitable for integration into portable devices.19,20
In a previous research, we have demonstrated that thermal detection methods offer an interesting alternative to traditional biosensor read-out platform (e.g. electrochemical detection and chromatographic techniques) since it requires the use of only two thermocouples and a heat-source, and the device can be used for measurements in real-time.21,22 MIP microparticles were functionalised on electrode surfaces, which were subsequently mounted into a patented thermocouple device.22,23 The changes in thermal resistance at the solid–liquid interface24 were then correlated to the biomolecule concentration in the sample. In a follow-up on this research, these MIP microparticles were directly deposited onto Screen-Printed Electrodes (SPEs) to determine neurotransmitter concentrations.25 SPEs are disposable electrodes with low batch-to-batch inconsistencies, which offer additional material advantages such as their flexibility and low-cost.26 However, algorithms are required to determine the heat-transfer resistance and the biomolecule concentration is determined in an indirect way. A straightforward approach is to directly deposit MIP particles onto thermocouples that are used for the determination of the temperature in the liquid.27 While this simplifies data analysis, it requires the use of additional layers to attach the MIP layers on the thermocouples. In addition, the microparticles hinders heat-flow through the thermocouple, thereby rendering detection less efficient and increasing the noise on the signal. In this manuscript, we will use nanoMIPs prepared by the solid-phase approach that can be directly applied onto thermocouples by means of dipcoating. Such nanoMIPs have been developed for a variety of targets that differ in size, going from a small molecule (biotin), to peptides (vancomycin and an epitope of the epidermal growth factor receptor) to a large protein (trypsin). We will demonstrate that we can measure these molecules at physiologically relevant concentrations with these thermal sensors. The developed sensors are a very powerful analytical tool due to their straightforward preparation process, simplicity to operate, rapid and low-cost detection. Molecular imprinting technology is versatile and the sensor platform can be extended to other biomolecules by simply varying the nanoMIPs. In addition, due to the biocompatibility of the sensors, this is the first step towards an in vivo diagnostic tool with potential clinical applications.
2. Materials and methods
2.1. Preparation of template-modified glass beads (solid-phase functionalization)
Glass beads (Spheriglass® 2429 CP00, 53–106 μm diameter, from Blagden Chemicals, UK) were first activated by boiling in a 2 M NaOH solution for 15 min, then washed with deionized water and rinsed with acetone, dried at 80 °C and subsequently incubated overnight in 2% v/v 3-aminopropyltrimethyloxysilane (APTMS) in a dry toluene solution (0.4 mL solution per g glass beads). This last step leads to beads bearing NH2 surface groups.For the preparation of EGFR-modified solid-phase, 60 g of glass beads were placed in a solution of succinimidyl iodoacetate linker, at 0.2 mg ml−1 in acetonitrile for 2 h in the dark (0.4 ml solution g per glass beads). The succinic moiety allows the linker to react with the amine-derivatised solid-phase, and the haloacetyl group enables subsequent coupling with the thiol group from cysteine added on the N-terminus of the peptide sequence (obtained from Ontores Biotechnologies, China). Afterwards, the beads were washed with 400 ml of acetonitrile in a sintered glass funnel and placed in a bottle containing 7 mg of the cysteine-modified peptide epitope in 40 ml of deoxygenated phosphate buffered saline (PBS) containing 5 mM EDTA, pH 8.2. After overnight incubation, the beads were washed with 700 ml of water in a sintered funnel and used for the synthesis of nanoMIPs.
For the preparation of biotin nanoMIPs, 60 g of APTMS-derivatised glass beads were incubated with a solution of biotin (Sigma Aldrich) containing 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) (both from Sigma Aldrich) at 0.5, 10 and 15 mg ml−1 respectively, in PBS pH 7.4. After overnight incubation, the beads were washed with 500 ml of water in a sintered funnel and used for the synthesis of nanoMIPs.
For the preparation of trypsin- and vancomycin-modified solid-phase, APTMS-derivatised glass beads were first incubated in a 7% glutaraldehyde solution in PBS for 2 h. Afterwards, the beads were washed with 700 ml of distilled water and placed in a bottle containing either trypsin (0.1 mg ml−1) or vancomycin (0.2 mg ml−1) in PBS, and incubated overnight at 4 °C. After overnight incubation, the beads were washed with 500 ml of water in a sintered funnel and used for the synthesis of nanoMIPs.
2.2. Synthesis of biotin, vancomycin, trypsin and EGFR-imprinted nanoMIPs
The procedure has been adapted from ref. 5 and 6. The following monomers were dissolved in PBS 5 mM, pH 7.4 (100 ml): N-isopropylacrylamide (39 mg), N,N′-methylenebisacrylamide (6 mg), N-tert-butylacrylamide (33 mg), N-(3-aminopropyl)methacrylamide hydrochloride (5.8 mg), acrylic acid (2.2 μl). N-tert-Butylacrylamide was previously dissolved in ethanol (1 ml) and then added to the aqueous solution.The solution containing the monomers was degassed under vacuum and sonicated for 5 min, and then purged with N2 for 20 min. Then, 60 g of template-derivatised glass beads were added to the solution. Polymerization was initiated by adding ammonium persulfate aqueous solution (800 μl, 60 mg ml−1) and N,N,N′,N′-tetramethylethylenediamine (24 μl). The headspace was flushed with N2 and the bottle sealed with a screw cap. Polymerization was carried-out at room temperature for 1 h. Subsequently, the content of the polymerization vessel was poured into a solid-phase extraction cartridge (60 ml) equipped with a frit (20 μm porosity). A total of 9 washes with 20 ml of distilled water at 20 °C was carried out to remove low affinity nanoMIPs, oligomers and unreacted monomer. Afterwards, the SPE cartridge containing the solid-phase was placed in a water bath at 70 °C for 15 min. An aliquot of 20 ml of distilled water pre-warmed at 65 °C was poured into the SPE to collect the high-affinity nanoMIPs. This procedure was repeated seven times, until about 140 ml of a solution of high-affinity nanoMIPs in water was collected. To ensure complete removal of all unreacted monomer from the bulk of the nanoparticles, aliquots of the collected solution were concentrated down to 2 ml using a centrifugal dialysis cartridge fitted with a membrane with 30 kDa molecular weight cut-off (Amicon Ultra, Merck Millipore) to remove unreacted monomers and other side products. This was followed by seven washes (14 ml) with deionized water on the same dialysis cartridge.
2.3. Size and concentration analysis of nanoMIPs
The particle size was measured with a Zetasizer Nano (Nano-S) particle-size analyser from Malvern Instruments Ltd (UK). An aliquot of the dispersion of nanoMIPs in distilled water was sonicated for 2 min and then analysed by DLS at 25 °C in a 3 cm3 disposable polystyrene cuvette. The instrument automatically chose attenuator position, measurement duration and number of runs. The values are reported as an average of 4 measurements. Transmission Electron Microscopy (TEM) images of nanoMIPs were taken using a JEOL JEM 1010, 100 kV high contrast TEM equipped with a Gatan SC1000 Orius CCD camera (Gatan, Abingdon Oxon, UK). Samples for the analysis have been prepared by depositing a drop of the nanoMIPs dispersion, previously filtered through a 1.2 μm PES syringe filter, on a carbon-coated TEM copper grid (400 mesh), and leaving them to dry at room temperature. The nanoMIP concentration was assessed by freeze-drying an aliquot (20 ml) of the particle solution and then weighing the solid.2.4. Thermal measurements of nanoMIPs sensors
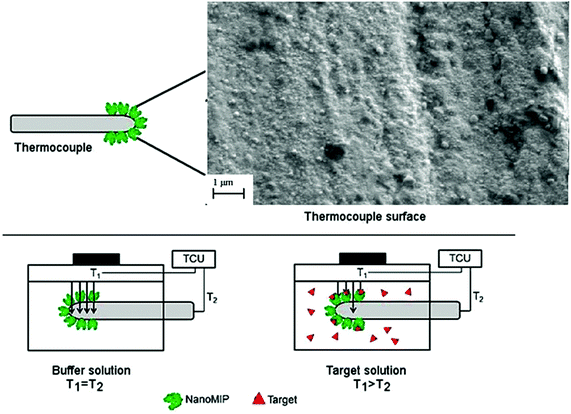
Thermocouples of type K from TC Direct (Uxbridge, United Kingdom) were dipped into a nanoMIP solution for 60 s and withdrawn at a rate of 5.1 cm min−1. The functionalised thermocouples were subsequently air-dried at room temperature. The presence of the particles on the tip of the thermocouples was determined by Scanning Electron Microscopy analysis with a Supra 40VP Field Emission Scanning Electron Microscope from Carl Zeiss Ltd (Oberkochen, Germany). The functionalised thermocouples were then inserted into a heat-transfer set up with an in-house design, which has been described in detail in previous work.23,24 This set up is coupled to a Perspex flow cell, which exposes the receptor layer to buffer and target solutions. A copper block underneath the sample serves as a heat-sink. Its temperature (T1) is actively steered with a proportional-integral-derivative (PID) controller that is attached to a power resistor. In all measurements, the temperature was kept constant at 37.00 ± 0.02 °C to mimic in vivo conditions. The functionalised MIP-thermocouple monitors the temperature in the flow cell, which is defined as T2 and is sampled every 1.0 s.In previous work,27 the authors described polylactic acid (PLLA) coated thermocouples that were impregnated with MIP particles. The PLLA-sheet (∼2 μm thickness) forms a thermally insulating layer around the thermocouple over the entire sheet within the flow cell. In that case, binding of target molecules to the MIP particles will increase the thermal resistance of the sheet, leading to a decrease in heat loss over the thermocouple and an overall increase of the measured temperature.
In this work, only the sensitive area of the tip of the thermocouple is covered by a very thin layer of nano-sized MIP particles (Fig. 1). The insulating effect of target rebinding has little to no effect on the thermocouple heat sink effect but does decrease the sensitivity of the tip in accurately determining T2. It was previously demonstrated21 that the thermal resistance at the solid–liquid interface increases when the analyte binds to MIP particles functionalised onto planar electrodes. With this method, the thermal resistance increases between the thermocouple tip and the liquid inside the flow cell and as a result, the measured temperature in the flow cell (T2) is significantly lowered. The benefit of this method is that a nanosized layer is obtained, which will enhance heat-flow through the surface and significantly increase effect size. The principle is demonstrated in Fig. 1, where TCU represents the temperature control unit of the heat sink (T1) and the temperature monitored in the flow cell (T2).
Prior to each measurement, the flow cell was filled with a solution of phosphate buffered saline (PBS) and left to stabilize for 45 min. Subsequently, solutions with increasing target concentration in PBS were added (0, 5, 10, 100, 500, 1000 nM) using an automatic syringe pump (ProSense, model NE-500, the Netherlands) at a constant rate of 0.25 mL min−1. The signal was left to stabilize for 30 min between each addition. The temperatures T1 and T2 were recorded throughout the measurement. Dose–response curves were constructed by plotting the target concentration vs. the relative change in thermal signal. The selectivity of the MIP sensor was determined by measuring the thermal response with a chemical compound that is similar to the original target. For these experiments, the signal was stabilised in PBS for 45 min and PBS solutions of increasing concentration (50, 100, 500, 1000 nM) of a similar analyte were added with 30 min intervals. Specificity tests were carried out with structural analogues or molecules which can act as interferents in a given matrix. In particular, pepsin was used in the case of trypsin nanoMIPs, teicoplanin for vancomycin nanoMIPs, ascorbic acid for biotin nanoMIPs and finally a related peptide sequence Beta-2-Microglobulin (hereafter referred to as B2MG) for EGFR nanoMIPs.
For proof-of-application in complex media, measurements were conducted with saliva samples spiked with EGFR. First, electrodes were stabilized in PBS for 30 min which was followed by addition of saliva diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with PBS. Subsequently, saliva samples (all diluted 1
1 with PBS. Subsequently, saliva samples (all diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with PBS to reduce viscosity) were spiked with 5, 10, 100, 1000 and 10
1 with PBS to reduce viscosity) were spiked with 5, 10, 100, 1000 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nM EGFR.
000 nM EGFR.
3. Discussion
3.1. Production and characterization of nanoMIPs
Herein we assess whether molecularly imprinted nanoparticles (nanoMIPs) can act as recognition elements once they are coated onto thermocouples, and whether they can improve the achievable limit-of-detection (LoD) and specificity of thermal MIP sensors. The innovative solid-phase approach here employed in the production of nanoMIPs relies on the covalent immobilisation of the template molecule on a solid support (e.g. glass beads, average diameter 75–90 μm). Polymerisation is then initiated in the presence of this support bearing the immobilised template. After polymerisation, the solid support acts as an affinity medium to isolate the high-affinity nanoMIPs from the remaining monomers, oligomers and low-affinity polymers (Fig. 2). Because of the affinity purification step, nanoMIPs possess high affinity and specificity toward their targets and exhibit a homogeneous distribution of binding site affinities.5 As previously demonstrated, the advantage of such solid-phase approach is the production of nanoMIPs virtually free from templates and in a short time-frame (few hours). Moreover, this approach can be easily automated to allow the manufacture of nanoMIPs with improved batch-to-batch consistency.6 Interestingly, nanoMIPs produced by the solid-phase approach here described have been successfully employed as imaging tools in vivo, thus demonstrating their potential in diagnostics.28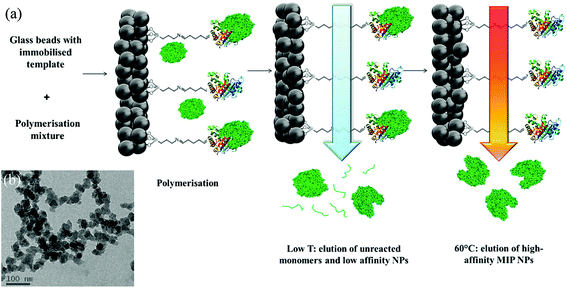 | ||
| Fig. 2 (a) Scheme of the solid-phase synthesis of nanoMIPs. In this example, a protein is shown as the template molecule. (b) Representative TEM of biotin MIPs. | ||
NanoMIPs against biotin, trypsin, vancomycin and an EGFR epitope were produced, therefore covering a wide range of template sizes and showing the general applicability of the method. To date, nanosized MIP structures have not been used in combination with thermal detection. In addition, this is the first report on the use of this platform with MIPs for the detection of large peptides and proteins. The imprinting of these compounds is notoriously difficult due to sensitive structural nature of proteins and the need to imprint in an aqueous environment, limiting the choice of monomers.29 Therefore, in combination with the biocompatibility of the recognition layer,5,28 this method holds great potential as a protein sensor that can be incorporated into e.g. a catheter for in vivo sensing.
The hydrodynamic size of the four different nanoMIPs in water was determined by DLS (Table 1). It should be noted that the hydrodynamic size can significantly differ from the dry size by TEM for several reasons: first, the polymer tends to hydrate and swell when in solution; secondly, nanoMIPs can undergo a certain level of agglomeration in solution, which leads to an apparent increase in the size by DLS; thirdly, it is worth noting that DLS analyses the light scattered from the sample as a whole. However, smaller particles scatter less light than larger particles (intensity of the scattering proportional to the sixth power of the particle diameter), therefore the scattered light of these latter (or even from aggregates) can cover the signal from smaller particles, and hence the apparent particle size distribution can be shifted towards larger sizes.
| Target | Size (nm) | PDI |
|---|---|---|
| Biotin | 161 ± 11 | 0.24 |
| EGFR | 239 ± 9 | 0.19 |
| Vancomycin | 275 ± 8 | 0.26 |
| Trypsin | 320 ± 7 | 0.22 |
3.2. Thermal experiments with thermocouples functionalized with nanoMIPs
Subsequently, these nanoMIPs were functionalized onto thermocouples using dipcoating.In an initial experiment, a blank thermocouple (not coated with nanoparticles) was exposed to solutions with increasing concentrations of the EGFR peptide (5, 10, 100, 500, 1000, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nM). It is clear that there is no significant effect on the temperature of the blank thermocouple upon stabilization in PBS compared to PBS with a high concentration (5–10
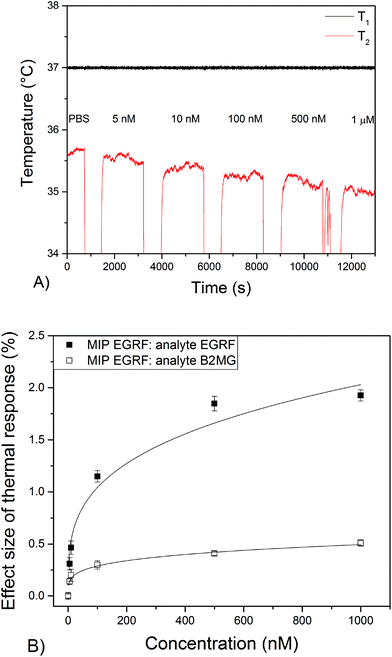
000 nM). It is clear that there is no significant effect on the temperature of the blank thermocouple upon stabilization in PBS compared to PBS with a high concentration (5–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nM) of EGFR (Appendix A†). Subsequently, thermocouples dipcoated with nanoMIPs for EGFR were inserted into the set up and the temperature was monitored when solutions with increasing concentrations of EGFR (5–1000 nM) were added to the flow cell. The temperature of T1 was strictly controlled at 37.00 ± 0.02 °C. In the case of T2, decreases in the temperature were measured after each injection due to the addition of fluid at room temperature to the set-up. The temperature control unit increased the power to regulate it back to the original temperature of T1, which requires a short stabilization time. In the case of the blank thermocouples where no binding occurs, T1 goes back to baseline level shortly after injection and T2 returns to this its initial level after a few minutes. When binding occurs at the MIP layer of the tip of the functionalised thermocouple, T1 is retained at its set temperature but T2 (which is not controlled but merely monitored) stabilizes at a value below the baseline (see Fig. 3a). Changes in T2 were recorded after addition of solutions with EGFR concentrations of 5 nM or higher, with the linear range of the sensor between 5–500 nM (see Fig. 3b). The thermal signal saturated at a maximum concentration of 1000 nM when T2 had dropped from 35.60 ± 0.05 °C to 35.00 ± 0.05 °C. Noise on the thermal signal was around 0.05 °C (∼0.15%) throughout the experiment. The difference in temperature was subsequently converted into an effect size by dividing the difference in temperature over the initial temperature, leading to a 1.90 ± 0.05% change in thermal signal at 1000 nM (Fig. 3b). Experiments performed with a non-related peptide sequence (B2MG) resulted in a 0.5 ± 0.04% change of the thermal signal at a concentration of 1000 nM. A minor response was expected due to the similarity that this compound bears with the target peptide EGFR. However, since this was nearly a factor of four lower compared to the original template (1.9%) at the same concentration, the experiment demonstrated the high selectivity of the developed biosensor platform.
000 nM) of EGFR (Appendix A†). Subsequently, thermocouples dipcoated with nanoMIPs for EGFR were inserted into the set up and the temperature was monitored when solutions with increasing concentrations of EGFR (5–1000 nM) were added to the flow cell. The temperature of T1 was strictly controlled at 37.00 ± 0.02 °C. In the case of T2, decreases in the temperature were measured after each injection due to the addition of fluid at room temperature to the set-up. The temperature control unit increased the power to regulate it back to the original temperature of T1, which requires a short stabilization time. In the case of the blank thermocouples where no binding occurs, T1 goes back to baseline level shortly after injection and T2 returns to this its initial level after a few minutes. When binding occurs at the MIP layer of the tip of the functionalised thermocouple, T1 is retained at its set temperature but T2 (which is not controlled but merely monitored) stabilizes at a value below the baseline (see Fig. 3a). Changes in T2 were recorded after addition of solutions with EGFR concentrations of 5 nM or higher, with the linear range of the sensor between 5–500 nM (see Fig. 3b). The thermal signal saturated at a maximum concentration of 1000 nM when T2 had dropped from 35.60 ± 0.05 °C to 35.00 ± 0.05 °C. Noise on the thermal signal was around 0.05 °C (∼0.15%) throughout the experiment. The difference in temperature was subsequently converted into an effect size by dividing the difference in temperature over the initial temperature, leading to a 1.90 ± 0.05% change in thermal signal at 1000 nM (Fig. 3b). Experiments performed with a non-related peptide sequence (B2MG) resulted in a 0.5 ± 0.04% change of the thermal signal at a concentration of 1000 nM. A minor response was expected due to the similarity that this compound bears with the target peptide EGFR. However, since this was nearly a factor of four lower compared to the original template (1.9%) at the same concentration, the experiment demonstrated the high selectivity of the developed biosensor platform.
Standard deviations were calculated by averaging out the data over 200 data points and are indicated as error bars in Fig. 3.
The LoD can be determined at the concentration at which the signal is equal to three times the standard deviation on the baseline level signal. With the dose–response curve and obtained fit, a LoD value of 3 ± 1 nM in PBS was estimated, which was an improvement of nearly three orders of magnitude compared to previous works on thermocouples.27 The explanation behind this is threefold; first, there is a ten-fold decrease in layer thickness that promotes heat-flow through the surface and increases the response to changes in the thermal signal and, second, the increased surface-to-volume ratio of the nanoparticles and, third, the higher affinity of the nanoMIPs towards its template. It is also an improvement compared to the more conventional approach where MIP particles were functionalised onto an electrode surface rather than directly on the thermocouple, which attained LoDs between 25–100 nM for small molecules in buffered solutions.23–25
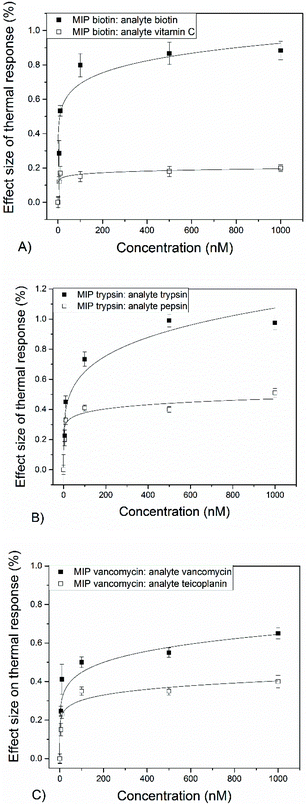
Subsequently, thermocouples were coated with nanoMIPs for, respectively, biotin, trypsin and vancomycin to determine the wide applicability of this method. For all these, a similar trend was observed and decreases in the temperature of the thermocouple were monitored after addition of spiked PBS solutions. The actual response in temperature for the three nanoMIPs is provided in the ESI B–D.† The measured dose–response curves are shown in Fig. 4-a. Measurements were performed with both the actual target molecules and similar biomolecules, which provides insight into the selectivity of the biosensor platform.
The four developed nanoMIPs all exhibit a reduction in the measured thermocouple temperature (decrease ∼0.5 °C) after exposure to solutions containing target molecules. With the use of the dose–response curves (Fig. 4a–c), LoDs of ∼3–5 nM were determined in buffered solutions. While LoDs were similar for the four nanoMIPs, there were minor differences in the maximum response of the thermal signal. For instance, for EGFR nanoMIPs a response of 1.90% was measured while for vancomycin 0.65% was attained. There could be several explanations, including the higher affinity of the EGFR nanoMIP, smaller size of vancomycin that leads to lower attenuation of the heat-transfer resistance, or differences in available surface area.
The selectivity of the thermal sensor was evaluated by comparing the signal of a given MIP over that of a similar molecule at a certain concentration (selectivity factor, see Table 2). For EGFR and biotin, selectivity factors (SF) of >4 were determined at concentrations of 100 nM. These are slightly higher at lower concentrations (∼5) compared to higher concentrations (∼3), which is because at a certain point saturation of the binding sites occurs. For vancomycin, the SF of 1.7 is lower compared to MIPs designed for other template molecules. This might be due to the fact that teicoplanin bears a very high structural similarity to vancomycin. Ascorbic acid was chosen as specificity target for biotin because they are both vitamins and therefore likely to compete with each other in the same sample matrix. These results are assembled in Table 2.
| Target | LoD (nM) | Response at 1000 nM (%) | Selectivity factor (c = 100 nM) | Linear range (nM) |
|---|---|---|---|---|
| Target/competitor | ||||
| EGFR | 3 ± 1 | 1.9 ± 0.1 | 4.5 | 0–500 |
| Biotin | 5 ± 2 | 0.8 ± 0.05 | 4.3 | 0–100 |
| Trypsin | 5 ± 2 | 0.98 ± 0.05 | 2.3 | 0–500 |
| Vancomycin | 4 ± 1 | 0.65 ± 0.03 | 1.7 | 0–100 |
LoDs that are obtained with this method are within the therapeutic range of antibiotic compounds in serum. Therefore, within a clinical setting, there is a demand to monitor in vivo pharmacokinetics of these drugs. Vancomycin is administered to treat severe infections and levels are monitored to maintain its concentration above a threshold but below levels that are associated with toxicity and side effects.30
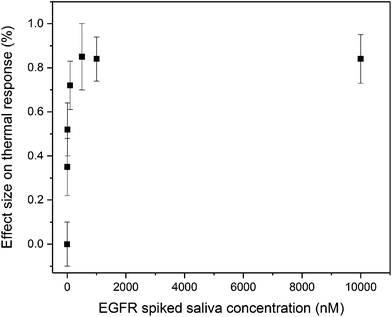
As proof-of-application for measurements in complex samples, an experiment was performed with spiked saliva samples. The electrodes were stabilized in PBS and after subsequent addition of saliva, the temperature decreased by 10% due to the matrix effect of the saliva. Subsequently, spiked saliva samples were administered each 30 min and these results are shown in Fig. 5.
Saliva is viscous and this complicates administration of samples by the automated syringe pump, which is why the samples were diluted with PBS. However, samples were diluted 50/50, meaning it remained an accurate representation of a complex matrix. While error bars are significantly higher due to the presence of matrix compounds, such as proteins, there are significant differences in thermal response observed to the saliva samples spiked with EGFR (linear range 0–500 nM). This is a clear indication that this methods hold promise for the detection of complex samples, and we could improve the noise on the significantly (from 0.15% in Fig. 5 to 0.05%) if we would apply a gentle percentile filter to the data.
There is also a need to measure protein concentrations for healthcare applications, and we present the first MIP-based sensor for the thermal detection of these compounds. This sensor platform could provide a viable alternative to existing vitamin tests, since serum biotin levels of <200 ng L−1 are associated with hair loss, dermatitis, or underlying metabolic disorders.31 This emerging thermal technology has room for improvement, for instance by adapting the temperature of the baseline to increase effect sizes. We have considered temperatures of 37 °C since this mimics in vivo conditions and has a significant difference with the ambient temperature in the lab, which promotes heat-flow and enhances stability of the signal. Measuring at lower temperatures could enhance the effect size which makes it of interest for biomarkers assays, but then the stability of the current temperature control unit would then need to be considered. Relevant biomarker assays include the EGFR test, used to predict the recurrence of breast and lung cancer.32 This method has the potential to be integrated in a portable device and can be transferred into a traffic light format, where patients would receive a warning if they will need to consult a doctor for further testing.
Another approach that can be considered to improve detection limits includes the use of Thermal Wave Transport Analysis.23 This method involves the analysis of modulated sinusoidal temperature signals that are imposed onto a linear temperature. It enhances overall stability of the signal and temperature adjustments of only 0.1 °C are required. Using TWTA could enhance specificity of the thermal sensors and is particularly interesting for in vivo applications since only minimal local heating (for instance a patch on a patient) is needed.
4. Conclusions
MIP nanoparticles were synthesised against biotin, vancomycin, an EGFR peptide and trypsin according to a solid-phase synthetic approach. The solid support was used as a medium to isolate the high affinity nanoMIPs after polymerisation. A homogeneous distribution of particles (in the range 161–320 nm) was obtained, which was confirmed by TEM analysis and measuring the hydrodynamic radius in water using DLS. Subsequently, the nanoMIPs were dissolved into water and dipcoated onto thermocouples of type K. The functionalised thermocouples were inserted into a device that measures the temperature of the liquid inside a flow cell developed according to a home-made design. To establish the baseline, the heat sink of the flow cell was kept at a constant temperature of 37.00 ± 0.02 °C. Significant drops in the temperature (∼0.50 ± 0.02 °C) measured by the MIP functionalised thermocouples were reported after addition of buffered solutions with template concentrations in the range of 5–500 nM. This decrease in the temperature is due to the attachment of the template to the MIP layer blocking the heat-transfer resistance at the interface of the thermocouple, leading to an overall lower temperature in the liquid of the flow cell. Dose–response curves were measured to determine the LoD of the sensors and correlate the template concentration to the decrease in measured temperature. In addition, selectivity of the thermal sensors was evaluated by exposing them to solutions of molecules that are similar in size, shape and function to the template.This demonstrates the general applicability of nanoMIPs as recognition elements in thermal sensing. LoDs are obtained in the low nanomolar range, which is an increase by three orders of magnitude compared to previous works when bulk MIP microparticles were attached onto thermocouples. This step change can be explained by the use of nanoMIPs, which are pre-selected to have high affinity binding sites, and have a smaller layer thickness that promotes heat-flow through the surface.
The use of thermal sensing in combination with MIP technology can offer sensors that are able to detect biomolecules in a straightforward, low-cost and fast (<5 min) manner. The described methodology is versatile and can be tailored to the requested target molecule by adjusting the nanoMIP layer. In this contribution, we have presented the first MIP-based sensor for thermal detection of proteins.
In light of the use of this biocompatible recognition layer, this represents the first step towards an in vivo sensor platform with great potential for a range of industries, as well as in biomedical and clinical research.
Conflicts of interest
J. Czulak and F. Canfarotta declare conflicts of interest because their company (MIP Diagnostics) produces those nanoparticles. Other authors do not declare conflict of interest.Acknowledgements
We would like to acknowledge the Engineering and Materials Research Centre of Manchester Metropolitan University for funding of the PhD of Kai Betlem and use of consumables. We are also grateful for funding provided by the Dutch province of Limburg through the “Limburg Meet” project. AS acknowledges the Urology Foundation and the Rosetree Trust for fellowship funding.References
- B. Sellergen and C. J. Allender, Molecularly Imprinted Polymers: A Bridge to Advanced Drug Delivery, Adv. Drug Delivery Rev., 2005, 57, 1733 CrossRef PubMed.
- M. J. Whitcombe, N. Kirsch and I. A. Nicholls, Molecular Imprinting Science and Technology: A Survey of the Literature for the Years 2004–2011, J. Mol. Recognit., 2014, 27, 297 CrossRef CAS PubMed.
- G. Wulf, Enzyme-like Catalysis by Molecularly Imprinted Polymers, Chem. Rev., 2002, 102, 1 CrossRef.
- K. Haupt and K. Mosbach, Molecularly Imprinted Polymers and Their Use in Biomimetic Sensors, Chem. Rev., 2002, 100, 2495 CrossRef.
- F. Canfarotta, A. Poma, A. Guerreiro and S. A. Piletsky, Solid Phase Synthesis of Molecularly Imprinted Nanoparticles, Nat. Protoc., 2016, 11, 443 CrossRef CAS PubMed.
- A. Poma, A. Guerreiro, M. J. Whitcombe, E. V. Piletska, A. P. F. Turner and S. A. Piletsky, Solid-Phase Synthesis of Molecularly Imprinted Polymer Nanoparticles with a Reusable Template – “Plastic Antibodies”, Adv. Funct. Mater., 2013, 23, 2821 CrossRef CAS PubMed.
- I. Basozabal, A. Guerreiro, A. Gomez-Caballero, M. A. Goicolea and R. J. Barrio, Direct Potentiometric Quantifcation of Histamine Using Solid-Phase Imprinted Nanoparticles as Recognition Elements, Biosens. Bioelectron., 2014, 58, 138–144 CrossRef CAS PubMed.
- S. Korposh, I. Chianella, A. Guerreiro, S. Caygill, S. Piletsky, S. W. James and R. P. Tatam, Selective Vancomycin Detection Using Optical Fibre Long Period Gratings Functionalised with Molecularly Imprinted Polymer Nanoparticles, Analyst, 2014, 139, 2229 RSC.
- I. Chianella, A. Guerreiro, E. Moczko, J. S. Caygill, E. V. Piletska, I. M. P. De Vargas Sansalvador, M. J. Whitcombe and S. A. Piletsky, Direct Replacement of Antibodies with Molecularly Imprinted Polymer Nanoparticles in ELISA—Development of a Novel Assay for Vancomycin, Anal. Chem., 2013, 85, 8462 CrossRef CAS PubMed.
- C. Cáceres, F. Canfarotta, I. Chianella, E. Pereira, E. Moczko, C. Esen, A. Guerreiro, E. Piletska, M. Whitcombe and S. Piletsky, Does size matter? Study of performance of pseudo-ELISAs based on molecularly imprinted polymer nanoparticles prepared for analytes of different sizes, Analyst, 2016, 141, 1405 RSC.
- K. Smolinska-Kempisty, A. Guerreiro, F. Canfarotta, C. Fuentes, M. Whitcombe and S. Piletsky, Performance of antibodies and imprinted polymer nanoparticles in ELISA - a direct comparison, Sci. Rep., 2016, 6, 37638 CrossRef CAS PubMed.
- S. P. Tang, F. Canfarotta, K. Smolinska-Kempisty, E. Piletska, A. Guerreiro and S. A. Piletsky, A Pseudo-ELISA based on Molecularly Imprinted Nanoparticles for Detection of Gentamicin in Real Samples, Anal. Methods, 2017, 9, 2853 RSC.
- F. Canfarotta, K. Smolinska-Kempisty and S. Piletsky, Replacement of Antibodies in Pseudo-ELISAs: Molecularly imprinted Nanoparticles for Vancomycin Detection, Methods Mol. Biol., 2017, 1575, 389 Search PubMed.
- J. O. Mahony, K. Nolan, M. R. Smyth and B. Mizaikoff, Molecularly Imprinted Polymers – Potential and Challenges in Analytical Chemistry, Anal. Chim. Acta, 2005, 534, 31 CrossRef CAS.
- L. Ye and K. Haupt, Molecularly Imprinted Polymers as Antibody and Receptor mimics for Assays, Sensors and Drug Discovery, Anal. Bioanal. Chem., 2004, 378, 1887 CrossRef CAS PubMed.
- D. Kriz and K. Mosbach, Competititive Amperometric Morphine Sensor Based on an Agarose Immobilised Molecularly Imprinted Polymer, Anal. Chim. Acta, 1995, 300, 71 CrossRef CAS.
- J. Svenson and I. A. Nicholls, On the Thermal and Chemical Stability of Molecularly Imprinted Polymers, Anal. Chim. Acta, 2001, 435, 19 CrossRef CAS.
- F. Canfarotta, A. Waters, R. Sadler, A. Guerreiro, P. McGill, D. Papkovsky, K. Haupt and S. Piletsky, Biocompatibility and Internalisation of Molecularly Imprinted Polymer Nanoparticles, Nano Res., 2016, 9, 11 CrossRef.
- C. Y. Huang, M. J. Syu, Y. S. Chang, C. H. Chang, T. C. Chou and B. D. Liu, A Portable Potentiostat for the Bilirubin-Specific Sensor Prepared from Molecular Imprinting, Biosens. Bioelectron., 2007, 22, 1694 CrossRef CAS PubMed.
- A. P. F. Turner, Biosensors – Sense and Sensitivity, Science, 2000, 290, 1315 CrossRef CAS PubMed.
- M. Peeters, P. Csipai, B. Geerets, A. Weustenraed, B. van Grinsven, J. Gruber, W. De Ceuninck, T. J. Cleij, F. J. Troost and P. Wagner, Heat-Transfer Based Detection of L-Nicotine, Histamine, and Serotonin Using Molecularly Imprinted Polymers as Biomimetic Receptors, Anal. Bioanal. Chem., 2013, 405, 6453 CrossRef CAS PubMed.
- B. van Grinsven, K. Betlem, T. J. Cleij, C. E. Banks and M. Peeters, Evaluating the Potential of Thermal Read-Out Techniques Combined with Molecularly Imprinted Polymers (MIPs) for the Sensing of Low-Weight Organic Molecules, J. Mol. Recognit., 2016, 30, e2563, DOI:10.1002/jmr.2563.
- M. Peeters, B. van Grinsven, C. W. Foster, T. J. Cleij, P. Wagner and C. E. Banks, Introducing Thermal Wave Transport Analysis (TWTA): a Thermal Technique for Dopamine Detection of Screen-Printed Electrodes Functionalized with Molecularly Imprinted Polymers (MIPs) Particles, Molecules, 2016, 21, 552 CrossRef PubMed.
- B. van Grinsven, N. Vanden Bon, H. Strauven, L. Grieten, M. Murib, K. Jiménez-Monroy, S. D. Janssens, K. Haenen, M. J. Schöning, V. Vermeeren, M. Ameloot, L. Michiels, R. Thoelen, W. De Ceuninck and P. Wagner, Heat-Transfer Resistance at Solid-Liquid Interfaces: A Tool for the Detection for Single-Nucleotide Polymorphisms in DNA, ACS Nano, 2012, 6, 2712 CrossRef CAS PubMed.
- S. Casadio, J. W. Lowdon, K. Betlem, J. T. Ueta, C. W. Foster, T. J. Cleij, B. van Grinsven, O. B. Sutcliffe, C. E. Banks and M. Peeters, Development of a Novel Flexible Polymer-Based Biosensor Platform for the Thermal Detection of Noradrenaline in Aqueous Solutions, Chem. Eng. J., 2017, 315, 459 CrossRef CAS.
- J. P. Metters, R. O. Kadara and C. E. Banks, New Directions in Screen-Printed Electroanalytical Sensors: An Overview of Recent Developments, Analyst, 2011, 136, 1067 RSC.
- H. Diliën, M. Peeters, J. Royakkers, J. A. W. Harings, P. Cornelis, P. Wagner, E. Steenredeker, C. E. Banks, K. Eersels, B. van Grinsven and T. J. Cleij, Label-Free Detection of Small Organic Molecules by MIP-functionalized Thermocouples: Towards in vivo Applications, ACS Sens., 2017, 2, 583 CrossRef PubMed.
- A. Cecchini, V. Raffa, F. Canfarotta, G. Signore, S. Piletsky, M. Macdonald and A. Cuschieri, In vivo Recognition of Human Vascular Endothelial Growth Factor by Molecularly Imprinted Polymers, Nano Lett., 2017, 17, 2307 CrossRef CAS PubMed.
- E. Verheyen, J. Schillemans, M. van Wijk, E. Hennink and C. F. van Nostrum, Challenges for the Effective Molecular Imprinting of Proteins, Biomaterials, 2011, 32, 3008 CrossRef CAS PubMed.
- J. H. Martin, R. Norris, M. Barras, J. Roberts, R. Morris, M. Doogue and G. R. D. Jones, Therapeutic Monitoring of Vancomycin in Adult Patients: A Consensus Review of the American Society of Health-System Pharmacists, the Infectious Diseases Society of America, and the Society of Infectious Diseases Pharmacists, Clin. Biochem. Rev., 2010, 31, 21 Search PubMed.
- R. M. Trüeb, Serum Biotin Levels in Women Complaining of Hair Loss, Int. J. Trichology, 2016, 8, 73 CrossRef PubMed.
- J. Richard, C. Sainsbury, K. Geoffrey, J. Needham, R. Farndon, A. J. Malcolm and A. L. Harris, A Epidermal-Growth Factor Receptor Status a as a Predictor or Early Recurrence of and Death from Breast Cancer, Lancet, 1987, 329, 1398 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7nr07785h |
| This journal is © The Royal Society of Chemistry 2018 |