Open Access Article
Open Access ArticleMarine natural products†
John W.
Blunt
 a,
Anthony R.
Carroll
a,
Anthony R.
Carroll
 *bc,
Brent R.
Copp
*bc,
Brent R.
Copp
 d,
Rohan A.
Davis
d,
Rohan A.
Davis
 c,
Robert A.
Keyzers
c,
Robert A.
Keyzers
 e and
Michèle R.
Prinsep
e and
Michèle R.
Prinsep
 f
f
aSchool of Physical and Chemical Sciences, University of Canterbury, Christchurch, New Zealand
bGriffith School of Environment, Griffith University, Gold Coast, Australia. E-mail: A.Carroll@griffith.edu.au
cGriffith Institute for Drug Discovery, Griffith University, Brisbane, Australia
dSchool of Chemical Sciences, University of Auckland, Auckland, New Zealand
eCentre for Biodiscovery, School of Chemical and Physical Sciences, Victoria University of Wellington, Wellington, New Zealand
fChemistry, School of Science, University of Waikato, Hamilton, New Zealand
First published on 16th January 2018
Abstract
Covering: 2016. Previous review: Nat. Prod. Rep., 2017, 34, 235–294
This review covers the literature published in 2016 for marine natural products (MNPs), with 757 citations (643 for the period January to December 2016) referring to compounds isolated from marine microorganisms and phytoplankton, green, brown and red algae, sponges, cnidarians, bryozoans, molluscs, tunicates, echinoderms, mangroves and other intertidal plants and microorganisms. The emphasis is on new compounds (1277 in 432 papers for 2016), together with the relevant biological activities, source organisms and country of origin. Reviews, biosynthetic studies, first syntheses, and syntheses that led to the revision of structures or stereochemistries, have been included.
1 Introduction
This review is of the literature for 2016 and describes 1277 new compounds from 432 papers, a 5% reduction from the 1340 new compounds in 429 papers reported for 2015.1 As in previous reviews, the structures are shown only for new MNPs, or for previously reported compounds where there has been a structural revision or a newly established stereochemistry. Previously reported compounds for which first syntheses or new bioactivities are described are referenced, but separate structures are generally not shown. Where the absolute configuration has been determined for all stereocentres in a compound, the identifying diagram number is distinguished by addition of the † symbol. The new format for this review introduced for the previous two reviews1,2 has been retained, with only a selection of highlighted structures (142) now shown in the review. Compound numbers for structures not highlighted in the review are italicised, and all structures are available for viewing, along with their names, taxonomic origins, locations for collections, and biological activities, in an ESI† document associated with this review. The Reviews section (2) contains selected highlighted reviews, with all other reviews referenced in a section of the ESI.† This review welcomes two new authors, Anthony Carroll and Rohan Davis, from Griffith University, Australia into the team.2 Reviews
A 65% increase in the number of MNP-related reviews appeared in 2016 compared to 2015. Of the 144 reviews published in this period, 34% focussed on specific compounds or compound classes, 28% related to the bioactivities of MNPs, 29% had an organism or geographic focus while the remainder were more general. The journal Marine Drugs accounted for 29% of all reviews published in the year. Eleven reviews that highlight significant aspects of MNP studies are discussed here and the remaining 133 are listed in the ESI.† MNPs reported in 2015 have been comprehensively reviewed.2 The definitive series of reviews by Newman and Cragg on NPs as drugs has been updated and expanded for the period 1981 to 2014.3 MNPs as drugs and drug candidates have also been reviewed by the same authors.4 With the increasing study of MNPs from China, it is timely that a review of the Chinese Marine Materia Medica (CMMM) has appeared.5 The definition of ‘marine derived fungi’ has been the subject of on-going debate and a review highlights this issue and proposes a new consensus definition.6 Algae are a major source of marine derived fungi and the chemistry of these organisms has been reviewed.7 A comprehensive review discusses the diversity of holothurian triterpene glycosides and their bioactivities reported over the last six decades.8 The diterpenoid chemistry of gorgonians and the diverse chemistry derived from Spongia species have been reviewed.9,10 A review of the role of natural product biosynthetic gene-clusters in bacterial ecology and evolution provides an interesting perspective on the drivers of chemical diversity within microorganisms.11 The challenges of dereplication in MNP studies and LCMS and NMR strategies to efficiently recognise known bioactive MNPs has been reviewed.12 Updating the MarinLit database13 continues to be an essential requirement for the preparation of this review and the structures and literature derived from this database form the basis for this review.3 Marine microorganisms and phytoplankton
3.1 Marine-sourced bacteria
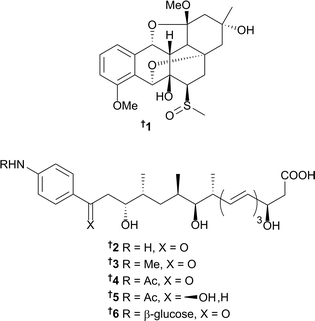
The number of new natural products from marine bacteria reported in 2016 (179) is a modest increase from the average for those reported in the previous three years (161), but a significant increase from the average for 2010–2012 (115). The genus Streptomyces continues to be the dominant force behind the discovery of new and exciting chemistry; numerous compounds from this prolific genus have also provided enticing bioactivities that will no doubt impact chemical biology and drug discovery/development in the future. A sediment-derived Streptomyces griseus M268 (Kiaochow Bay, China) was the source of grisemycin 1, which contains an unusual ether-bridged system and a methylsulfinyl moiety; this is the first sulphur-containing angucyclinone to be reported.14 Strain M268 had previously been shown to produce the epoxybenz[a]anthracene derivative, kiamycin;15 and both compounds had their absolute configurations assigned following analysis of X-ray diffraction data.14 Large-scale fermentation of a Streptomyces strain (SNM31) (from intertidal mudflats, Buan, Republic of Korea) and extensive chemical investigations yielded the new p-aminoacetophenonic acid derivatives mohangic acids A–E 2–6. Mohangic acid E 6 is the first glycosylated derivative for this particular chemotype. No significant in vitro cytotoxicity towards various human cancer cell lines nor antimicrobial activity against pathogenic bacteria and fungi was identified. When all five congeners were tested for cancer chemoprevention, using a quinone reductase (QR) assay at 20 μM, only mohangic acid E 6 displayed any activity (causing a 2.1-fold increase in QR activity compared to the control), suggesting the glucose moiety is important for QR activity in this chemotype.16Additional chemical investigations on the fermentation culture derived from a Streptomyces strain CNH189,17 yielded the ansalactams B–D 7–9 along with the previously identified metabolite, ansalactam A.17,18 Compounds 7–9 represent three new carbon skeletons and illustrate the plasticity within the ansamycin biosynthetic pathway. Ansalactams B–D displayed moderate antibacterial activity towards MRSA.
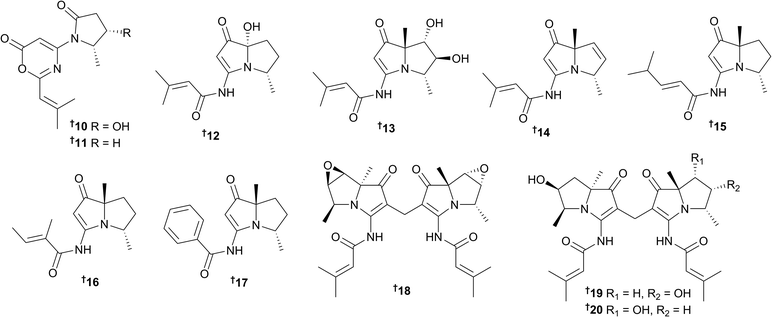
Spinoxazines A 10 and B 11, which both possess the rare structural motif, 1,3-oxazin-6-one, along with six new bohemamine analogues bohemamines D–I 12–17 were isolated from the extract of Streptomyces sp. SNB-048 (sand sample, Bahamian tidal flat).19 Additional research on another Bahamian-collected bacterial strain, Streptomyces spinoverrucosus SNB-032 by the same research group, yielded three new dimeric bohemamines, dibohemamines A–C 18–20 that were subsequently shown to be formed via a non-enzymatic process with formaldehyde, which was detected in the culture.20 This mild dimerisation process was exploited to generate a series of semi-synthetic analogues for biological evaluation. Dibohemamines B and C displayed potent activity against the non-small cell lung cancer cell line A549, with IC50 values of 140 and 145 nM, respectively.20
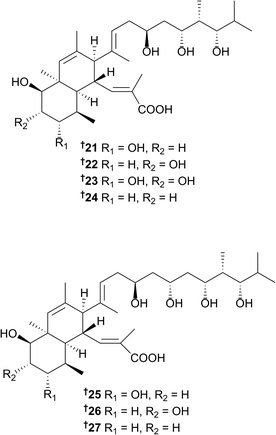
Two separate studies on two different Streptomyces strains (one collected in Papua New Guinea and the other in China) each yielded new nahuoic acid metabolites.21,22 Nahuoic acids B–E, along with the known polyketide natural product, nahuoic acid A 21,23 were identified and reported from the Chinese strain.21 Shortly after the first publication of nahuoic acids B–E,21 another research group published a study22 using the same trivial names to identify several different congeners. The first total synthesis of one of the new nahuoic acid congeners, nahuoic acid Ci (Bii),24 presented the authors with the opportunity to correct this situation.24 The analogues reported in the second study22 have subsequently been named nahuoic acids Bi–Ei22, 23, 25 and 26 while the congeners from the original study21 have been named nahuoic acids Bii–Eii23, 24, 27, and 25. Thus, a total of seven analogues (including nahuoic acid A 21) of this architecturally intriguing polyketide structure class have been isolated to date. It should be noted that based on the new terminology, nahuoic acid Ci is the same as nahuoic acid Bii, and nahuoic acid Di has the same structure as nahouic acid Eii. Several of these polyketides have been shown to display selective inhibition of SETD8 (a lysine methyl transferase) in U20S osteosarcoma cells.22 With methylation events playing important roles in the epigenetic regulation of gene expression, the discovery of new small molecules that selectively modulate enzymes controlling these events, may impact future cancer research.
The glycosidic spirotetronates tetrocarcin N 28 and O 29 were isolated from a Micromonospora sp. culture (sediment sample, Bohai Bay, Dalian, China) following a PCR-based genetic screening method that targeted the dTDP-glucose-4,6-dehydratase gene.25 Compounds 28 and 29 displayed modest antibacterial activity against Bacillus subtilis. Preliminary structure–activity relationship data indicated that sugar moieties at C-9 and the C-32 formyl moiety found in the previously isolated tetrocarcin congeners (not shown)26,27 are important for the antibacterial activity of this unique compound class.
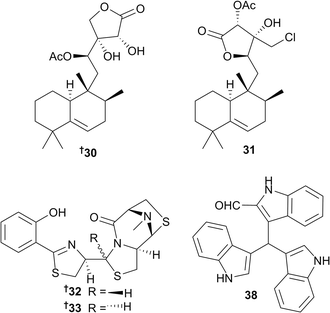
The halimane-type diterpenoids micromonohalimane A 30 and B 31 (Micromonospora sp. from the ascidian Symplegma brakenhielmi, Florida, USA) showed modest antibacterial activity against MRSA. Additional biological evaluation of 31 suggested that micromonohalimane B is bacteriostatic, rather than bacteriocidal.28 Ulbactins F 32 and G 33 are two polycyclic thiazoline congeners isolated from a culture extract of a sponge-derived Brevibacillus sp. (TP-B0800), collected off the coast of Japan. These compounds both inhibited migration of tumour cells in the submicromolar to micromolar range.29 From the gastrointestinal tract of a fish dredged near the South Orkney Islands (Antartica), a psychrotolerant bacterial strain identified as Vibrio splendidus was the source of 15 bis- and tris-indole derivatives, six of which 34–39 were new indole alkaloids. Another new bisindole analogue 40 was obtained from an additional psychrotolerant strain, Arthrobacter psychrochitiniphilus, that was isolated from the excrement of penguins. While some of these indole metabolites showed moderate activity towards several Gram-negative and -positive bacteria, trisindolal 38 was shown to be the most cytotoxic congener from the series following in vitro screening against an 11 human tumour cell line (HTCL) panel, being most active towards human breast cancer (MAXF401) and melanoma (MEXF 462) cell lines.30
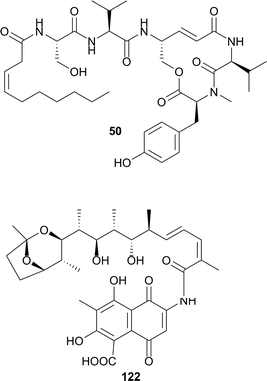
A total of 130 α-proteobacteria belonging to the Rhodospirillaceae family, which were sampled from oceans around the world, have been cultured and investigated for their ability to produce a specific class of lipopeptides, collectively known as the thalassospiramides.31 Twenty-one new thalassospiramides A6–A11 41–46, B3–B5 47–49, C2 50, E2 51, F1–F3 52–54, H 55, H1–H3 56–58, I 59, I1 60 and J 61 were identified and evaluated for not only their ability to inhibit human calpain but also as potential neuroprotective agents. In the calpain in vitro assay, 12 thalassospiramides were tested, and several single- and double-digit nanomolar inhibitors were revealed; thalassospiramide C2 50 was the most potent lipopeptide with an IC50 of 1.6 nM. Subsequently, nine of the best calpain inhibitors were chosen for testing in a murine neuroprotective assay; thalassospiramides A4 (previously reported analogue), H 55 and H1 56 significantly reduced the neurotoxic response in the mouse model. These studies indicated that this unique class of lipopeptides may have potential use in neurodegenerative conditions such as Alzheimer's disease. Furthermore, the authors of this paper also investigated the diversity of biosynthetic gene cluster (BGC) architectures by sequencing the genomes of 28 Rhodospirillaceae strains, which identified three types of dysfunctional BGCs and four functional BGCs that corresponded to the four thalassospiramide production patterns. This family-wide genome sequencing identified seven BGCs in total, five of which were new to science. This study, which successfully applied biochemical and genomic approaches to the discovery of genes and mechanisms associated with thalassospiramide biosynthesis, will greatly assist scientists that research these novel bioactive lipopeptides in the future.
Additional new metabolites were also identified from the genera Streptomyces62–121,32–62Salinispora122 and 123,63,64Bacillus124–130,65–69Pseudoalteromonas131 and 132,70Haliangium133,71Thalassotalea134–138,72Pseudomonas139 and 140,73Shewanella141,74Pontibacter142,75Brevibacterium143–145,76Streptosporangium146–148,77Streptomonospora149–152,78Verrucosispora153,79Micromonospora154 and 155,80Pseudonocardia156–163,81,82Nocardiopsis164–176,83,84Williamsia177,85 and an unidentified metagenome clone 178 and 179.86 Several structure revisions were reported during 2016, including thiasporine A 180,87 xiamenmycin A 181,88 and xiamenmycin C 182,89 halichomycin 183,90 and iso-naseseazine B 184.91 Furthermore, total syntheses of a number of bioactive or architecturally attractive MNP scaffolds have also been successfully completed and reported in 2016. For example, the isolation (from Salinispora arenicola), structure elucidation and subsequent total synthesis of rifsaliniketal 122 (ref. 63) along with the known and related rifamycin congeners salinisporamycin,92 and salinketals A and B,93 were reported. Other total syntheses of marine bacterial metabolites include (±)-spiroindimicins B and C,94 bacilosarcin C,95 cyclomarins A96 and D,97 cyclomarazines A and B,98 nitropyrrolins A, B and D,99 actinophenanthroline A,100,101 fijiolide A,102 heronamides A–C,103 dermacozines A–C.104 The proposed chemical structure of marineosin A was also successfully synthesised; the structure of the target compound was established following X-ray data analysis.105 While the NMR data of the synthetic compound showed a high degree of similarity with the previously reported natural product, there were also noticeable differences. Additionally, differences in the physical properties (white solid vs. colourless oil) and specific rotation data (opposite signs in the same solvent) indicated that the isolated and synthetic compounds were not the same. It was postulated that these data discrepancies could be potentially attributed to atropisomers, diastereoisomers or structural isomers, however further structure elucidation studies on these unusual spiroaminals are required.105
3.2 Marine-sourced fungi (excluding from mangroves)
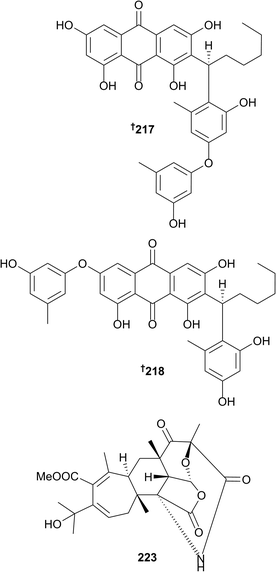
The number of new compounds reported from marine fungi (not associated with mangroves) has decreased slightly with 328 new compounds reported in 2016 compared to 369 in 2015. A number of new metabolites have been obtained from the genera Acaromyces (naphtha-[2,3-b]pyrandione analogue 185 and thiazole 186),106Acremonium (steroid 187 and cyclopentanone derivative 188,107 diterpene glycosides 189–192,108 sesquiterpenoids 193–207 (ref. 109) and benzophenone 208 (ref. 110)), Alternaria (cyclopentenone 209 (ref. 111)), Arthirinium ((+)-5-chlorogriseofulvin 210 (ref. 112)) and Ascotricha (polyketide-derived linear 211–213 and macrocyclic 214 polyesters, hexaketide 215 and (−)-orthosporin 216 (ref. 113)). The racemate of 5-chlorogriseofulvin has been synthesised but this is the first isolation of (+)-5-chlorogriseofulvin 210 from a natural source (simultaneously obtained from Penicillium canescens below)114 and although (+)-orthosporin has been obtained from a terrestrial fungus and (−)-orthosporin has been synthesised, the current report represents the first isolation of (−)-orthosporin from a natural source.113 The genus Aspergillus has again been well studied and has yielded many new metabolites. A deep-sea strain of Aspergillus versicolor yielded a series of phenolic compounds aspergilols A–F 217–222, of which 217 and 218 possess a new scaffold with a carbon–carbon fusion of an orcinol unit to an anthraquinone.115Production of aspergstressin 223, an unusual hybrid polyketide–terpenoid metabolite was induced in an Aspergillus sp. obtained from a hydrothermal vent by cobalt ion stimulation.116 Other metabolites produced by Aspergillus species included meroterpenoids 224–226,117 butenolide derivatives 227–229,118 aromatic nucleoside 230,119 terpenoids 231 and 232,120 2-benzylpyridin-4-one derivatives 233 and 234,121 dimeric naphthopyrone 235,122 naphthopyranone 236,123 diorcinol 237 (through cocultivation with Ircinia felina),124 α-pyrone polyene 238 (ref. 125) (the structure appears in a screening library but no source is given for the compound), chlorinated depsidone 239, folipastatin 240 and 2-chlorounguinol 241 (the last two as first time marine isolates)126 and sesterterpenoids 242 and 243.127 Fumiquinazoline-type alkaloids 244–254 (ref. 128) were isolated along with cottoquinazolines B–D, the structures of which were revised to 255–257, enantiomers of the structures originally reported.128 Other compounds isolated from Aspergillus species include 20-nor-isopimarane diterpenoids 258–262,129 tetranorlabdane diterpenoids 263 and 264,130 cyclic dipeptide 265,131 asteltoxins 266 and 267 and chromone 268,132 eremophilane sesquiterpenes dihydrobipolaroxin (the configuration of which was determined as 269), 270–273,133 phenolic bisabolanes 274 and 275 (ref. 134) and (R)-(−)-hydroxysydnoic acid 276 (ref. 135) and the methyl naphthoate 277,136 these last two being obtained as first time marine isolates. Reisolation of pseurotin A2, from a different strain of Aspergillus fumigatus to that from which it was originally obtained, led to revision of the structure to 278.137
New metabolites were obtained from the genera Biscogniauxia (isopyrrolonaphthoquinone 279 and sansalvamide A amide 280; the latter a first time MNP),138Chaetomium (dioxopiperazine alkaloid 281 (ref. 139) and cytochalasins 282 and 283 (ref. 140)), Chondrostereum (new triquinane-type sesquiterpenoids 284–286 and the sesquiterpenoid anhydroarthrosporone 287 as a first time MNP),141Cladosporium (azaphilones 288 and 289, bicyclic diol 290 and 1-(3,5-dihydroxy-4-methylphenyl)propan-2-one 291, the last two as new NPs),142Clonostachus (isocoumarin 292 (ref. 143)) and Cochliobolus diethylene glycol phthalate ester oligomers 293–299.144Coniothyrium cereale yielded the unusual nitrogenous heterodimers 300 and 301, comprising sterol and polyketidic phenalenone portions, triketone 302 (obtained as an acetone adduct) and phenalenone derivative 303, containing an oxepane–imine–dione ring formed via an unprecedented imine functionality between two carbonyl groups.145 Although 301 had previously been isolated from a terrestrial fungus as a C-15 epimeric mixture, it was isolated here as a first time MNP and in an epimeric pure form (15S).145
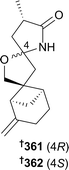
Naphthalenones 304 and 305 and depsidone 306 were obtained from the genus Corynespora,146 diketopiperazines 307–309 from a Dichotomomyces species,147 asteltoxin derived dimers 310–312 from a diethyl sulfate (DES) mutated Emericella strain148 and an indole alkaloid 313 (ref. 143) and anthraquinone–xanthone polyketides 314–319 (ref. 149) from Engyodontium species. The absolute configurations of the known JBIR-97/98 320 and JBIR-99 321 co-isolated from the Engyodontium species were also determined.149 New metabolites were obtained from the genera Fusarium (polycyclic quinazoline alkaloid 322,150 β-resorcylic macrolides 323–326 (ref. 151) and relgro (absolute configuration determined as 327 (ref. 151)), octahydronaphthalene derivative 328 (ref. 151) and cyclic hexadepsipeptide 329 (ref. 152)), Hypocrea (tyrosol derivative 330 (ref. 153) and trichodenol B 331, the latter as a first time marine isolate (ref. 153)), Neosartorya (cyclotetrapeptides 332 and 333,154 diketopiperazine derivative 334,154 terpenoids 335–339 (the last two of which are reported from a marine source for the first time),155 2-naphthoic acid derivative 340,155 isocoumarin 341,155 polyketides 342 and 343,156 and benzoic acid derivatives 344–350 (ref. 156)), Nigrospora (cyclohexadepsipeptides 351–353,157 scopularide A (the configuration of which was determined as 354 (ref. 157)) and hydroanthraquinone dimer 355 (ref. 158)) and Paecilomyces (pyrrolooxazine 356,159 bicyclic fatty acids 357 and 358 (ref. 160) and indole alkaloids 359 and 360 (ref. 161)). A compound with the same planar structure as 360 was previously obtained from a terrestrial fungus. Sporulaminals A 361 and B 362, epimeric spiroaminal derivatives, were isolated from Paraconiothyrium sporulosum and successfully separated. The epimerisation was found to be induced by temperature, pH and water addition.162
As always, the genus Penicillium has been a prolific source of new metabolites, including meroterpenoids 363–366,163 flavone 367,164 spiroketal 368 (ref. 165) and macrolides 369 and 370.166 A bioinformatics tool, MeHaloCoA (Marine Halogenated Compound Analysis) was developed and included in the software R. Integration of the tool into a dereplication approach resulted in identification of known and new halogenated compounds including (+)-5-chlorogriseofulvin 210 (ref. 112) and griseophenone I 371 from Penicillium canescens.114 Chrysamides A–C 372–374 are dimeric nitrophenyl trans-epoxyamides obtained from Penicillium chrysogenum (deep sea sediment) of which 372 and 373 possess an unprecedented centrosymmetric dimer skeleton and 374 suppresses production of the pro-inflammatory cytokine interleukin-17.167 A number of metabolites have been obtained from strains of Penicillium purpurogenum mutated by either exposure to neomycin (cyclopentachromone 375 (ref. 168) and cyclic dipeptide 376 (ref. 169)) or DES (oxaphenalenones 377 (ref. 170) and bacillosporin C 378,170 and alkaloids 379–382, with 382 representing a structural revision of asperverin171). Of these, chromosulfine 375 is the first cyclopentachromone reported to possess a sulfide chain168 and although bacillosporin C had been isolated previously from both terrestrial and marine fungi, the absolute configuration was reported for the first time.170
Other metabolites isolated from the Penicillium genus include tanzawaic acid derivative 383,172 meroterpenoid 384,173 furan-2-carboxylic acid derivatives 385–387,173 polyketides 388–398,174399 and 400 (ref. 175) and cerebrosides 401 and 402.176 The planar structures of 399 and 400 (ref. 175) appear in a screening library but no source information is given. Cerebrosides 401 and 402 (ref. 176) were named penicillosides, but that name had already been assigned to some unrelated compounds, previously isolated from a succulent plant.177 Different culture conditions for a sponge-derived Penicillium sp. led to variation in production of some macrocyclic polyketides. Under standard growth conditions, the culture yielded 12-keto-10,11-dehydrocurvularin 403 (ref. 178) but application of a previously published procedure of experimental design and chemometric analysis to optimise production of related metabolites, led to production of 15(S)-cis-10,11-epoxycurvularin 404 (ref. 178) and cyclothiocurvularins 405–407 (405 and 406 diastereoisomeric) and cyclosulfoxicurvularins 408 and 409; curvularins condensed with a thiolactate residue.178 Results of biosynthetic feeding experiments utilising [U-13C315N]-L-cysteine and spontaneous formation of cyclothiocurvularins from 10,11-dehydrocurvularin indicated that formation of cyclothiocurvularins may be a detoxification process for the fungus.178
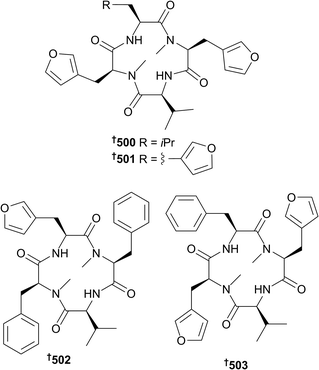
Epidithiodiketopiperazines 410–412,179 verrucosidin derivative 413,180 macrolides 414–417,181 azaphilones 418–421,182 diene aldehyde 422 (ref. 183) and tetrahydrofuran derivatives 423–426,183 and fatty acid esters 427 and 428 (ref. 184) were also obtained from Penicillium strains as were alkaloids haenamindole and citreoindole, the structures of which were revised to 429 and 430, respectively based on NMR data and C3 Marfey's methodology.185 Citreoindole had previously been obtained from a terrestrial Penicillium species but the current isolation185 was the first from the marine environment. 6-Methylcurvulinic acid 431, a known terrestrial plant metabolite, was also isolated from the marine environment for the first time.186 Reexamination of the published NMR spectral data of (22E)-24-methylcholesta-8(14),22-diene-3β,5α,6β,7α-tetraol, isolated from a sponge-associated Penicillium sp., indicated that the structure was in fact 5α,6α-epoxy-(22E,24R)-24-methylcholesta-8(14),22-diene-3β,7α-diol 432.187 New natural products were isolated from the genera Pestalotiopsis (chlorinated diphenylmethanes 433–436),188Peyronellaea (isocoumarin derivatives 437–441),189Phoma (isocoumarin derivative 442,190 steroid 443 (ref. 191) and 444–450 (ref. 192)), Pleosporales (azaphilone 451),193Pseudallescheria (diasteroisomeric sesquiterpenes 452 and 453,194 cyclopiazonic acid analogue 454,194 diketopiperazines 455 and 456 (ref. 194) and 457 (ref. 195) and lactone derivative 458 (ref. 195)). Chromone derivatives 459 and 460 were obtained from Rhinocladiella sp.,143 and aromadendrane sesquiterpenoids 461–467 (ref. 196) from the genus Scedosporium. The genera Scopulariopsis (xanthones 468 and 469 and the known AGI-B4, the configuration of the last determined as 470, phenolic sesquiterpenes 471 and 472, alkaloid 473 and α-pyrone derivative 474),197Simplicillium (linear peptides 475–482),198Spiromastix (phenolic lactones 483–495),199Sporidesmium (diphenyl ether derivative 496)200 and Stachybotrys (isoindolone 497 (ref. 201) and phenylspirodrimanes 498 and 499 (ref. 202)) also yielded new metabolites. Endolides A 500 and B 501, unusual N-methylated tetrapeptides that contain the rare amino acid 3-(3-furyl)-alanine, were isolated from a sponge-derived Stachylidium sp.203 Neither demonstrated any cytotoxicity in a range of bioassays but endolide A 500 showed binding affinity to the vasopressin receptor whilst endolide B 501 possessed selective affinity for the serotonin receptor.203 Supplementation of the culture medium with phenylalanine produced two further analogues, endolides C 502 and D 503 and negatively affected endolide A 500 production.204 A series of biosynthetic feeding experiments established that all of the carbon atoms of the rare amino acid were derived from the shikimate pathway except for the N-methyl group which arose from the methyl group of methionine.204
The genus Talaromyces yielded isocoumarin derivative 504,200 diphenylketones 505 and 506 (ref. 205) and xanthones 507 and 1,4,5-trihydroxy-2-methyl-9H-xanthen-9-one 508 (the latter a known synthetic compound but isolated here as a new NP)205 whilst Tolypocladium sp. yielded decalin-tetramic acids 509 and 510 (ref. 206) and the Trichobotrys genus was the source of macrodiolides 511–513.207 A number of metabolites were isolated from the genus Trichoderma including disulfide-bridged dipeptide 514,208 diterpene 515,209 norditerpene 516,210 decalin derivatives 517 and 518 (ref. 211) and long-chain peptaibols 519–521.212 An azaphilone derivative 522 was obtained from Xylariales sp.213 and an unidentified fungus isolated from a brown alga yielded diterpenes 523–525.214 Synthesis of the proposed structure of trichoderin A, an anti-tuberculosis aminolipopeptide originally obtained from a sponge-derived Trichoderma sp., has revised the configuration at C-6 from (S) to (R) as in 526.215 Total synthesis of the originally assigned structure of cereoanhydride C (Coniothyrium cereale, green alga Enteromorpha sp.), led to reassignment of the structure as spiroketal 527.216 Total synthesis of the tetracyclic cyclopiane diterpenes conidiogenol (Penicillium chrysogenum, red alga, Laurencia sp.) and conidiogenone B (Penicillium sp., sediment), led to revision of their configurations to 528 and 529 respectively.217 Total synthesis of the cyclodepsipeptides clavatustide A and B (Aspergillus clavatus, crab Xenograpsus testudinatus) has been achieved via a seven step method from (R)-phenyllactic acid.218 The indole alkaloids notoamides F and R (Aspergillus sp., mussel Mytilus edulis galloprovincialis) and the related spirooxindole (−)-sclerotiamide, have been synthesised in a convergent manner from commercially available Seebach acetal.219 A first synthesis of notoamide I was also claimed219 but the synthesis had already been reported.220 Denrodolides C and D (Dendrodochium sp., sea cucumber Holothuria nobilis) have been prepared from commercially available materials.221 The macrolide gliomasolide C (Gliomastix sp., sponge Phakellia fusca) has been synthesised,222 as has the lipopeptide fellutamide A (Penicillium fellutanum, fish Apogon endekataenia)223 and the alkaloids penipanoid C and 2-(4-hydroxybenzyl)quinazolin-4(3H)-one (Penicillium paneum, sediment).224 A number of new activities were reported for known fungal metabolites. Diorcinol exhibited acetylcholinesterase inhibition,225 naphthopyrone TMC-256C displayed neuroprotective and anti-neuroinflammatory effects226 and pannorin, alternariol and alternariol-9-methylether were inhibitors of the signal pathway modulating enzyme glycogen-synthase-kinase 3B, (pannorin 530 also a first time MNP).227 A study which analysed two Penicillium strains at different time points throughout the culture period by HPLC-DAD-HRMS indicated that a greater chemodiversity of metabolites may be obtained from the one strain by performing extractions at different times throughout the culture period including early and late growth phases.228 Studies on the effect of light on the biosynthesis of the anticancer polyketide 1403C (haloroquinone, SZ-685C) in Halorosellinia sp., indicated that light significantly increased production of this metabolite (74%) over production in the dark.229 Comparison of the secondary metabolite profile of a monoculture of Aspergillus flavipes with that of a strain cocultured with a Streptomyces strain isolated from the same marine sediment, indicated that physical contact with the bacterium induced production of six cytotoxic cytochalasans (such as aspochalasin P) in the fungus.230
3.3 Fungi from mangroves
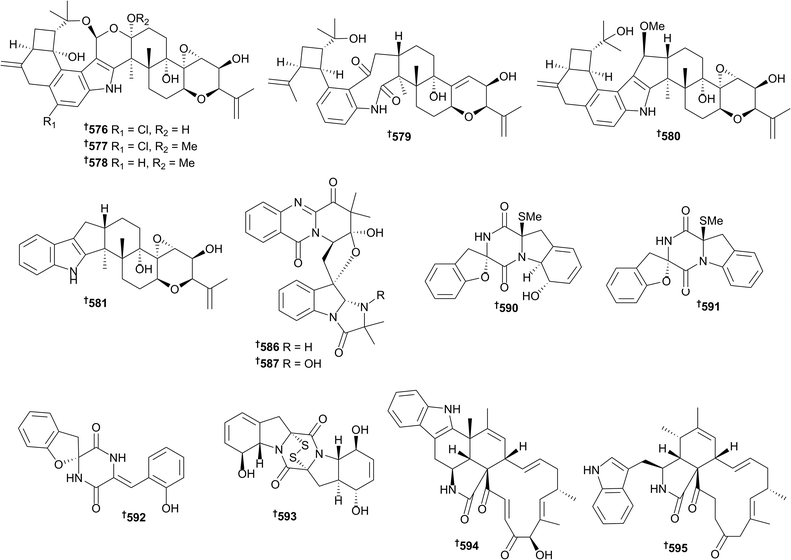
There has been a continued increase in the number of new metabolites reported from mangrove-associated fungi (142 in 2016, 126 in 2015 and 108 in 2014), with the majority coming from endophytic species. An Alternaria sp. yielded altenusin derivatives 531–535 (ref. 231) and a species denoted “Ascomycota sp.” (although this is a phylum not a genus) was the source of 2,3-diaryl indone derivatives 536–538 (as racemates of M- and P-helicity enantiomers) and isobenzofuran derivatives 539 and 540 (also as racemates).232 Endophytic fungi of the genus Aspergillus were the source of sesterterpenoids 541 and 542 (ref. 233) and isocoumarin derivatives 543–548, of which 545 and 546 are known synthetic intermediates but were obtained as new NPs.234 Pyrone 549, naphthoquinone 550 and cyclic urea 551 were isolated from Astrosphaeriella nypae235 whilst phenyl derivatives 552–555 were obtained from a Botryosphaeria sp.236Capnodium sp. yielded the eremophilane sesquiterpene 556,237 dichlororesorcinol derivatives 557–559 were obtained from Cosmospora vilior238 and endophytic Eurotium rubrum was the source of enantiomers of a 2-benzofuran-1(3H)-one, 560 and 561, the racemate of which exhibited potent 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging activity.239 Endophytic Lasiodiplodia species yielded polyketides 562–565 (ref. 240) and a series of preussomerin analogues, of which the chlorinated 566 and 567 were new compounds, 568 was a new natural product and 569–575 were obtained from the marine environment for the first time.241 The knowledge that genome mining of a related fungal strain indicated the presence of biosynthetic gene clusters for terpene production, prompted researchers to investigate an endophytic strain of Mucor irregularis for secondary metabolite content. This resulted in isolation of rhizovarins A–F 576–581, structurally unique indole diterpenes.242 Rhizovarin A 576 possesses an unprecedented linkage of an acetal and a hemiketal whilst rhizovarins B 577 and C 578 possess an acetal linked to a ketal functionality. All three compounds contain an unprecedented eight-membered cyclic ether moiety coupled to five other rings.242Nectriacids A–C 582–584 and 12-epicitreoisocoumarinol 585 are polyketides obtained from an endophytic Nectria sp. of which nectriacids B 583 and C 584 exhibited potent inhibition of α-glucosidase.243 Endophytic Neosartorya udagawae was the source of four quinazoline-containing alkaloids, neosartoryadins A 586 and B 587 and fiscalins E 588 and F 589 of which 586 and 587 are representatives of a new class of such alkaloids containing an unique 6/6/6/5 quinazoline ring system directly connected to a 6/5/5 imidazoindolone ring system which was speculated to be biosynthesised from L-tryptophan, anthranilic acid, L-valine and 2-aminoisobutyric acid.244 Neosartoryadins A 586 and B 587 also exhibited activity against the H1N1 influenza virus (more potent than the positive control).244 The Penicillium genus has been the source of a number of mangrove fungus-associated metabolites including the diketopiperazines spirobrocazine A–C 590–592 and bisthiodiketopiperazine derivative brocazine G 593, of which brocazine G 593 displayed potent cytotoxicity to sensitive and cisplatin-resistant HTCLs and strong activity against S. aureus.245 Chaetoglobosins penochalasin I 594 and penochalasin J 595 were obtained from endophytic P. chrysogenum of which penochalasin I 594 possessed an unprecedented hexacyclic 6/5/6/5/6/13 fused ring system.246
Other metabolites obtained from Penicillium strains include benzopyran derivative 596,247 dihydroisocoumarins 597–599 (ref. 248) and 600–602,249 polyketides 603 (ref. 250) and 604 and 605,251 azaphilone 606 (ref. 252) and meroterpenoid 607.253 Other genera of mangrove-associated fungi to yield new metabolites included Pestalotiopsis (polyketides 608–613 (ref. 254) and macrolides 614–620 (ref. 255)), Phomopsis (chromone derivatives 621–624 (ref. 256) and cytochalasins 625–628 (ref. 257)), Pseudolagarobasidium (sesquiterpenes 629–648),258Rhytidhysteron (chromone 649–653),259Stemphylium (tetrahydroanthraquinone derivatives 654–656),260Talaromyces (isocoumarins 657–662 and benzofurans 663 and 664)261 and Trichoderma (diterpenoids 665 and 666).262 Some chamigrane sesquiterpenes 667–672 were isolated from a basidiomycetous endophyte which was only identified to family (Meruliaceae).263 Total synthesis of the purported structures of cephalosporolides H and I and penisporolide B (all obtained from Penicillium spp.) and their possible diastereoisomers has indicated that the relative configuration of the tricyclic core should be revised to 673–675, and that the sidechains also require an unknown constitutional structure revision.264 Total synthesis of the proposed structure of a phenylethanol derivative obtained from a Penicillium sp. and comparison of the optical rotation and NMR data of the synthetic product with those reported for the natural product, has indicated that the proposed structure must be incorrect,265 whilst total synthesis of the purported structure of phomolide G (Phomopsis sp.) has indicated that the configuration should be revised to 676, the C-3 epimer of that originally proposed.266 Five gene cluster knockout mutants of a strain of Apergillus ustus were obtained and experiments involving the enzymes relevant to terpene synthesis from each gene cluster indicated that biosynthesis of the ophiobolin skeleton involved these five gene clusters and that they are responsible for C15, C20, C25 and C30 terpenoid biosynthesis.267
3.4 Cyanobacteria
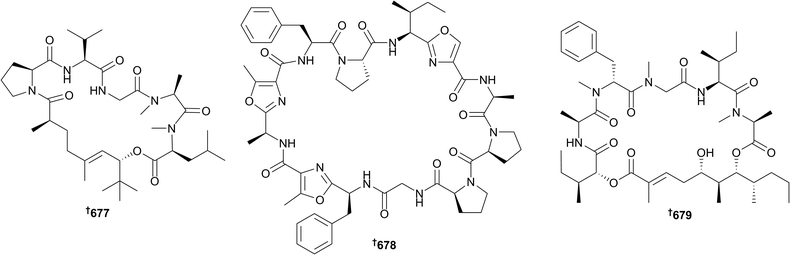
There has been a slight decrease in the number of new natural products from cyanobacteria since 2015, however the numbers of reports over the past 5 years have been consistently low overall (28 in 2016: 31 in 2015; 20 in 2014; 9 in 2013; 31 in 2012). As in previous years, the majority of new metabolites from this phylum are peptidic in nature and most have some biological activity recorded; significant examples include janadolide 677,268 wewakazole B 678,269 and odoamide 679.270Janadolide 677 (Okeania sp., Janado, Japan), a cyclic peptide–polyketide hybrid possessing a rare tert-butyl moiety, showed potent activity towards Trypanosoma brucei brucei (IC50 47 nM) which was superior to the commonly used therapeutic drug suramin. Furthermore, significant selectivity (>212 fold) towards the trypanosome parasite was identified since no in vitro cytotoxicity towards the human cell lines MRC-5, HL60 and HeLa cells was noted at 10 μM.268 The cyanobactin wewakazole B 678 was obtained using MS-guided isolation from an extract of a Red sea specimen of Moorea producens (formerly Lyngbya majuscula), and was cytotoxic towards human MCF7 (IC50 0.58 μM) and H460 (IC50 1.0 μM) cancer cell lines.269 The most potent cytotoxin reported from cyanobacteria in 2016 was odoamide 679, obtained from bioassay-guided fractionation studies on an extract from a Japanese Okeania species. This new 26-membered cyclodepsipeptide had an IC50 of 26.3 nM against HeLa S3 human cervical cancer cells.270 Further biological evaluation on the previously identified cyanobacterial depsipeptide coibamide A271,272 has shown that this molecule inhibits Vascular Endothelial Growth Factor (VEGF) A/VEGF Receptor 2 (VEGFR2) expression and suppresses tumour growth in a nude mouse xenograft model of glioblastoma (U87-MG). Similarities between coibamide A and apratoxin A273 biological outputs (cell morphology, VEGFR2 expression, macroautophagy signalling) suggests these two cyanobacterial metabolites share a common mechanism of action.274 Other new metabolites of either peptidic or non-peptidic nature were obtained from the genera Hydrocoleum (680 and 681),275Leptolyngbya (682),276Lyngbya (683),277Moorea (684–686,278 and 687 (ref. 279)), Nodularia (688–692)280Synechocystis (693–699),281Nostoc (700),282Okeania (701),283Caldora (702),284 and Symploca (703)285 and one potentially new genus (704)286 that is closely related to Trichodesmium, Okeania and Oscillatoria based on 16S rRNA sequence analysis. The structure of caylobolide B 705 was corrected; the E-configuration between C-2/C-3 in the original 2010 paper was amended to a Z-configuration.287 The first total syntheses of several cyanobacteria metabolites, such as biselyngbyolide B,288,289 wewakazole B 678,290 odoamide 679,291 lyngbyastatin 7 (ref. 292) and cocosolide 703 (ref. 285) have been achieved. In order to establish the absolute configuration of the γ-pyrone containing polyketide, yoshinone A, and provide larger quantities of this bioactive compound for additional testing, the total synthesis of two possible diastereomers was conducted. NMR and optical rotation data comparison of the natural product and synthetic isomers enabled the absolute configuration of yoshinone A to be assigned as 706.293 Koshikalide, a cytotoxic 14-membered macrolide, had its relative configuration reported in 2010, with the absolute configuration unattainable due to scarcity of material (0.3 mg).294 The first total synthesis of this molecule has confirmed the relative configuration initially assigned, and enabled the assignment of the absolute configuration for this molecule.295 Studies towards the synthesis of specific structural motifs or fragments (e.g. polyhydroxylated moieties common to oscillariolide and phormidolides A–C,296 the aglycone of lyngbouilloside297), and analogues of bioactive products (for example largazoles,298 calothrins,299 and aeruginosins300) have been undertaken. Collectively, these studies will hopefully expedite and impact synthetic and medicinal chemistry efforts on biologically active cyanobacterial metabolites, and lead to suitable candidates in adequate quantities for future drug development.
3.5 Dinoflagellates
The number of new metabolites reported from dinoflagellates has dropped slightly with 11 compounds reported in 2016 compared with 15 in each of 2015 and 2014. Alexandrium ostenfeldii was the source of gymnodimine D 707 with two tetrahydrofuran rings in the macrocycle.301 The macrolides iriomoteolide-10a 708 and -12a 709 were obtained from an Amphidinium sp.302 Three new brevisulcatic acids (BSXs), ladder-frame polyethers 710–712 were isolated from Karenia brevisculcata,303 two new karlotoxins, polyketides 713 and 714 were obtained from Karlodinium veneficum,304 and the ladder-frame polyether prymnesin-B1 715 was obtained from the microalga Prymnesium parvum.305 The relative configuration of the C-3–C-12 portion of amphirionin-5 716 was assigned by stereodivergent synthesis of six diastereomeric model compounds and comparison of their NMR spectroscopic data with those reported for the natural product.306,307 The results of stereoselective and stereodivergent synthesis of the proposed C-79–C-104 fragment of symbiodinolide and stereodivergent syntheses of the C-79–C-97 and C-94–C-104 fragments have led to the reassignement of the relative configuration of this portion of 717.308,309 The complete biosynthetic pathway to the polyketide 13-desmethylspirolide C in Alexandrium ostenfeldii has been established by feeding experiments with 13C methyl-labelled methionine and application of the odd-even methylation rule.310 Feeding 13C-labelled acetates to Amphidinium sp. established that all of the carbons of amphidinin A and amphidinolide P were derived from acetates. The polyketide chain of amphidinin A was formed from one triketide and two diketide chains and three unusual isolated C1 units derived from C-2 of cleaved acetates, while the polyketide chain of amphidinolide P was formed from one pentaketide chain, two acetate units, and three isolated C1 units also derived from C-2 of cleaved acetates.311 Feeding of 15N-labelled intermediates into paralytic shellfish toxins (PSTs) in Alexandrium circinalis indicated that biosynthetic intermediates predicted from gene sequences are genuine precursors of PSTs and that a related compound derived from one of these precursors is a shunt product released from cells to reduce PST production.3124 Green algae
Only two new MNPs were reported from the phylum Chlorophyta in 2016 and only a further eight papers and four reviews have described studies on compounds previously reported from green algae. As part of a study of the edible green micro-alga, Chlorella sorokiniana, to measure the concentration of the macular protective compound lutein, two new MNPs, sesquiterpene chlorellatin A 718 and ergosterol derivative chlorellatin B 719 were identified.313 Both had previously been reported as synthetic derivatives. Studies on the bioactivity of known green algal compounds include the inhibition of red tide algae by extracts and compounds derived from Ulva intestinalis and U. prolifera with fatty acid esters and carotenoid metabolites being the most active components.314,315 4-Hydroxy-2,3-dimethyl-2-nonen-4-olide isolated from U. pertusa inhibited pro-inflammatory cytokine production in CpG-stimulated bone marrow-derived dendritic cells in the μM range.316 A proton NMR-based metabolomics study identified that dimethylsulfoniopropionate and acrylate derived from an unidentified Ulva species were sequestered by the sea hare Aplysia juliana and used as defensive compounds.317 The carotenoid astaxanthin318,319 and sulfolipids320 continue to be investigated for various biological activities.5 Brown algae
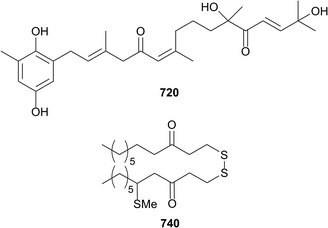
Twenty eight new compounds were reported from six species of brown algae in 2016 while a total of 46 papers and 11 reviews on brown algae were published. Twelve new meroditerpenoids, cystodiones G–L 720–725 and cystones A–F 726–731 along with eight known meroditerpenes have been reported from the Spanish brown algae Cystoseira usneoides.321 The compounds were tested for antioxidant and anti-inflammatory activity and all show radical scavenging activity and some (such as 720) were inhibitors of the production of the pro-inflammatory cytokine TNF-α in LPS-stimulated THP-1 human macrophages at 10 μM. Dictyopteris divaricata contained a cadinane sesquiterpene cadinan-4(15)-ene-1β,5α-diol 732 along with its known C-5 epimer and the norcadinane trans-3-norisocalamenen-4-ol 733.322 An additional sesquiterpene 734 was isolated from Taonia atomaria along with known compounds; five sesquiterpenes, a unsaturated fatty acid and a unsaturated fatty glycerol ether. A number of the compounds (including 734) inhibited bacterial adhesion and barnacle settlement.323 A study of the edible brown algae Cladosiphon okamuranus led to the characterisation of two steroids mozukulin A 735 and B 736, however neither compound showed activity in an anti-inflammatory assay.313 A series of six disulfides 737–742 and two known disulfides were isolated from the Greek alga, Dictyopteris membranacea. None of the compounds showed antibacterial activity against multidrug resistant S. aureus, but 740 showed anti-inflammatory activity, inhibiting NO production in a LPS stimulation assay (IC50 3.8 μM).324Extracts from eight New Caledonian Lobophora species (L. rosacea, L. nigrescens, L. crassa, L. abscondita, L. dimorpha, L. undulata, L. hederacea and L. monticola) were shown to bleach the scleractinan corals, Acropora muricata and Stylophora pistillata. Three C21 polyunsaturated alcohols lobophorenols A–C 743–745 were subsequently isolated from L. rosacea and shown to bleach A. muricata on contact.325 Known brown algal-derived meroterpenes have been the focus of several biological studies with atomaric acid active against Leishmania amazonensis,326 sargaquinoic and sargahydroquinoic acids possessing anti-adipogenic and pro-osteoblastogenic activities327 and 11-hydroxy-1′-O-methylamentadione reducing the effect of dextran sulfate-induced colitis in mice.328 Dolastane diterpenes from Canistrocarpus cervicornis inhibited HIV-1,329 and spartane diterpenes from Stoechospermum marginatum induced apoptosis in melanoma cells.330 The first total synthesis of dictyoxetane331,332 and the unnatural enantiomer (−)-erogorgiaene,333 were reported. Additionally, there were many papers dealing with the biological properties of brown algal lipids,334–337 polyphenolics338–348 and carotenoids.349–360
6 Red algae
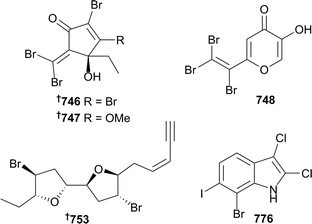
The reporting of thirty-seven new compounds from red algae in eight papers is close to the average for each of the previous five years. Four polybrominated C4–C8 hydrocarbons 746–749 (Plocamium australasica) include the ptilones A–C 746–748 which possess skeletons not previously seen in algal metabolites.361 Three C15-acetogenins 750–752 were isolated from Laurencia marilzae.362 The absolute configuration of elatenyne 753 (Laurencia elata363) was previously determined somewhat ambiguously by synthesis of a proposed diastereoisomer,364 but the absolute configuration has now been unambiguously determined by the crystalline sponge method, which enables the crystallographic analysis of non-crystalline compounds on the microgram scale. In this case, only ∼100 μg of elatenyne was required with ∼95% being recovered after the experiment. This represents the first real application of the crystalline sponge method to the determination of the absolute configuration of a natural product.365 A synthesis of (−)-bisezekayne A 754 (Aplysia oculifera366) has led to a structural revision and the determination of absolute configuration.367Sesquiterpenoids continue to be commonly encountered in the red algae, with nine brominated chamigrene and cuparane sesquiterpenoids 755–763 being obtained from Laurencia tristicha,368 the eudesmane 764 from L. obtusa,369 three brominated eudesmanes (selinanes) 765–767 and a brominated cycloeudesmane 768 from L. pinnata.370 Three meroterpenoids 769–771 with variable anti-oxidative activities were characterised from Hypnea musciformis.371 Halogenated indoles are another commonly encountered structural type, with eleven polyhalogenated indoles 772–782 from Rhodophyllis membranacea, including six which were the first isolation of bromo–chloro–iodo secondary metabolites, as exemplified by 776.372 Further related compounds were four brominated indole related alkaloids 783–786 (Laurencia similis),373 and bromophenol 787 (Odonthalia corymbifera).374
Papers describing work on new bioactivities for known compounds from red algae include the reporting of the larvicidal potential of obtusol (Laurencia dendroidea)375 against the dengue fever mosquito Aedes aegypti,376 bis(2,3-dibromo-4,5-dihydroxybenzyl) ether (BDDE) (Odonthalia corymbifera)377 as a novel PTP1B inhibitor and potential anti-diabetic agent,378 immunomodulatory effects of the mycosporine-like amino acids shinorine and porphyra-334 from Porphyra sp.,379 and the insecticidal activity guided isolation of palytoxin380 from Chondria armata. This report also presented refined NMR spectral assignments for palytoxin and claimed this as the first isolation of the toxin from a marine plant.381 Other studies included the diel variation of sesquiterpene elatol382 production: a chemical defense mechanism in Laurencia dendroidea,383 the geographic distribution along the SE Brazilian coast of natural products produced by L. dendroidea,384 a study on the sterol biosynthesis pathway in L. dendroidea by the cloning and functional characterisation of a cycloartenol cyclase,385 the biological activities of known compounds from three edible Gracilaria spp.,386 and the design and synthesis of analogues of galaxamide (Galaxaura filamentosa)387 as potential antitumor agents.388 Other synthetic studies included the first total synthesis of an antibacterial polyhalogenated monoterpene, (−)-anverene (Plocamium cartilagineum389),390 and the total syntheses391 of the Laurencia marilzae metabolites 12-epoxyobtusallene IV,392 obtusallene X,392 and marilzabicycloallenes C and D.393
7 Sponges
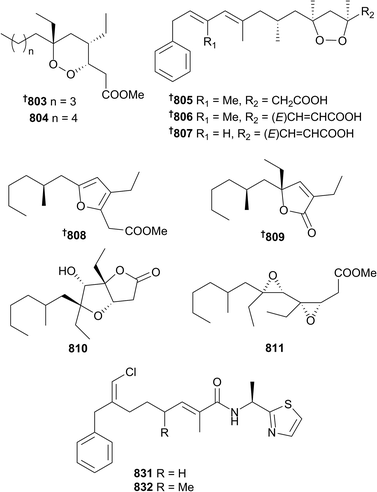
With 224 new and new to marine compounds reported in 2016, compared with 291 in 2015,1 the impacts of changing focus away from sponges towards microorganisms and other phyla are dramatic; the number of sponge metabolites reported in 2016 is the lowest in over a decade. Nonetheless, sponges are still an important taxonomic group for the discovery of new, bioactive natural products. A number of new glyceride (788 and 789) and ceramide (790–793) lipids have been reported from species of Theonella and Erylus,394,395 while branched fatty acids (794 and 795), polyoxygenated fatty acid amides (796 and 797) and N-acyl-dopamine glycosides 798–801 were sourced from Asteropus, Melonanchora, Myxilla and Theonella sponges.396–399 Unusually, only one new polyacetylene 802 was reported in 2016, from Halichondria panicea.400 Examination of the two-sponge association between Xestospongia deweerdtae and Plakortis halichondrioides, in addition to P. zyggompha, resulted in the isolation of 6-epi-7,8-dihydroplakortide K 803, plakortide AA 804, and three chemically unstable plakinic acids N–P 805–807. A combination of both Mosher's acid analysis, as well as comparison of density functional theory (DFT) calculated and experimental ECD spectra were used to establish the absolute configurations of these compounds. The new plakinic acids have inverted configurations compared with other members of the series, although consideration of the likely biosynthesis would suggest they are all generated from a single unified phenylacetic acid (likely phenylalanine derived) starter unit followed by standard iterative ketide additions. Surprisingly, given the sponge extract exhibited antifungal activity against Cryptococcus gattii, none of the new compounds showed any bioactivity.401A Chinese (Xisha Islands) P. simplex was the source of four plakortin-metabolites, plakorsin D methyl ester 808, plakilactone I 809, plakortone Q 810 and the first vicinal diepoxide-containing polyketide plakdiepoxide 811. Stability testing of the endoperoxide functionality of the most abundant Plakortis metabolite reported suggests that many of the polyketides isolated from the genus are likely artefacts from reductive or basic sensitivities. Plakdiepoxide 811 is a potent and selective inhibitor of peroxisome proliferator-activated receptor (PPAR)-γ and hence may have promise as a lead for treatment of Type II diabetes.402 A large number of quinones have been reported from Australian Clathria (812–815)403 and Indonesian Petrosia (816–830)404 sponges. Conulothiazoles A 831 and B 832 are chlorinated NRPS/PKS hybrids isolated from a Bahamian Smenospongia conulosa that combine features from both the well-known cyanobacterial barbamide and jamaicamide compound classes. As less than 45 μg of each compound was isolated, Marfey's method was used to establish the configuration of the thiazole amide using only 4 μg of isolated compound for ozonolysis and hydrolysis. Unsurprisingly, a severe paucity of material prevented profiling of bioactivity.405
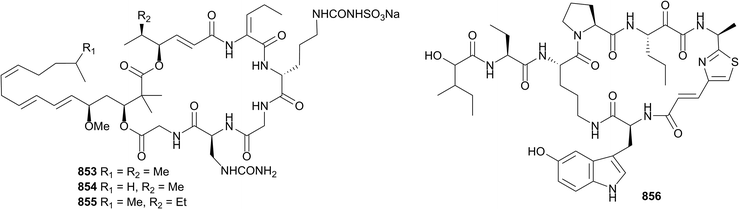
As always, a large number of peptidic and peptide-hybrid natural products have been reported from sponges in 2016. Metagenomic analysis has shown that kasumigamide 833, a known freshwater cyanobacterial product, is produced by an Entotheonella symbiont in a Discodermia sponge,406,407 while other peptides were sourced from the Theonella (834–836),408Stylissa (837–841),409,410Cribrochalina (842),411Callyspongia (843),412Asteropus (845–849)413,414 and Petrosia (850–852)415 genera. A Japanese (Nakagi, Shizuoka Prefecture) specimen of Discodermia kiiensis was the source of sulfolipodiscamides A–C 853–855, unprecedented N-sulfouridyl containing-compounds and essentially the sulfonated analogues of the known lipodiscamides A–C,416 isolated again concurrently from the same sponge. The sulfonated analogues are approximately two-times more active against P388 murine leukaemia cells than the non-sulfonated, the latter likely to be degradative artefacts of isolation.417 Only 33 μg of jamaicensamide A 856 was isolated from Plakina jamaciensis collected at Plana Cays in the Bahamas, however this was enough material to establish the structure of the cyclic hexapeptide. The configurations of the amino acid portion were secured by Marfey's analysis. The structural similarity of 856 with peptides isolated from other lithistid sponges of disparate genera collected from geographically distinct sites suggest that these compounds are likely produced by cosmopolitan symbionts of the genus Entotheonella. A paucity of material prevented biological evaluation of the compound.418
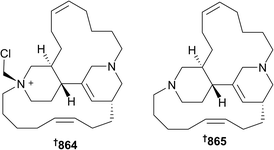
A new diketopiperazine 857 was isolated from Callyspongia sp.,419 while macrocyclic alkaloids and their precursors were isolated from Callyspongia (858 and 859)420 and Halichondria (860–863)421 sponges, respectively. Chloromethylhalicyclamine B 864 (Acanthostrongylophora ingens, Makassar, Sulawesi, Indonesia) is a selective inhibitor of protein kinase CK1 (IC50 = 6 μM vs. both γ/ε isoforms) while being inactive against eight other kinases. The chlorinated compound was found to be an artefact of extraction with dichloromethane, reacting with the known compound halicyclamine B 865,422 which was itself inactive in the same kinase assays, indicating the extreme sensitivity of observed activity versus the structure of the compounds assayed. This study also established the absolute configuration of the known compound 865.423
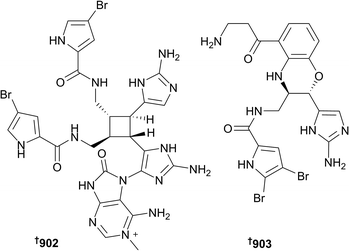
3-Alkylpyridine compounds 866 and 867 have been isolated from Xestospongia and Halichondria species, respectively; the synthesis of the former compound was also achieved.424,425 Indole containing natural products 868 and 869 (known synthetically),426 and 870–874 (the last of which is known synthetically),427–430 including β-carbolines 875 and 876,431 have been isolated from species of Acanthostrongylophora, Geodia, Haliclona, Hyrtios and Lipastrotethya, and sourced from Asia, the Middle East and Scandinavia. An aromatic alkaloid 877 has been sourced from Aaptos aaptos (Vietnam),432 while two isoquinoline compounds 878 and 879 were reported from Xestospongia sp. (Hainan Province, China).433 As always, sponges have been rich sources of guanidine-derived metabolites. Halichondria panicea (Okinawa) yielded 6-epi-monancorin 880,421 while Monanchora sponges were the sources of various monanchorin- and crambescin-type compounds 888–889.434,435 In addition, pyrrole and imidazole compounds were sourced from Agelas sp. (890–898),436Pericharax heteroraphis (899),437 and Lissodendoryx sp. (900 and 901).438 Although inactive in anticancer, antibacterial and antifungal screens, unlike other members of the series, 15′-oxoadenosceptrin 902 and decarboxyagelamadin C 903 (Agelas sceptrum) are chemically interesting. Like many other congeners, these compounds are highly hydrogen deficient and hence the use of 1H–13C and 1H–15N HMBC experiments were of limited utility for structural elucidation. Rather, DFT calculations of NMR chemical shift data were used to corroborate with those of the detected resonances. Moreover, 902 is the first pyrroloimidazole compound to incorporate adenine.439
Given their normal prevalence, there were only two reports of new bromotyrosine metabolites in 2016, the first from an unidentified Indonesian Aplysinellidae sponge (904–910)440 and the other from Acanthodendrilla sp. collected in Thailand (911), the total synthesis of which was also accomplished.441 Lanesoic acid 912 is a novel zwitterionic acid isolated from a Theonella sponge collected at Lanes, Indonesia. The structure of the compound, including an unusual tetrahydropyrimidine cation motif, was established from standard spectroscopic techniques and confirmed by DFT-calculated properties, including confirmation by the DP4+ parameter. Interestingly, 912 was moderately active against the pancreatic cancer PSN1 cell line (IC50 = 8.9 μg mL−1) but was inactive against colon, breast and lung tumour cells and hence shows cytotoxic specificity.442Jaspis splendens was the source of five new alkaloids jaspterin 913, splendamide 914, jaspnin A 915, jaspamycin 916 and 6-bromo-1H-indole-3-carboximidamide 917, the latter two of which are known synthetically.443 These compounds were discovered through the application of a novel, unbiased, phenotypic screen using a human olfactory neurosphere-derived cellular model of Parkinson's disease. Jaspnin A 915 contains a rare methyl thioimidazole functionality, while jaspamycin 916 in particular displayed a distinctive perturbation of the Parkinson's derived cells and hence may be a useful probe of the molecular basis for the neurodegenerative disease. Additionally compound 917 contains the rare indole-3-carboximidamide motif.444
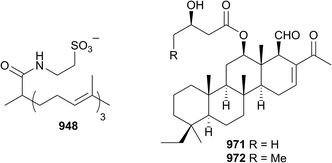
Each year the vast majority of sponge-derived compounds fall into the isoprenoid class and 2016 was no exception. A specimen of Spongia yielded two merosesquiterpenoids 918 and 919 (ref. 445) as did three different collections of Dysidea (920–930).446–448 Only two meroditerpenoids 931 and 932 were reported in 2016,449 while sesquiterpenoids, their formamides, isocyanides, thiocyanates, isothiocyanates and dimers 933–947 (ref. 450–452) were reported from Axinyssa, Dysidea and Halichondria sponges; 944 is a known synthetic compound.453 A novel taurinated norditerpene geodiataurine 948 was reported from a sample of Geodia macandrewii dredged in both Nordland and Trøndelag, Norway. The molecule was discovered following a comprehensive LCMS-based metabolomics survey of Norwegian Geodia specimens that discovered a statistically different chemotype from the more common barettin-containing species. All four samples of G. macandrewii contained 948 and no barettin. Geodiataurine exhibited weak activity (IC50 = 8.5 μM) against A2058 melanoma cells but with no antibacterial effects.454 Other diterpenoids, 949 and 950 (total syntheses also achieved),455 especially based upon the ubiquitous spongian diterpene skeleton 951–962,456–458 were isolated from Theonella, Dysidea, Spongia and Dendrilla sponges, all with varying activities. Several norscalarane 963–967 (ref. 459 and 460) and scalarane 968–970 (ref. 461 and 462) sesterterpenoids were reported in 2016 from sites in Asia and Egypt. New scalaranes 971 and 972 were isolated from Carteriospongia sp. collected at Taitung, Taiwan, as part of a campaign to identify new topoisomerase inhibitors. Compound 971 was a potent disruptor of mitochondrial membrane potential in Molt 4 (leukaemia) cells (IC50 = 125 nM), inducing apoptosis, and significantly more potent than the other isolates. Detailed mechanistic investigation indicated that the scalaranes inhibited topoisomerase IIα expression, but also induced expression of heat shock protein (Hsp) 70, acetylated tubulin and activated caspase 3, suggesting that the proapoptotic activity of the compounds is due to dual inhibition of Hsp90 and topoisomerase activities.463
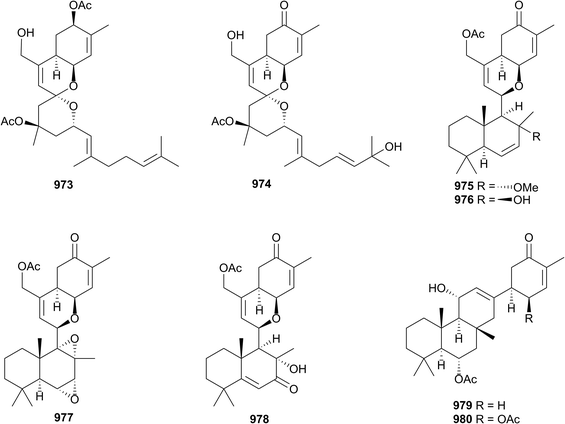
A comprehensive study of a Canadian Phorbas sponge (Howe Sound, British Columbia) yielded eight new sesterterpenoids with various carbon skeletons. Alotaketals A 973 and B 974, and ansellones D–G 975–978 were new congeners of known scaffolds, while anvilones A 979 and B 980 belong to a new class. In addition, 977 possesses the rare 1,2-3,4-bisepoxide moiety, rarely seen amongst natural products. Several of the isolates induced HIV proviral gene expression, through activation of protein kinase C signalling.464
A report of pregnanes 981–984 (ref. 465) from a Myrmekioderma sponge and four reports of sterols and sterol esters 985–1000 (ref. 466–469) were made from Xestospongia and Topsentia sponges in 2016; compounds 994 and 995 had been synthesised previously.470,471 Only two triterpenoid structures were published in 2016, one 1001 from Jaspis stellifera,472 and the other 1002 from a Lipastrotethya sp.473 Synthetic callipeltins B, E,474 and M475 were found to be inactive, whilst natural isolates (Latrunculia sp.) were toxic to HeLa cells, suggesting the observed bioactivity was from low levels (∼15%) of callipeltins C and H.476 Leiodermatolide, a polyketide macrolide and potent cytotoxin,477 has been found to disrupt spindle formation during mitosis, although it does not cause polymerisation or depolymerisation of microtubules and hence has a unique mode of action as an antimitotic agent. Rather, the molecular target of leiodermatolide is likely to be microtubule-associated proteins.478 Stryphnusin (Stryphnus fortis)479 was discovered as a weak inhibitor of acetylcholinesterase activity. Given its structural simplicity and similarity to other natural acetylcholinesterase agonists, a synthetic campaign produced multiple analogues with strong competitive activity to the target enzyme and additionally butylcholinesterase. Studies of the physiological effects of the library upon muscle function and neuromuscular transmission suggest a novel mode of action and hence stryphnusin represents a new lead for neurodegenerative diseases.480 Pyrroloiminoquinone metabolites including discorhabdin B481 and makaluvamine F482 isolated from an Australian Latrunculia sp. inhibited Hypoxia Inducible Factor-1α (HIF-1α) transcription and hence inhibited angiogenesis,483 while makaluvamine J (Zyzzya sp.)484 was a potent antioxidant.485 The quintessential MNPs oroidin486 and sceptrin487 had antibacterial adjuvant activity and suppressed antibiotic resistance,488 while the former is also a new lead for HIV antiviral development.489 Isofistularin-3 (Aplysina aerophoba)490 was found to induce apoptosis, with a novel mode of action via inhibition of DNA methyltransferase 1, making it a new anticancer lead.491 Internet-based tools for scientific research continue to grow in scope and availability. Submission of 71 MNP structures to the Eli Lilly Open Innovation Drug Discovery492 platform identified several potential leads as inhibitors of VEGF, including araguspongine C493 and puupehenone,494 hence both these could be attractive targets for further investigation as angiogenesis inhibitors.495 The ubiquitous meroterpenoid avarone496 and several congeners497–499 enhanced root growth in various terrestrial plants and could have application as agricultural growth stimulants,500 while a dimeric bisabolenyl urea501 was found to inhibit protein tyrosine phosphatase 1B (PTP1B) at sub-toxic concentrations, as well as enhancing insulin-stimulated phosphorylation of Akt and hence may be a new lead for Type II diabetes.502 The common spongian diterpenoids gracilin A,503,504 H,505 and L,506 and additionally tetrahydroaplysulphurin-1,507 bound strongly to cyclophilin A in a manner similar to cyclosporine A, and hence are new immunosuppressants.508,509 The highly oxygenated diterpenoid gagunin D510 inhibited melanin production through inhibition of tyrosinase expression, making it a potential skin whitening cosmetic agent.511 A large number of first syntheses have been reported in 2016, including vesparioside B,512,513 strongylodiols C and D,514,515 lembehyne B,516,517 the previously unreported xestospongenyne,518 and petrosiol A.519,520 The relative configuration of the C-13–C-25 section of hemicalide 1003 (ref. 521) has been determined by synthesis.522 Plakilactone B, C, des-hydroxyplakilactone B,523 and des-hydroxygracilioether C524 have been synthesised,525 as has gracilioether E.526,527 Peptides are common sponge metabolites and hence are targets of synthetic campaigns. Gombamide A,528–530 stylissamide G,531,532 and phakellistatin-15 (ref. 533) were all synthesised in 2016, with the latter compound being inactive while the natural version inhibited the growth of several HTCLS, indicating the presence of a highly potent contaminant in the initial extract.534 The structures of solomonamide B (Theonella swinhoei) 1004 (ref. 535 and 536) and microsclerodermin J (Microscleroderma herdmani) 1005 (ref. 537) were revised following synthesis, the latter study also accomplished the synthesis of the dehydro derivative of microsclerodermin B, dehydromicrosclerodermin B 1006.538 Three separate syntheses established the absolute configuration of callyspongiolide A (Callyspongia sp.) 1007.539–542 Aurantoside G543 has been synthesised,544 while a synthesis of haliclonin A,545 consistent with the written description of the configuration of the molecule but opposite to that drawn (incorrectly) in the original publication, has been reported.546 Renieramycin T547 has been made,548,549 while the absolute configurations of isowondonins A 1008 and B 1009,550 isolated from Poecillastra wondoensis, were established following their syntheses.551 Other first total syntheses of hyrtinadine B,552,553 dictyodendrin H and I,554,555 crambescin A and C,556,557 clavatadine C,558,559 cyclosmenospongine,560,561 strongylophorine 1, 2, 3,562 8,563 and 9,564,565 arenaran A and B,566,567 polyrhaphin D,568,569 hamigeran D,570 G, L,571 N–Q,572,573 and luffarin P574,575 have been achieved. Both the relative and absolute configurations of furospongin-1 1010 (ref. 576 and 577) and dihydrofurospongin-2 1011 (ref. 578) have been established by synthesis,579 as has the side-chain configuration
of clathsterol 1012.580,581 A detailed combination of genomic and metagenomic analyses has shown that the biosynthesis of polytheonamide (Theonella swinhoei),582 a member of the growing class of Ribosomally synthesised and Post-translationally modified Peptides (RiPPs), includes the first example of a B12-dependent radical S-adenosylmethionine C-methylation step.583 A cautionary tale has been published detailing the need for care when reporting new metabolites from dual sponge associations. Ianthelline is a member of the bromotyrosine compounds commonly found from Verongid sponges, originally Ianthella ardis,584 and later Stryphnus fortis.585 A comprehensive LCMS investigation of a Norwegian (Svalbard) S. fortis, and additionally Hexadella dedritifera that is commonly found encrusting upon it, has shown that it is actually the latter that is the true producer of ianthelline, and hence great care must be made to try and avoid contamination of one sponge sample with another.586 An HPLC-based chemical investigation using both diode array and evaporative light scattering detection of compounds from Aplysina cavernicola collected around Maire Is., France, has shown that the reproductive state of the sponge has very little impact upon the secondary metabolism of the sponge. Instead, water temperature seems to play the largest role in controlling the bromotyrosine chemodiversity, and hence chemical defence, of the sponge.587 An analysis of the chemical reactivity of the merosesquiterpenoid siphondictyal B (Siphondictyon coralliphagum)588 under aerobic conditions to produce corallidictyals A and B (Aka coralliphagum, synonymous with S. coralliphagum) and via acid to produce corallidictyals C and D was undertaken.589,590 The oxidative cyclisation proceeds to give the same 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio of corallidictyals A and B as found in nature, suggesting that they may be products of an autooxidative chemical process rather than enzymatically controlled; the corallidictyals may therefore also be artefacts of isolation.591 Finally, a study has been undertaken to compare the sterol biosynthetic potential of four fossilised sponge species (Salpingoeca rosetta, Capsaspora owczarzaki, Sphaeroforma artica, Creolimax fragrantissima) to try and identify key fossil biomarkers of eukaryote development. In particular, 24-isopropylcholesterol has been suggested as a “molecular fossil” and as the key historical metabolite as evidence of life, but questions arise whether ancient sponges or algae were the source of the compound in previous reports. Genomic data suggest that sponges independently developed genes encoding for C30 sterol biosynthesis, including 24-isopropylcholesterol, before other ancient phyla, especially pelagophyte algae. This indicates that it is unlikely that algae are the source of ancient 24-isopropylcholesterol and is consistent with the hypothesis that sponges evolved long before the Cambrian explosion 542 million years ago.592
2 ratio of corallidictyals A and B as found in nature, suggesting that they may be products of an autooxidative chemical process rather than enzymatically controlled; the corallidictyals may therefore also be artefacts of isolation.591 Finally, a study has been undertaken to compare the sterol biosynthetic potential of four fossilised sponge species (Salpingoeca rosetta, Capsaspora owczarzaki, Sphaeroforma artica, Creolimax fragrantissima) to try and identify key fossil biomarkers of eukaryote development. In particular, 24-isopropylcholesterol has been suggested as a “molecular fossil” and as the key historical metabolite as evidence of life, but questions arise whether ancient sponges or algae were the source of the compound in previous reports. Genomic data suggest that sponges independently developed genes encoding for C30 sterol biosynthesis, including 24-isopropylcholesterol, before other ancient phyla, especially pelagophyte algae. This indicates that it is unlikely that algae are the source of ancient 24-isopropylcholesterol and is consistent with the hypothesis that sponges evolved long before the Cambrian explosion 542 million years ago.592
8 Cnidarians
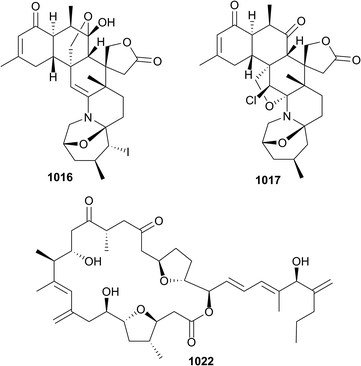
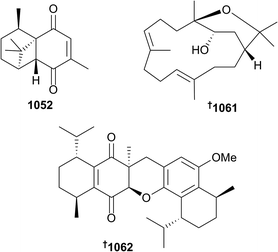
The 202 new compounds reported from cnidarians in 2016 are just below the previous decadal average. Of this total, ten nitrogenous metabolites were reported from hydroids, zoanthids and soft corals, including the cytotoxic ceramide 1013 (Heteroxenia ghardaqensis),593 brominated-indoles 1014 and 1015 (hydroid Abietinaria abietina) which were found to activate NF-kB-dependent transcription,594 and zoanthamines 1016–1021 from the zoanthid coral Zoanthus kuroshio.595 Of note were 5α-iodozoanthamine 1016 and 11β-chloro-11-deoxykuroshine A 1017 which are the first reported examples of halogenated zoanthamine alkaloids. New examples of amphidinolides, C4 1022, B8 1023 and B9 1024, in addition to the known dinoflagellate-sourced T1, were reported from Brazilian specimens of the octocoral Stragulum bicolor.596 Modest cytotoxicity was observed for 1022 and 1023. The simple dinormonoterpenes 1025 and 1026 were reported from Sinularia mollis.597Thirty-one sesquiterpenes were reported, comprised of capgermacrene C 1027 (Capnella sp.),598 lochmolin H 1028 (Sinularia lochmodes),599 cadinane-type endo- and hydro-peroxides 1029–1031 (Sinularia sp.),6001032 and 1033 from S. verruca,601 from which was also reported a rare pyrroloindoline alkaloid 1034 in addition to two simpler cyclopentenones 1035 and 1036, bisabolanes 1037–1045, cadinanes 1046–1051 and a sesquiterpene bearing a new tricyclic skeleton 1052 (Pseudopterogorgia rigida),602 cadinane 1053 (Menella sp.),603 eudesmane-type 1054 (Subergorgia suberosa),604 and guaiane lactones 1055 and 1056 (Menella woodin).605 The final example of a sesquiterpene, cadinane 1057, isolated from Sinularia vanderlandi, was included in a publication that also reported spartane diterpenoid 1058, lipids 1059 and 1060, and cembrane isodecaryiol 1061 from S. gravis.606 The structure of the latter metabolite, which is a diastereomer of the often reported cnidarian cembranoid decaryiol, was rigorously established and absolute configuration assigned by X-ray crystallography. X-ray crystallography was also used to establish the structure and absolute configuration of the unusual bis-sesquiterpene 1062 (Rhytisma fulvum fulvum) – the metabolite is likely derived from dimerisation of the natural product 5-hydro-8-methoxycalamenene.607 Of six assorted diterpenes 1063 and 1064 (Sinularia sp.),608 and 1065–1068 (Gersemia fruticosa),609 lobane 1063 and eunicellane 1068 exhibited modest antibacterial activity.
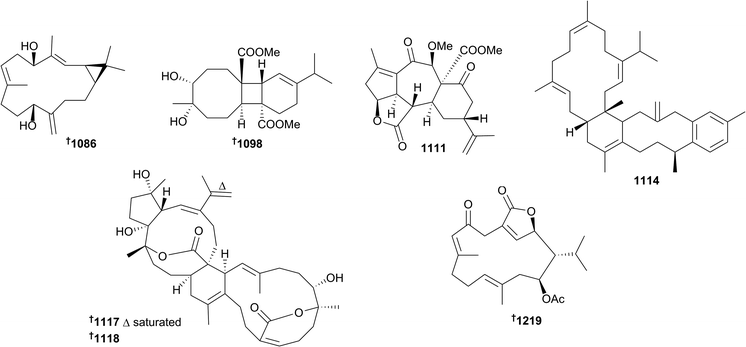
Nearly fifty cembrane-related metabolites were reported from cnidarians in 2016. Amongst these were the 15-acetoxy examples 1069–1071 (Nephthea sp.),610 pyranose-containing 1072 (Sarcophyton trocheliophorum),611 a mildly anti-Staphylococcal 16-hydroxyl cembranoid 1073 (Sarcophyton sp.),612 epoxides trocheliolide B 1074 (S. trocheliophorum)613 and sarcophytonoxides A–E 1075–1079 (S. ehrenbergi),614 sinulerectols A–C 1080–1082 and degraded cembranoid sinulerectadione 1083 (Sinularia erecta).615 The latter study also led to a revision of relative configuration of the previously reported cembranoid sinularectin 1084. Further examples of cembranoids or related metabolites included sinularcasbanes M–O 1085–1087 (Sinularia polydactyla),616 where the absolute configuration of 1086 was secured by X-ray crystallography, anti-inflammatory uprolides N–P 1088–1090 (Eunicea succinea),617 cubitanoids (a scaffold previously speculated as being photochemically-derived from cembranoids) nanoculones A 1091 and B 1092 and cembranoids nanolobols A–C 1093–1095 (Sinularia nanolobata),618 locrassumins A–G 1095–1102, (−)-laevigatol B 1103, (−)-isosarcophine 1104 and sarcophytoxide analogue 1105 (Lobophytum crassum),619 casbane-type diterpenoid 1106 and diepoxycembranes 1107 and 1108 (Lobophytum sp.),620 and pambanolides A–C 1109–1112 and a seco-sinulochmodin analogue 1113 (Sinularia inelegans).621 While 1110 and 1111 were initially isolated as an inseparable mixture, an X-ray structure of 1111 (pambanolide B2) was eventually secured. This study also established the configuration of rameswaralide via X-ray crystallography. Bis-cembrene trocheliane 1114 and pyran-cembranes sarcotrocheldiol A 1115 and B 1116 were isolated from Sarcophyton trocheliophorum and strong antibacterial activity was reported for 1114.622 Finally for cembranoids, the structures of unusual cembrane–capnosane heterodimers bissubvilides A 1117 and B 1118 (S. subviride) were secured by combinations of spectroscopic analysis and time dependent DFT (TDDFT)/ECD and NMR calculations.623 Also of note amongst the cembranoid natural products was the unprecedented carbon skeleton present in locrassumin C 1098 and the use of analysis of an induced CD spectrum resulting from an in situ Mo2(OAc)4 complex to determine absolute configuration.
Twenty-one new briarane diterpenes were reported in 2016, comprised of briarenolides M–T 1119–1126 (ref. 624) and ZI–ZVI 1127–1132 (ref. 625) (both sets of metabolites from the same collection of Briareum sp.), and gemmacolides AZ–BF 1133–1139 from Dichotella gemmacea.626 Briarenolides M, P, S, T, ZII and ZVI were found to inhibit production of the pro-inflammatory inducible nitric oxide synthase (iNOS), briarenolides N, P and T inhibited the product of COX-2 in LPS-stimulated macrophage cells, and the gemmacolides exhibited low to no cytotoxicity towards two HTCLs and moderate antibacterial activity. The remaining eight diterpenes, all eunicellins, included klymollins Y 1140 and Z 1141 and klyxumollins A–D 1142–1145 (Klyxum molle),627 and cladieunicellins R 1146 and S 1147 (Cladiella tuberculosa).628 Diterpenes 1141 and 1142 and 1147 exhibited modest cytotoxicity towards HTCLs. Cnidarians, comprising both zoanthids and soft corals, also yielded a variety of steroids including seco-sterols pinnigorgiol A–C 1148–1150,629 and D 1151 and E 1152,630 pinnisterol A–C 1153–1155,631 (Pinnigorgia sp.) and haebaruol 1156 (Clavularia sp.),632seco-sterol 1157 and epoxy-sterol 1158, (Pinnigorgia sp.),633seco-sterol sibogol D 1159 and cross-conjugated sterol dienones sibogol E 1160 and F 1161 (Muricella sibogae),634 pregnanes subergorgol T–X 1162–1166 (Subergorgia suberosa),635 cross-conjugated dienones petasitosterone A–C 1167–1169 (Umbellulifera petasites),636 ecdysones zoanthone A 1170 (Zoanthus sp.)637 and palythone A 1171 (Palythoa mutuki),638 18-oxygenated sterol 1172 (Nephthea sp.),639 19-oxygenated sterol 1173 (Pacifigorgia senta),640 sterols including cyclopropylated (gorgosterol) examples 1174–1177 (Pinnigorgia sp.),6411178 (Lobophytum lobophytum),642 klyflaccisteroids G–J 1179–1182 (Klyxum flaccidum),643 and 1183 and 1184 (Sinularia microspiculata),644 4α-methylated sterols 1185–1191 (Litophyton mollis),645 polyhydroxylated sterols 1192–1209 (Gorgonia sp.),646 24-methylene sterols 1210 (Nephthea columnaris)6471211 (Sinularia sp.)6481212 and 1213 (Nephthea erecta)649 and 1214 (Sinularia nanolobata),650 24-epoxy sterol 1215 (S. nanolobata),650 and steroidal glycosides 1216–1218 (Astrogorgia dumbea).651 The structure of sarcophytonolide H has been confirmed by total stereospecific synthesis: the same study led to revision of the relative configuration of the 14-acetoxy substituent of isosarcophytonolide D to 1219.652
An enantioselective synthesis of (1S)-suberosanone led to configurational assignment of the related suberosenol A.653 Unfortunately the potent HTCL cytotoxicity reported for naturally occurring (1S)-suberosanone was not observed for the synthetic material. An organocatalytic system was used to prepare non-natural enantiomer (1R)-suberosanone,654 and first syntheses of malonganenone J655 and seco-pseudopteroxazole656 have been reported. Further biological studies have identified that hydrogen peroxide derived from peroxy sesquiterpenoids induced apoptosis in HCT116 cells,657 pseudopterosin protects synaptic function during oxidative stress suggesting a potential role as a neuromodulatory agent,658 a metabolomics approach has identified that flexibilide-treated HCT116 cells suffer TCA cycle down-regulation and up-regulated levels of sphingosine-1-phosphate,659 sinularin suppresses the proliferation of gastric tumour cells and induces apoptosis via mitochondrial dysfunction,660 5-epi-sinuleptolide inhibits early biofilm formation of the human pathogenic Gram negative bacterium Acinetobacter baumannii,661 11-dehydrosinulariolide exerts antiapoptotic and anti-inflammatory effects at different time points after spinal cord injury662 and also exerts a neuroprotective effect in models of Parkinson's disease,663 11-epi-sinulariolide acetate exhibits cytotoxicity towards HA22T hepatocellular carcinoma cells inducing apoptosis through mitochondrial dysfunction and endoplasmic reticulum stress pathways,664 a sterol from the hard coral Acropora formosa induces p53-mediated apoptosis in A549 human non-small cell lung cancer cell-line,665 and the carotenoid peridinin exerts anti-proliferative and pro-apoptotic effects against human T-cell leukemia virus type 1-infected T-cell lines by suppressing NF-kB and Akt signalling666 and anti-viral properties.638 Comprehensive LCMS analysis of organisms living in close proximity to Palythoa tuberculosa on an Okinawan reef, has detected the presence of palytoxin and 42-hydroxy-palytoxin exclusively in the coral.667
9 Bryozoans
In contrast to 2015 when nine new compounds were reported, there were no new metabolites reported from this understudied phylum in 2016. There were two reports relating to compounds previously isolated from the phylum however. The cytotoxicity to HeLa cells of the alkaloid pterocellin A, a metabolite of the New Zealand endemic species Pterocella vesiculosa, was shown to occur via mitochondrial apoptotic processes.668 Amathaspiramides A–F originally isolated from the Australasian endemic species Amathia wilsoni, and four analogues have been synthesised and tested for antiproliferative activity against four HTCLs.669 In contrast to the original report in which all amathaspiramides were shown to be inactive towards P338 murine tumour cells at 25 μM,670 amathaspiramide C was the most active compound showing weak (IC50 5.8 μM) activity towards the MiaPaCa-2 pancreatic cancer cell line. Furthermore only amanthaspiramindes A, C and E containing a pyrrolidine and 8R configuration of the N-acyl hemiaminal moiety showed antiproliferative activity. A series of simplified bryostatin analogues671 and salicylate-derived bryostatin analogues672 were prepared and exhibited very potent anti-Chikungunya virus activity.671,67210 Molluscs
The 24 new metabolites reported in 2016 from molluscs is close to the average number reported per year over the past decade. New metabolites reported from molluscs included polypropionates exiguapyrone 1220 and exiguaone 1221 (cephalaspidean (bubble snail) mollusc Haminoea exigua),673 kahalalide analogues Z11222 and Z21223 (sea hare Elysia ornata),674 rearranged diterpenes and norditerpenes 1224–1238 from Australian specimens of the nudibranchs Goniobranchus verrieri, G. splendidus, G. cf. splendidus and G. daphne,675,676 diterpenes 1239–1242, including one 1239 which was identified as being a potent fish feeding deterrent, from the aeolidoidean Phyllodesmium longicirrum,677 and two new secosterols 1243 and 1244 from the sea hare Aplysia kurodai.678Of note were the structure elucidations of the Goniobranchus metabolites, many of which were isolated in sub-milligram quantities. In a beautiful exposition of the utility of total synthesis, the putative linear contiguous polyketide precursor of the mollusc metabolite dolabriferol has been shown to undergo regioselective retro-Claisen fragmentation to form the natural product under mild conditions.679 Total syntheses of (+)-panacene,680 brominated chamigrene sesquiterpenes, including aplydactone,681,682 the protein synthesis inhibitor chlorolissoclimide,683 and (−)-chromodorolide B684 have been reported. Carotenoid mytiloxanthin, originally reported from the sea mussel Mytilus californianus, quenches singlet oxygen and inhibits lipid peroxidation.685 The mode of binding of aplysiatoxin to protein kinase Cδ C1B domain in a model bilayer membrane has been studied using molecular dynamics simulation,686 while surface plasmon resonance was used to analyse the actin-tubulin protein–protein interaction induced by aplyronine A.687 Latrunculin A is selectively accumulated in the rim of the mantle of Chromodoris nudibranchs, with the implication that, being the most exposed part of the mantle, the natural product is used as a predator deterrent.688 Latrunculin A demonstrated potent antifeedant activity towards rock pool shrimps (Palaemon serenus). New examples of conotoxins purified from venom of Conus species molluscs were reported: PiVIIA,689 a 25-mer producing significant increases in Ca2+ currents (C. princeps); Im10A,690 an 11-mer bearing four cysteine residues (C. imperialis); Lo6/7a and Lo6/7b,691 24- and 27-mers, respectively, which are examples of framework VI/VII conotoxins from C. longurionis; Asi3a a framework III 15-mer and Asi14a a framework XIV 17-mer, both691 from C. asiaticus; and AusB, an unsual 18-mer example bearing only one disulfide bond, isolated from C. australis.691 Screening of a library of synthetic conotoxin variants has identified a peptide that upon further structure–activity relationship tuning, led to analogues that were potent and selective agonists of the κ-opioid receptor.692 Large quantities of the nAChR antagonist, α-conotoxin LvIA, have been prepared via an E. coli recombinant expression system.693 Some carnivorous cone snails, including worm-hunting snails of the subgenus Rhizoconus, have evolved two distinct venoms – one set of peptides for defence, and another set for predation.694 A phylogenetic and molecular evolution analysis of αD-conotoxins suggests that they evolved as part of a defensive strategy in this subgenus.
11 Tunicates (ascidians)
The ten new tunicate-derived natural products presented in this review is the second lowest number reported in one year since 2002. The metabolites reported included a phosphorylated polyketide 1245,695 a new glycosylated macrolide mandelalide E 1246,696 an acetylenic amino alcohol 1247 and N-hydroxylated-1,2,3,4-tetrahydro-β-carboline 1248,697 neuroactive guanidine alkaloids 1249–1252,698 another example 1253 of the unusual tetracyclic-cored eudistidine alkaloid family,699 and a disulfated steroid 1254.700 Noteworthy was the added complexity, compared with congeners reported in 2015, of eudistidine C 1253 reported from Eudistoma sp., collected in Palau.699 The racemic natural product was synthesised by a straight-forward nucleophilic substitution reaction using eudistidine A as starting material, simple extension of which led to preparation of a library of un-natural analogues. Chiral HPLC allowed for resolution of the two enantiomers of the natural product, with absolute configuration assigned to each by comparison of experimental ECD spectra with those calculated using TDDFT calculations. (−)-(S)-Eudistidine C was a modest inhibitor of p300-HIF-1α protein interaction.Also of note in 2016, was a report on the effectiveness of chemical shift calculations in the structure elucidation of the complex marine natural products the mandelalides (tunicate) and coibamide A (cyanobacterium).701 Total syntheses of obscuraminol A,702 crucigaterin 277,703 polycarpathiamines A and B,704 and iheyamine A705 have been reported, and in all cases, the structures of the reported natural products were confirmed. Novel rigidin analogues targeting β-tubulin have been found to exhibit potent antitumour activity in vitro and in vivo,706 while clavaminol A and diacetyl clavaminol H exhibit antibacterial, antifungal and biofilm inhibiting activities.707 The chemistry and biology of the mandelalides continues to attract attention with new synthetic routes and biological activities reported.708–710 Semi-synthesis of a library of 2′-N-acyl derivatives of ecteinascidin 770 has identified two analogues, an isoxazole amide and an 4-methoxyphenyl amide, to be more potent in vitro cytotoxins than the natural product.711 Analogues of the amino-imidazole alkaloids polycarpine and polycarpaurines A and C have been identified as antivirals (against tobacco mosaic virus) and fungicidal against a set of plant pathogens.712 Copper(II) complexes of cyclic peptides related to patellamide exhibit glycosidase and β-lactamase-like activity,713 while the effects of unnatural amino acid variation on the open/folded conformations of ascidiacyclamide have been investigated by NMR, CD and X-ray crystallography.714 A yeast-based chemical genetics approach was used to identify the cellular target of eudistomin C as being the uS11-containing ribosomal subunit and that it is a potent inhibitor of protein translation.715 Eudistomin U is a weak DNA binder.716
12 Echinoderms
The forty-six new metabolites reported from echinoderms in this review is about average for the number reported per annum over the last decade. A new radical-scavenging aminonaphthoquinone, spinamine E 1255, was reported from the urchins Strongylocentrotus pallidus and Mesocentrotus nudus,717 while trace amounts of curacin E 1256 were isolated from the brittle star Ophiocoma scolopendrina using bioassay-guided fractionation.718 What is unusual about the latter metabolite is that it is structurally related to curacin A,719 an antimitotic reported from a cyanobacterium. Absolute configuration about the thiazoline ring was related to the known configuration of the same ring system in curacin A via analysis of ECD data.The remaining metabolites reported from echinoderms were dominated by anthraquinones 1257 and 1258 (crinoid Comatula rotalaria),403 pyrrole oligoglycosides 1259 and 1260 (starfish Acanthaster planci)720 and sterols 1261–1263 (starfish Archaster typicus)721 and their glycosides including 1264–1267 (starfish Protoreaster lincki),7221268–1277 (starfish Anthenea aspera),7231278 and 1279 (starfish Acanthaster planci),7241280–1282 (starfish Pentaceraster regulus),7251283–1285 (starfish Craspidaster hesperus),7261286 (sea cucumber Colochirus robustus),7271287 (starfish Aphelasterias japonica),7281288–1291 (sea cucumber Colochirus robustus),7291292–1295 (ref. 730) and 1296–1299 (ref. 731) (both sets from sea cucumber Cucumaria fallax) and 1300 (sea cucumber Astichopus multifidus).732 Two synthetic routes to spinamine E 1255 (2-amino-3,6,7-trihydroxynaphthazarin) have been reported733 as has the first synthesis of the pentacyclic naphthazarin-derived dimer mirabiquinone A.734,735 The structure of the complex sialic acid embedded octasaccharide ganglioside GP3 (Asterina pectinfera)736 has been confirmed by total synthesis.737 The sea cucumber triterpenoid glycoside frondoside A738 exhibits activity in a human pancreatic mouse xenograft model but only when given intraperitoneally (oral gavage delivery had no effect).739 It also exhibits activity towards prostate cancer cell lines in vitro and in vivo.740 A related sea cucumber triterpenoid glycoside, echinoside A,741 exhibited diurnal time-dependent inhibition of fatty acid biosynthesis in mice while enhancing mitochondrial fatty acid β-oxidation.742 The tetrasaccharide saponin echinoside A743 has higher bioavailability than the hexasaccharide saponin holotoxin A1 (ref. 744) as measured using the Caco-2 cell model and single-pass intestinal perfusion.745 Pentasaccharide saponin, cucumarioside A2-2,746 activates cellular immunity by interacting with P2X receptors on macrophages.747
13 Mangroves
Mangroves or their associates were the sources of a moderately antiplasmodial embelin analogue 1301 (Aegiceras corniculatum),748 two dolabranes tagalsin V 1302 and W 1303 (Ceriops tagal),749 a new tirucallane 1304 and two tetranortriterpenes 1305 and 1306 (seeds of Xylocarpus moluccensis),750 a further eight tetranortriterpenoids 1307–1314 (leaves and twigs of X. granatum),751 and limonoids 1315–1318 (X. moluccensis).752 The structures of xylogranatumin A 1308 and thaixylomolins O 1315 and P 1316 are notable, with the former containing an unusual oxygen-bridging arrangement, and the latter two with their orthoacetate bridge. Absolute configurations were assigned to 1307 and 1318 by TDDFT calculated ECD analysis, to 1315 and 1307 by single crystal X-ray diffraction studies and to 1316 by comparison of ECD spectra.14 Miscellaneous
Norwegian sea herring caviar (Clupea harengus) contain arsenolipids 1319–1324.753 The metabolites were characterised solely by MS/MS analysis, meaning that the structures are only proposals, with the glycerol ester point of substitution and position and geometry of alkene double bonds not determined.15 Conclusion
The rich and unique chemical diversity of NPs have long been an important source of drugs and drug leads.3 Comparisons of the chemical diversity of drugs compared to NPs and synthetic libraries have shown that NP chemical diversity is more closely aligned with drugs than synthetic libraries.754 This is unsurprising since over half of all drugs are either NPs, NP-derived or NP-inspired.3 Chemical diversity has been reported to align with biological diversity and marine collection efforts over the years have focused on obtaining diverse biota to maximise the potential chemical diversity that they might contain.755 It has also been recognised that some MNPs isolated from macro-invertebrates may actually be produced by micro-organisms and in recent years this has, in part, led to an increasing trend towards the study of marine micro-organisms with reduced efforts focusing on other taxa.755 With 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 609 MNPs having now been reported from a variety of marine sources13 it is timely to assess the chemical diversity found within these organisms and to compare this diversity to that of approved drugs. In this conclusion 11 physico-chemical descriptors, clog
609 MNPs having now been reported from a variety of marine sources13 it is timely to assess the chemical diversity found within these organisms and to compare this diversity to that of approved drugs. In this conclusion 11 physico-chemical descriptors, clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, molecular weight (MW), polar surface area, counts of stereogenic centres, small rings, aromatic atoms, sp3 atoms, hydrogen bond donors and acceptors, basic nitrogens and acidic oxygens, were calculated for 28
P, molecular weight (MW), polar surface area, counts of stereogenic centres, small rings, aromatic atoms, sp3 atoms, hydrogen bond donors and acceptors, basic nitrogens and acidic oxygens, were calculated for 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 609 MNPs reported since 1957 and 2908 approved drugs derived from the SWEETLEADS database.756 Principal component analysis (PCA) of these descriptors was then undertaken. Pairwise cluster analysis of the MNPs from the 15 phyla (refer to y axis in Fig. 1) from which the majority of MNPs have been reported was also carried out using the FragFp (substructure fragment dictionary) descriptor generated in the free chemiformatics software Osiris Datawarrior.757 Two of the Lipinski criteria for “drug-likeness”, MW and log
609 MNPs reported since 1957 and 2908 approved drugs derived from the SWEETLEADS database.756 Principal component analysis (PCA) of these descriptors was then undertaken. Pairwise cluster analysis of the MNPs from the 15 phyla (refer to y axis in Fig. 1) from which the majority of MNPs have been reported was also carried out using the FragFp (substructure fragment dictionary) descriptor generated in the free chemiformatics software Osiris Datawarrior.757 Two of the Lipinski criteria for “drug-likeness”, MW and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, are presented in Fig. 1 and 2. Although on average the clog
P, are presented in Fig. 1 and 2. Although on average the clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P for MNPs are shifted three log units higher compared to approved drugs, the trend is different for different phyla, with compounds reported from Actinobacteria, Ascomycota, Chordata (predominantly tunicates) and Bryozoa possessing the most similar profile to approved drugs. The MW histograms for MNPs and approved drugs are very similar with approved drugs containing a slightly higher proportion of low MW compounds. Comparison at the phylum level tells a different story with Echinodermata, Dinophyta, Cyanobacteria and Tracheophyta (mangroves) containing proportionately higher MW compounds. A distinguishing feature of NPs compared to synthetic compounds is their abundance of stereogenic centres. On average 58% of approved drugs have one or no stereogenic centre compared to only 25% of MNPs (Fig. 3). Bryozoa, Chordata and Proteobacteria show the closest match to approved drugs, while Echinodermata, Dinophyta, Cnidaria, Mollusca and Tracheophyta on average contain molecules with a larger number of stereogenic centres. A plot of principal component 1 (PC1) vs. PC2 (accounting for 80% of the variability in the PCA data) (Fig. 4 and 5) reinforces the above conclusions indicating that compounds reported from Echinodermata and Dinophyta occupy complementary but different areas of chemical space compared to the other 13 phyla (and approved drugs). Ochrophyta (brown algae) compounds occupy a unique area of chemical space, while Cnidaria, Cyanobacteria, Mollusca, Cyanophyta and Rhodophyta (green and red algae) contain a higher proportion of molecules with either a large number of stereogenic centres, sp3 carbons and/or rotatable bonds. The myriad compounds reported from Porifera show a range of chemical properties and this is reflected in the broad area of chemical space occupied by these compounds compared to those reported from the other 14 phyla. But how useful is a PCA comparison in defining chemical diversity? Pairwise cluster analysis of the compounds isolated from the top 15 phyla and similarity analysis of their shared clusters indicates that although compounds often occupy a similar area of chemical space, their structures are different, with on average less than an 8% overlap of clusters shared between two phyla (Fig. 6). An exception was the compounds reported from Mollusca, organisms that are known to sequester compounds from dietary sources, and these compounds shared a higher (10–17%) similarity with most other phyla except Bryozoa, Tracheophyta and Proteobacteria. Finally, MNPs are diverse since 6264 clusters were generated from the 28
P for MNPs are shifted three log units higher compared to approved drugs, the trend is different for different phyla, with compounds reported from Actinobacteria, Ascomycota, Chordata (predominantly tunicates) and Bryozoa possessing the most similar profile to approved drugs. The MW histograms for MNPs and approved drugs are very similar with approved drugs containing a slightly higher proportion of low MW compounds. Comparison at the phylum level tells a different story with Echinodermata, Dinophyta, Cyanobacteria and Tracheophyta (mangroves) containing proportionately higher MW compounds. A distinguishing feature of NPs compared to synthetic compounds is their abundance of stereogenic centres. On average 58% of approved drugs have one or no stereogenic centre compared to only 25% of MNPs (Fig. 3). Bryozoa, Chordata and Proteobacteria show the closest match to approved drugs, while Echinodermata, Dinophyta, Cnidaria, Mollusca and Tracheophyta on average contain molecules with a larger number of stereogenic centres. A plot of principal component 1 (PC1) vs. PC2 (accounting for 80% of the variability in the PCA data) (Fig. 4 and 5) reinforces the above conclusions indicating that compounds reported from Echinodermata and Dinophyta occupy complementary but different areas of chemical space compared to the other 13 phyla (and approved drugs). Ochrophyta (brown algae) compounds occupy a unique area of chemical space, while Cnidaria, Cyanobacteria, Mollusca, Cyanophyta and Rhodophyta (green and red algae) contain a higher proportion of molecules with either a large number of stereogenic centres, sp3 carbons and/or rotatable bonds. The myriad compounds reported from Porifera show a range of chemical properties and this is reflected in the broad area of chemical space occupied by these compounds compared to those reported from the other 14 phyla. But how useful is a PCA comparison in defining chemical diversity? Pairwise cluster analysis of the compounds isolated from the top 15 phyla and similarity analysis of their shared clusters indicates that although compounds often occupy a similar area of chemical space, their structures are different, with on average less than an 8% overlap of clusters shared between two phyla (Fig. 6). An exception was the compounds reported from Mollusca, organisms that are known to sequester compounds from dietary sources, and these compounds shared a higher (10–17%) similarity with most other phyla except Bryozoa, Tracheophyta and Proteobacteria. Finally, MNPs are diverse since 6264 clusters were generated from the 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 609 compounds analysed and the majority of the compounds were represented in clusters of less than ten members (Fig. 7). Only the Cnidaria, Echinodermata, Cyanobacteria and Tracheophyta contain individual clusters that represent >30% of the total compounds reported from each phylum. The take home messages from these analyses are that the chemical space occupied by approved drugs overlaps with that of MNPs, chemical diversity within marine organisms is high and that targeting one group of marine organism over another is perilous if one is to trying to maximise the exploration of chemical diversity. Likewise the trend towards targeted micro-organism collections at the expense of invertebrate and algae collections is likely to lead to unique chemical diversity being overlooked.
609 compounds analysed and the majority of the compounds were represented in clusters of less than ten members (Fig. 7). Only the Cnidaria, Echinodermata, Cyanobacteria and Tracheophyta contain individual clusters that represent >30% of the total compounds reported from each phylum. The take home messages from these analyses are that the chemical space occupied by approved drugs overlaps with that of MNPs, chemical diversity within marine organisms is high and that targeting one group of marine organism over another is perilous if one is to trying to maximise the exploration of chemical diversity. Likewise the trend towards targeted micro-organism collections at the expense of invertebrate and algae collections is likely to lead to unique chemical diversity being overlooked.
 | ||
| Fig. 5 Loadings plot for the PCA in Fig. 4. | ||
 | ||
| Fig. 7 Compound cluster analysis. Proportion of compounds in each phylum that are members of clusters of varying size. | ||
16 Conflicts of interest
There are no conflicts to declare.17 Acknowledgements
We thank Dr Helen Potter (Royal Society of Chemistry) for the provision of data used in this review, adapted from the MarinLit database with permission from the Royal Society of Chemistry.13 This review closes a chapter of dedicated and insightful contribution by Professor John Blunt and we wish him well for the future. Professor Blunt has been the corresponding author for the last fifteen issues and has provided exceptional guidance, oversight and mentorship to the authorship team. John has been the engine room of the review. He has liaised with the editorial team at NPR and RSC, coordinated the contributions of the co-authors, and with his longtime colleague Professor Murray Munro compiled the text and all of the structures and generated the ESI† files as well as writing individual review sections. His ability to coordinate the contributions of the co-authors, seamlessly compile their individual contributions and organise the 1000+ new structures each year, to produce such a quality annual review is testament to John's high standards. The database, MarinLit, was the “brainchild” of Professors Blunt and Munro and it is this database (now administered by RSC) that has provided the backbone upon which the annual review has been compiled. It has been a true pleasure working with John and we wish to thank him for his leadership in MNP research and his outstanding contributions to these reviews.18 References
- J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. G. Munro and M. R. Prinsep, Nat. Prod. Rep., 2017, 34, 235–294, 10.1039/c6np00124f.
- J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. G. Munro and M. R. Prinsep, Nat. Prod. Rep., 2016, 33, 382–431, 10.1039/c5np00156k.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2016, 79, 629–661, DOI:10.1021/acs.jnatprod.5b01055.
- D. Newman and G. Cragg, Planta Med., 2016, 82, 775–789, DOI:10.1055/s-0042-101353.
- X.-M. Fu, M.-Q. Zhang, C.-L. Shao, G.-Q. Li, H. Bai, G.-L. Dai, Q.-W. Chen, W. Kong, X.-J. Fu and C.-Y. Wang, Mar. Drugs, 2016, 14, 46, DOI:10.3390/md14030046.
- K.-L. Pang, D. P. Overy, E. B. Gareth Jones, M. d. L. Calado, G. Burgaud, A. K. Walker, J. A. Johnson, R. G. Kerr, H.-J. Cha and G. F. Bills, Fungal Biol. Rev., 2016, 30, 163–175, DOI:10.1016/j.fbr.2016.08.001.
- N.-Y. Ji and B.-G. Wang, Fungal Divers., 2016, 80, 301–342, DOI:10.1007/s13225-016-0358-9.
- Y. Bahrami and C. M. M. Franco, Mar. Drugs, 2016, 14, 147, DOI:10.3390/md14080147.
- H. Lei, Chem. Biodiversity, 2016, 13, 345–365, DOI:10.1002/cbdv.201500030.
- P. Máximo, L. M. Ferreira, P. Branco, P. Lima and A. Lourenço, Mar. Drugs, 2016, 14, 139, DOI:10.3390/md14080139.
- P. R. Jensen, Trends Microbiol., 2016, 24, 968–977, DOI:10.1016/j.tim.2016.07.006.
- I. Pérez-Victoria, J. Martín and F. Reyes, Planta Med., 2016, 82, 857–871, DOI:10.1055/s-0042-101763.
- http://pubs.rsc.org/marinlit, accessed July 2017.
- Z. Xie, L. Zhou, L. Guo, X. Yang, G. Qu, C. Wu and S. Zhang, Org. Lett., 2016, 18, 1402–1405, DOI:10.1021/acs.orglett.6b00332.
- J. Xie, B. Liu, H. Wang, S. Yang, H. Zhang, Y. Wang, N. Ji, S. Qin and H. Laatsch, Mar. Drugs, 2012, 10, 551–558, DOI:10.3390/md10030551.
- M. Bae, K. Moon, J. Kim, H. Joo Park, S. Kook Lee, J. Shin and D.-C. Oh, J. Nat. Prod., 2016, 79, 332–339, DOI:10.1021/acs.jnatprod.5b00956.
- M. C. Wilson, S. J. Nam, T. A. M. Gulder, C. A. Kauffman, P. R. Jensen, W. Fenical and B. S. Moore, J. Am. Chem. Soc., 2011, 133, 1971–1977, DOI:10.1021/ja109226s.
- T. Cam Le, I. Yang, Y. Joon Yoon, S.-J. Nam and W. Fenical, Org. Lett., 2016, 18, 2256–2259, DOI:10.1021/acs.orglett.6b00892.
- P. Fu, S. La and J. B. MacMillan, J. Nat. Prod., 2016, 79, 455–462, DOI:10.1021/acs.jnatprod.5b00604.
- P. Fu, A. Legako, S. La and J. B. MacMillan, Chem.–Eur. J., 2016, 22, 3491–3495, DOI:10.1002/chem.201600024.
- X.-H. Nong, X.-Y. Zhang, X.-Y. Xu, J. Wang and S.-H. Qi, J. Nat. Prod., 2016, 79, 141–148, DOI:10.1021/acs.jnatprod.5b00805.
- D. E. Williams, F. Izard, S. Arnould, D. S. Dalisay, C. Tantapakul, W. Maneerat, T. Matainaho, E. Julien and R. J. Andersen, J. Org. Chem., 2016, 81, 1324–1332, DOI:10.1021/acs.joc.5b02569.
- D. E. Williams, D. S. Dalisay, F. Li, J. Amphlett, W. Maneerat, M. A. G. Chavez and Y. A. Wang, Org. Lett., 2013, 15, 414–417, DOI:10.1021/ol303416k.
- Q. Liu, Y. Deng and A. B. Smith III, J. Am. Chem. Soc., 2017, 139, 13668–13671, DOI:10.1021/acs.jacs.7b08683.
- Y. Tan, Y. Hu, Q. Wang, H. Zhou, Y. Wang and M. Gan, RSC Adv., 2016, 6, 91773–91778, 10.1039/c6ra17026a.
- F. Tomita and T. Tamaoki, J. Antibiot., 1980, 33, 940–945, DOI:10.7164/antibiotics.33.940.
- T. Tamaoki, M. Kasai, K. Shirahata, S. Ohkubo, M. Morimoto, K. Mineura, S. Ishii and F. Tomita, J. Antibiot., 1980, 33, 946–950, DOI:10.7164/antibiotics.33.94.
- Y. Zhang, N. Adnani, D. R. Braun, G. A. Ellis, K. J. Barns, S. P. Nance, I. A. Guzei and T. S. Bugni, J. Nat. Prod., 2016, 79, 2968–2972, DOI:10.1021/acs.jnatprod.6b00555.
- Y. Igarashi, D. Asano, M. Sawamura, Y. In, T. Ishida and M. Imoto, Org. Lett., 2016, 18, 1658–1661, DOI:10.1021/acs.orglett.6b00531.
- V. Nair, I. Schuhmann, H. Anke, G. Kelter, H.-H. Fiebig, E. Helmke and H. Laatsch, Planta Med., 2016, 82, 910–918, DOI:10.1055/s-0042-108204.
- W. Zhang, L. Lu, Q. Lai, B. Zhu, Z. Li, Y. Xu, Z. Shao, K. Herrup, B. S. Moore, A. C. Ross and P.-Y. Qian, J. Biol. Chem., 2016, 291, 27228–27238, DOI:10.1055/s-0042-108204.10.1074/jbc.M116.756858.
- M. Luo, G. Tang, J. Ju, L. Lu and H. Huang, Nat. Prod. Res., 2016, 30, 138–143, DOI:10.1080/14786419.2015.1045509.
- S.-H. Kim, T.-K.-Q. Ha, W. Keun Oh, J. Shin and D.-C. Oh, J. Nat. Prod., 2016, 79, 51–58, DOI:10.1021/acs.jnatprod.5b00634.
- Z. Chen, J. Hao, L. Wang, Y. Wang, F. Kong and W. Zhu, Sci. Rep., 2016, 6, 20004, DOI:10.1038/srep20004.
- D.-S. Lee, C.-S. Yoon, Y.-T. Jung, J.-H. Yoon, Y.-C. Kim and H. Oh, J. Nat. Prod., 2016, 79, 1105–1111, DOI:10.1021/acs.jnatprod.6b00009.
- Y. Liang, X. Xie, L. Chen, S. Yan, X. Ye, K. Anjum, H. Huang, X. Lian and Z. Zhang, Mar. Drugs, 2016, 14, 10, DOI:10.3390/md14010010.
- S. Ç. Aksoy, A. Uzel and E. Bedir, J. Antibiot., 2016, 69, 51–56, DOI:10.1038/ja.2015.72.
- U. Mangamuri, V. Muvva, S. Poda, K. Naragani, R. Kumar Munaganti, B. Chitturi and V. Yenamandra, 3 Biotech, 2016, 6, 63, DOI:10.1007/s13205-016-0398-6.
- S. S. Thomasi, C. Ladeira, D. Ferreira, R. d. F. Sprenger, A. C. Badino, A. G. Ferreira and T. Venâncio, Helv. Chim. Acta, 2016, 99, 281–285, DOI:10.1002/hlca.201500038.
- Y.-M. Zhang, H.-Y. Li, C. Hu, H.-F. Sheng, Y. Zhang, B.-R. Lin and G.-X. Zhou, Mar. Drugs, 2016, 14, 84, DOI:10.3390/md14050084.
- M. Pérez, C. Schleissner, R. Fernández, P. Rodríguez, F. Reyes, P. Zuñiga, F. de la Calle and C. Cuevas, J. Antibiot., 2016, 69, 388–394, DOI:10.1038/ja.2015.121.
- C. Cheng, E. M. Othman, A. Reimer, M. Grüne, V. K. -Pavlovic, H. Stopper, U. Hentschel and U. R. Abdelmohsen, Tetrahedron Lett., 2016, 57, 2786–2789, DOI:10.1016/j.tetlet.2016.05.042.
- S. R. Park, A. Tripathi, J. Wu, P. J. Schultz, I. Yim, T. J. McQuade, F. Yu, C.-J. Arevang, A. Y. Mensah, G. T. Castillo, C. Xi and D. H. Sherman, Nat. Commun., 2016, 7, 10710, DOI:10.1038/ncomms10710.
- Y.-x. Ou, J.-f. Huang, X.-m. Li, Q.-j. Kang and Y.-t. Pan, Nat. Prod. Res., 2016, 30, 1771–1775, DOI:10.1080/14786419.2015.1137570.
- Q. Che, J. Li, D. Li, Q. Gu and T. Zhu, J. Antibiot., 2016, 69, 467–469, DOI:10.1038/ja.2015.133.
- Q. Che, H. Tan, X. Han, X. Zhang, Q. Gu, T. Zhu and D. Li, Org. Lett., 2016, 18, 3358–3361, DOI:10.1021/acs.orglett.6b01485.
- H. M. Hassan, C. Boonlarppradab and W. Fenical, J. Antibiot., 2016, 69, 511–514, DOI:10.1038/ja.2016.56.
- C. Cheng, E. M. Othman, A. Fekete, M. Krischke, H. Stopper, R. E. Ebel, M. J. Mueller, U. Hentschel and U. R. Abdelmohsen, Tetrahedron Lett., 2016, 57, 4196–4199, DOI:10.1016/j.tetlet.2016.08.005.
- Y. Han, E. Tian, D. Xu, M. Ma, Z. Deng and K. Hong, Molecules, 2016, 21, 970, DOI:10.3390/molecules21080970.
- A. Ninomiya, Y. Katsuyama, T. Kuranaga, M. Miyazaki, Y. Nogi, S. Okada, T. Wakimoto, Y. Ohnishi, S. Matsunaga and K. Takada, ChemBioChem, 2016, 17, 1709–1712, DOI:10.1002/cbic.201600350.
- S. Fu, F. Wang, H. Li, Y. Bao, Y. Yang, H. Shen, B. Lin and G. Zhou, Nat. Prod. Res., 2016, 30, 2460–2467, DOI:10.1080/14786419.2016.1201668.
- L. Shaala, D. Youssef, J. Badr and S. Harakeh, Molecules, 2016, 21, 1116, DOI:10.3390/molecules21091116.
- O. Sekurova, I. Pérez-Victoria, J. Martín, K. Degnes, H. Sletta, F. Reyes and S. Zotchev, Molecules, 2016, 21, 1131, DOI:10.3390/molecules21091131.
- Y.-H. Chen, M.-C. Lu, H.-M. Chung, C.-F. Weng, J.-H. Su, Y.-T. Yang, Y.-D. Su, Y.-C. Chang, J. Kuo, Y.-C. Wu and P.-J. Sung, Tetrahedron Lett., 2016, 57, 4863–4865, DOI:10.1016/j.tetlet.2016.09.066.
- X. Zhang, X. Ye, W. Chai, X.-Y. Lian and Z. Zhang, Mar. Drugs, 2016, 14, 181, DOI:10.3390/md14100181.
- H. Huang, H. Li, Y. Qiu, L. Hou, J. Ju and W. Li, Mar. Drugs, 2016, 14, 184, DOI:10.3390/md14100184.
- R. Lacret, I. P. Victoria, D. O. Costales, M. de la Cruz, E. Domingo, J. Martín, C. Díaz, F. Vicente, O. Genilloud and F. Reyes, Mar. Drugs, 2016, 14, 188, DOI:10.3390/md14100188.
- C.-X. Wang, R. Ding, S.-T. Jiang, J.-S. Tang, D. Hu, G.-D. Chen, F. Lin, K. Hong, X.-S. Yao and H. Gao, J. Nat. Prod., 2016, 79, 2446–2454, DOI:10.1021/acs.jnatprod.6b00200.
- N. Yang and C. Sun, Front. Microbiol., 2016, 7, 1467, DOI:10.3389/fmicb.2016.01467.
- X. Zhang, Z. Li, L. Du, G. E. Chlipala, P. C. Lopez, W. Zhang, D. H. Sherman and S. Li, Tetrahedron Lett., 2016, 57, 5919–5923, DOI:10.1016/j.tetlet.2016.11.080.
- A. Agena, I. Hermawan, T. Fujiwara, A. Kanamoto and J. Tanaka, Nat. Prod. Commun., 2016, 11, 219–240 Search PubMed.
- K. A. Shaaban, M. Shaaban, V. Nair, I. Schuhmann, H. Y. Win, L. Lei, B. Dittrich, E. Helmke, A. Schüffler and H. Laatsch, Z. Naturforsch., B: J. Chem. Sci., 2016, 71, 1191–1198, DOI:10.1515/znb-2016-0143.
- Y. Feng, J. Liu, Y. P. Carrasco, J. B. MacMillan and J. K. De Brabander, J. Am. Chem. Soc., 2016, 138, 7130–7142, DOI:10.1021/jacs.6b03248.
- A. M. Í. Martínez, F. C. Martínez, J. de la Rosa, M. Cueto, A. D. Marrero, J. Darias, A. B. Espinosa, L. J. P. Rosas and I. E. S. Mercado, Rev. Biol. Mar. Oceanogr., 2016, 51, 161–170, DOI:10.4067/S0718-19572016000100015.
- C. Gao, X. Yi, L. Yu, L. Pan, X. Yang, M. Qin and R. Huang, Chem. Nat. Compd., 2016, 52, 368–369, DOI:10.1007/s10600-016-1648-x.
- S. Son, S.-K. Ko, M. Jang, J. Kim, G. Kim, J. Lee, E. Jeon, Y. Futamura, I.-J. Ryoo, J.-S. Lee, H. Oh, Y.-S. Hong, B. Kim, S. Takahashi, H. Osada, J.-H. Jang and J. Ahn, Mar. Drugs, 2016, 14, 72, DOI:10.3390/md14040072.
- W. Li, X.-X. Tang, X. Yan, Z. Wu, Z.-W. Yi, M.-J. Fang, X. Su and Y.-K. Qiu, Nat. Prod. Res., 2016, 30, 2777–2782, DOI:10.1080/14786419.2016.1155576.
- X. Yan, Y.-X. Zhou, X.-X. Tang, X.-X. Liu, Z.-W. Yi, M.-J. Fang, Z. Wu, F.-Q. Jiang and Y.-K. Qiu, Mar. Drugs, 2016, 14, 195, DOI:10.3390/md14110195.
- Y.-J. Choi, H.-W. Shin, Y.-S. Chun, A. Simplice Leutou, B. Wha Son and J.-W. Park, Oncotarget, 2016, 7, 62107–62122, DOI:10.18632/oncotarget.11529.
- W. Kim, Y. Kim, J. Kim, B.-H. Nam, D.-G. Kim, C. An, J. Lee, P. Kim, H. Lee, J.-S. Oh and J. Lee, Mar. Drugs, 2016, 14, 24, DOI:10.3390/md14010024.
- Y. Sun, T. Tomura, J. Sato, T. Iizuka, R. Fudou and M. Ojika, Molecules, 2016, 21, 59, DOI:10.3390/molecules21010059.
- R. W. Deering, J. Chen, J. Sun, H. Ma, J. Dubert, J. L. Barja, N. P. Seeram, H. Wang and D. C. Rowley, J. Nat. Prod., 2016, 79, 447–450, DOI:10.1021/acs.jnatprod.5b00972.
- P. Tedesco, I. Maida, F. Palma Esposito, E. Tortorella, K. Subko, C. Ezeofor, Y. Zhang, J. Tabudravu, M. Jaspars, R. Fani and D. de Pascale, Mar. Drugs, 2016, 14, 83, DOI:10.3390/md14050083.
- S. L. Moore, L. Berthomier, C. D. Braganza, J. K. MacKichan, J. L. Ryan, G. Visnovsky and R. A. Keyzers, Bioorg. Med. Chem. Lett., 2016, 26, 3086–3088, DOI:10.1016/j.bmcl.2016.05.002.
- S. S. Balan, C. Ganesh Kumar and S. Jayalakshmi, Process Biochem., 2016, 51, 2198–2207, DOI:10.1016/j.procbio.2016.09.009.
- B.-X. Liu, Q. Guo, G.-T. Peng, X.-X. He, X.-J. Chen, L.-F. Lei, Y. Deng, X. Jun Su and C.-X. Zhang, Nat. Prod. Res., 2016, 30, 7–12, DOI:10.1080/14786419.2015.1026340.
- J. Tian, H. Chen, Z. Guo, N. Liu, J. Li, Y. Huang, W. Xiang and Y. Chen, Appl. Microbiol. Biotechnol., 2016, 100, 4189–4199, DOI:10.1007/s00253-015-7248-z.
- J. Lee, C. Han, T. Gu Lee, J. Chin, H. Choi, W. Lee, M. Jeong Paik, D. Hwan Won, G. Jeong, J. Ko, Y. Joon Yoon, S.-J. Nam, W. Fenical and H. Kang, Tetrahedron Lett., 2016, 57, 1997–2000, DOI:10.1016/j.tetlet.2016.03.084.
- P. Huang, F. Xie, B. Ren, Q. Wang, J. Wang, Q. Wang, W. M. Abdel-Mageed, M. Liu, J. Han, A. Oyeleye, J. Shen, F. Song, H. Dai, X. Liu and L. Zhang, Appl. Microbiol. Biotechnol., 2016, 100, 7437–7447, DOI:10.1007/s00253-016-7406-y.
- Q. Vu Thi, V. H. Tran, H. D. Thi Maia, C. V. Le, M. L. Thi Hong, B. T. Murphy, V. M. Chau and V. C. Pham, Nat. Prod. Commun., 2016, 11, 49–51 Search PubMed.
- X.-M. Zhang, D.-F. Zhang, W.-J. Li and C.-H. Lu, Helv. Chim. Acta, 2016, 99, 191–196, DOI:10.1002/hlca.201500109.
- X. Ye, K. Anjum, T. Song, W. Wang, S. Yu, H. Huang, X.-Y. Lian and Z. Zhang, Nat. Prod. Res., 2016, 30, 1156–1161, DOI:10.1080/14786419.2015.1047775.
- H. Zhang, K. Saurav, Z. Yu, A. Mándi, T. Kurtán, J. Li, X. Tian, Q. Zhang, W. Zhang and C. Zhang, J. Nat. Prod., 2016, 79, 1610–1618, DOI:10.1021/acs.jnatprod.6b00175.
- Q. Vu Thi, V. H. Tran, H. D. Thi Mai, C. Vinh Le, M. L. Thi Hong, B. T. Murphy, V. M. Chau and V. C. Pham, Nat. Prod. Commun., 2016, 11, 401–405 Search PubMed.
- C.-L. Xie, S.-W. Niu, T.-T. Zhou, G.-Y. Zhang, Q. Yang and X.-W. Yang, Biochem. Syst. Ecol., 2016, 67, 129–133, DOI:10.1016/j.bse.2016.06.004.
- X. Yan, X.-X. Tang, D. Qin, Z.-W. Yi, M.-J. Fang, Z. Wu and Y.-K. Qiu, Mar. Drugs, 2016, 14, 107, DOI:10.3390/md14060107.
- T. Seitz, P. Fu, F.-L. Haut, L. Adam, M. Habicht, D. Lentz, J. B. MacMillan and M. Christmann, Org. Lett., 2016, 18, 3070–3073, DOI:10.1021/acs.orglett.6b01166.
- X. Jiao, Y. Yao, B. Yang, X. Liu, X. Li, H. Yang, L. Li, J. Xu, M. Xu and P. Xie, Org. Biomol. Chem., 2016, 14, 1805–1813, 10.1039/c5ob02476e.
- Y.-Y. Yao, X.-Y. Liu, X.-Y. Li, H.-G. Yang, L. Li, X.-Z. Jiao and P. Xie, J. Asian Nat. Prod. Res., 2016, 18, 976–987, DOI:10.1080/10286020.2016.1188808.
- Z. Yu, L. Wang, J. Yang, F. Zhang, Y. Sun, M. Yu, Y. Yan, Y.-T. Ma and S.-X. Huang, Tetrahedron Lett., 2016, 57, 1375–1378, DOI:10.1016/j.tetlet.2016.02.061.
- L. Buedenbender, T. Grkovic, S. Duffy, D. Ipek Kurtböke, V. M. Avery and A. R. Carroll, Tetrahedron Lett., 2016, 57, 5893–5895, DOI:10.1016/j.tetlet.2016.11.071.
- S. Matsuda, K. Adachi, Y. Matsuo, M. Nukina and Y. Shizuri, J. Antibiot., 2009, 62, 519–526, DOI:10.1038/ja.2009.75.
- P. G. Williams, R. N. Asolkar, T. Kondratyuk, J. M. Pezzuto, P. R. Jensen and W. Fenical, J. Nat. Prod., 2007, 70, 83–88, DOI:10.1021/np0604580.
- L. M. Blair and J. Sperry, Chem. Commun., 2016, 52, 800–802, 10.1039/c5cc09060a.
- K. Kurasawa, S. Kuwahara and M. Enomoto, Tetrahedron Lett., 2016, 57, 4997–4999, DOI:10.1016/j.tetlet.2016.09.090.
- P. Barbie and U. Kazmaier, Org. Lett., 2016, 18, 204–207, DOI:10.1021/acs.orglett.5b03292.
- P. Barbie and U. Kazmaier, Org. Biomol. Chem., 2016, 14, 6036–6054, 10.1039/c6ob00800c.
- P. Barbie and U. Kazmaier, Org. Biomol. Chem., 2016, 14, 6055–6064, 10.1039/c6ob00801a.
- H. Mitani, T. Matsuo, T. Kodama, K. Nishikawa, Y. Tachi and Y. Morimoto, Tetrahedron, 2016, 72, 7179–7184, DOI:10.1016/j.tet.2016.09.058.
- P. M. Garcia-Barrantes, J. R. Harp and C. W. Lindsley, Tetrahedron Lett., 2016, 57, 2194–2196, DOI:10.1016/j.tetlet.2016.04.019.
- S. Kr Ghosh and R. Nagarajan, Tetrahedron Lett., 2016, 57, 4009–4011, DOI:10.1016/j.tetlet.2016.06.045.
- C. Heinz and N. Cramer, Chimia, 2016, 70, 258–262, DOI:10.2533/chimia.2016.258.
- N. Kanoh, S. Itoh, K. Fujita, K. Sakanishi, R. Sugiyama, Y. Terajima, Y. Iwabuchi, S. Nishimura and H. Kakeya, Chem.–Eur. J., 2016, 22, 8586–8595, DOI:10.1002/chem.201600569.
- V. R. Ghanta, A. Pasula, L. Soma and B. Raman, ChemistrySelect, 2016, 1, 1296–1299, DOI:10.1002/slct.201600415.
- B. Xu, G. Li, J. Li and Y. Shi, Org. Lett., 2016, 18, 2028–2031, DOI:10.1021/acs.orglett.6b00632.
- X.-W. Gao, H.-X. Liu, Z.-H. Sun, Y.-C. Chen, Y.-Z. Tan and W.-M. Zhang, Molecules, 2016, 21, 371, DOI:10.3390/molecules21040371.
- X. An, B.-M. Feng, G. Chen, S.-F. Chen, H.-F. Wang and Y.-H. Pei, Chin. J. Nat. Med., 2016, 14, 934–938, DOI:10.1016/S1875-5364(17)30019-5.
- S. S. Afiyatullov, A. I. Kalinovsky, A. S. Antonov, O. I. Zhuravleva, Y. V. Khudyakova, D. L. Aminin, A. N. Yurchenko and M. V. Pivkin, Phytochem. Lett., 2016, 15, 66–71, DOI:10.1016/j.phytol.2015.11.010.
- Z. Cheng, J. Zhao, D. Liu, P. Proksch, Z. Zhao and W. Lin, J. Nat. Prod., 2016, 79, 1035–1047, DOI:10.1021/acs.jnatprod.5b01103.
- J.-T. Yeon, H. Kim, K.-J. Kim, J. Lee, D. Hwan Won, S.-J. Nam, S. Hwan Kim, H. Kang and Y.-J. Son, J. Nat. Prod., 2016, 79, 1730–1736, DOI:10.1021/acs.jnatprod.6b00004.
- L.-H. Zhang, H.-W. Wang, J.-Y. Xu, J. Li and L. Liu, Nat. Prod. Res., 2016, 30, 2305–2310, DOI:10.1080/14786419.2016.1166498.
- M.-Y. Wei, R.-F. Xu, S.-Y. Du, C.-Y. Wang, T.-Y. Xu and C.-L. Shao, Chem. Nat. Compd., 2016, 52, 1011–1014, DOI:10.1007/s10600-016-1849-3.
- J.-L. Zhang, W.-J. Wang, X.-M. Xu, D.-Y. Li, H.-M. Hua, E.-L. Ma and Z.-L. Li, Tetrahedron, 2016, 72, 4895–4901, DOI:10.1016/j.tet.2016.06.062.
- C. Roullier, Y. Guitton, M. Valery, S. Amand, S. Prado, T. Robiou du Pont, O. Grovel and Y. F. Pouchus, Anal. Chem., 2016, 88, 9143–9150, DOI:10.1021/acs.analchem.6b02128.
- Z. Wu, Y. Wang, D. Liu, P. Proksch, S. Yu and W. Lin, Tetrahedron, 2016, 72, 50–57, DOI:10.1016/j.tet.2015.10.038.
- C. Ding, X. Wu, B. Auckloo, C.-T. Chen, Y. Ye, K. Wang and B. Wu, Molecules, 2016, 21, 105, DOI:10.3390/molecules21010105.
- J. Peng, X. Zhang, W. Wang, T. Zhu, Q. Gu and D. Li, Mar. Drugs, 2016, 14, 131, DOI:10.3390/md14070131.
- C. Wang, L. Guo, J. Hao, L. Wang and W. Zhu, J. Nat. Prod., 2016, 79, 2977–2981, DOI:10.1021/acs.jnatprod.6b00766.
- O. I. Zhuravleva, N. N. Kirichuk, V. A. Denisenko, P. S. Dmitrenok, M. V. Pivkin and S. S. Afiyatullov, Chem. Nat. Compd., 2016, 52, 266–268, DOI:10.1007/s10600-016-1610-y.
- Y. Wang, D.-H. Li, Z.-L. Li, Y.-J. Sun, H.-M. Hua, T. Liu and J. Bai, Molecules, 2016, 21, 31, DOI:10.3390/molecules21010031.
- X. Zhou, W. Fang, S. Tan, X. Lin, T. Xun, B. Yang, S. Liu and Y. Liu, Bioorg. Med. Chem. Lett., 2016, 26, 361–365, DOI:10.1016/j.bmcl.2015.12.005.
- D.-H. Li, T. Han, L.-P. Guan, J. Bai, N. Zhao, Z.-L. Li, X. Wu and H.-M. Hua, Nat. Prod. Res., 2016, 30, 1116–1122, DOI:10.1080/14786419.2015.1043553.
- A. S. Leutou, K. Yun and B. W. Son, Arch. Pharmacal Res., 2016, 39, 806–810, DOI:10.1007/s12272-016-0764-2.
- O. I. Zhuravleva, N. N. Kirichuk, V. A. Denisenko, P. S. Dmitrenok, E. A. Yurchenko, E. M. Min'ko, E. V. Ivanets and S. S. Afiyatullov, Chem. Nat. Compd., 2016, 52, 227–230, DOI:10.1007/s10600-016-1601-z.
- D. A. Adpressa and S. Loesgen, Chem. Biodiversity, 2016, 13, 253–259, DOI:10.1002/cbdv.201500310.
- R. Uchida, K. Nakajyo, K. Kobayashi, T. Ohshiro, T. Terahara, C. Imada and H. Tomoda, J. Antibiot., 2016, 69, 647–651, DOI:10.1038/ja.2016.27.
- G. K. Oleinikova, V. A. Denisenko, D. V. Berdyshev, M. A. Pushilin, N. N. Kirichuk, N. I. Menzorova, A. S. Kuzmich, E. A. Yurchenko, O. I. Zhuravleva and S. S. Afiyatullov, Phytochem. Lett., 2016, 17, 135–139, DOI:10.1016/j.phytol.2016.07.002.
- Z. Cheng, L. Lou, D. Liu, X. Li, P. Proksch, S. Yin and W. Lin, J. Nat. Prod., 2016, 79, 2941–2952, DOI:10.1021/acs.jnatprod.6b00801.
- X.-D. Li, X.-M. Li, X. Li, G.-M. Xu, Y. Liu and B.-G. Wang, J. Nat. Prod., 2016, 79, 1347–1353, DOI:10.1021/acs.jnatprod.5b01153.
- X.-D. Li, X. Li, X.-M. Li, G.-M. Xu, P. Zhang, L.-H. Meng and B.-G. Wang, Planta Med., 2016, 82, 877–881, DOI:10.1055/s-0042-102965.
- X. Zhou, P. Fang, J. Tang, Z. Wu, X. Li, S. Li, Y. Wang, G. Liu, Z. He, D. Gou, X. Yao and L. Wang, Nat. Prod. Res., 2016, 30, 52–57, DOI:10.1080/14786419.2015.1033623.
- Y.-Q. Tian, X.-P. Lin, Z. Wang, X.-F. Zhou, X.-C. Qin, K. Kaliyaperumal, T.-Y. Zhang, Z.-C. Tu and Y. Liu, Molecules, 2016, 21, 34, DOI:10.3390/molecules21010034.
- L. Wang, M. Li, J. Tang and X. Li, Molecules, 2016, 21, 473, DOI:10.3390/molecules21040473.
- S. Liu, H. Dai, B. Konuklugil, R. S. Orfali, W. Lin, R. Kalscheuer, Z. Liu and P. Proksch, Phytochem. Lett., 2016, 18, 187–191, DOI:10.1016/j.phytol.2016.10.015.
- C.-Y. Wang, Y.-F. Liu, F. Cao and C.-Y. Wang, Chem. Nat. Compd., 2016, 52, 1129–1132, DOI:10.1007/s10600-016-1885-z.
- L. Guo, C. Wang, W.-c. Zhu and F.-q. Xu, Biotechnol. Biotechnol. Equip., 2016, 30, 602–606, DOI:10.1080/13102818.2016.1146635.
- T. Yamada, M. Ohshima, K. Yuasa, T. Kikuchi and R. Tanaka, Mar. Drugs, 2016, 14, 74, DOI:10.3390/md14040074.
- B. Wu, J. Wiese, R. Schmaljohann and J. F. Imhoff, Mar. Drugs, 2016, 14, 204, DOI:10.3390/md14110204.
- K. Yun, T. Thang Khong, A. Simplice Leutou, G.-D. Kim, J. Hong, C. HwanLee and B. W. Son, Chem. Pharm. Bull., 2016, 64, 59–62, DOI:10.1248/cpb.c15-00525.
- Z. Zhang, X. Min, J. Huang, Y. Zhong, Y. Wu, X. Li, Y. Deng, Z. Jiang, Z. Shao, L. Zhang and F. He, Mar. Drugs, 2016, 14, 233, DOI:10.3390/md14120233.
- L. Huang, W.-J. Lan, R. Deng, G.-K. Feng, Q.-Y. Xu, Z.-Y. Hu, X.-F. Zhu and H.-J. Li, Mar. Drugs, 2016, 14, 157, DOI:10.3390/md14090157.
- Z. Fan, Z.-H. Sun, H.-X. Liu, Y.-C. Chen, H.-H. Li and W.-M. Zhang, J. Asian Nat. Prod. Res., 2016, 18, 1024–1029, DOI:10.1080/10286020.2016.1181623.
- L.-H. Meng, H.-Q. Chen, I. Form, B. Konuklugil, P. Proksch and B.-G. Wang, Nat. Prod. Commun., 2016, 11, 1293–1296 Search PubMed.
- M. Chen, W. Zhang, C.-L. Shao, Z.-M. Chi and C.-Y. Wang, Mar. Biotechnol., 2016, 18, 409–417, DOI:10.1007/s10126-016-9703-y.
- M. Elsebai, H. Ghabbour and M. Mehiri, Molecules, 2016, 21, 178, DOI:10.3390/molecules21020178.
- D.-L. Zhao, C.-L. Shao, C.-Y. Wang, M. Wang, L.-J. Yang and C.-Y. Wang, Molecules, 2016, 21, 160, DOI:10.3390/molecules21020160.
- Z. Fan, Z.-H. Sun, Z. Liu, Y.-C. Chen, H.-X. Liu, H.-H. Li and W.-M. Zhang, Mar. Drugs, 2016, 14, 164, DOI:10.3390/md14090164.
- H. Long, Z. Cheng, W. Huang, Q. Wu, X. Li, J. Cui, P. Proksch and W. Lin, Org. Lett., 2016, 18, 4678–4681, DOI:10.1021/acs.orglett.6b02313.
- B. Wu, J. Wiese, A. Wenzel-Storjohann, S. Malien, R. Schmaljohann and J. F. Imhoff, Chem.–Eur. J., 2016, 22, 7452–7462, DOI:10.1002/chem.201600430.
- V. Nenkep, K. Yun and B. W. Son, J. Antibiot., 2016, 69, 709–711, DOI:10.1038/ja.2015.137.
- P. Saetang, V. Rukachaisirikul, S. Phongpaichit, J. Sakayaroj, X. Shi, J. Chen and X. Shen, Tetrahedron, 2016, 72, 6421–6427, DOI:10.1016/j.tet.2016.08.048.
- X. Xu, S. Zhao, Y. Yu, Z. Chen, H. Shen and L. Zhou, Nat. Prod. Commun., 2016, 11, 1825–1826 Search PubMed.
- L.-J. Ding, W. Yuan, Y.-X. Li, X.-J. Liao, H. Sun, Q. Peng, B.-N. Han, H.-W. Lin, Z.-Y. Li, F. Yang and S.-H. Xu, Nat. Prod. Res., 2016, 30, 1633–1638, DOI:10.1080/14786419.2015.1129333.
- W. W. M. Zin, S. Buttachon, T. Dethoup, C. Fernandes, S. Cravo, M. M. M. Pinto, L. Gales, J. A. Pereira, A. M. S. Silva, N. Sekeroglu and A. Kijjoa, Mar. Drugs, 2016, 14, 136, DOI:10.3390/md14070136.
- W.-J. Lan, S.-J. Fu, M.-Y. Xu, W.-L. Liang, C.-K. Lam, G.-H. Zhong, J. Xu, D.-P. Yang and H.-J. Li, Mar. Drugs, 2016, 14, 18, DOI:10.3390/md14010018.
- C. Prompanya, T. Dethoup, L. Gales, M. Lee, J. A. C. Pereira, A. M. S. Silva, M. M. M. Pinto and A. Kijjoa, Mar. Drugs, 2016, 14, 134, DOI:10.3390/md14070134.
- L.-J. Ding, W. Yuan, X.-J. Liao, B.-N. Han, S.-P. Wang, Z.-Y. Li, S.-H. Xu, W. Zhang and H.-W. Lin, J. Nat. Prod., 2016, 79, 2045–2052, DOI:10.1021/acs.jnatprod.6b00349.
- W.-F. Xu, X.-M. Hou, K.-L. Yang, F. Cao, R.-Y. Yang, C.-Y. Wang and C.-L. Shao, Mar. Drugs, 2016, 14, 51, DOI:10.3390/md14030051.
- K. Yun, A. Simplice Leutou, J.-R. Rho and B. W. Son, Bull. Korean Chem. Soc., 2016, 37, 103–104, DOI:10.1002/bkcs.10615.
- H. Wang, J. Hong, J. Yin, J. Liu, Y. Liu, J. Sue Choi and J. H. Jung, Bioorg. Med. Chem. Lett., 2016, 26, 2220–2223, DOI:10.1016/j.bmcl.2016.03.057.
- P. Zhang, X.-M. Li, X.-X. Mao, A. Mándi, T. Kurtán and B.-G. Wang, Beilstein J. Org. Chem., 2016, 12, 2012–2018, DOI:10.3762/bjoc.12.188.
- L.-H. Zhang, B.-M. Feng, G. Chen, S.-G. Li, Y. Sun, H.-H. Wu, J. Bai, H.-M. Hua, H.-F. Wang and Y.-H. Pei, RSC Adv., 2016, 6, 42361–42366, 10.1039/c6ra01401a.
- H. Liu, X.-M. Li, Y. Liu, P. Zhang, J.-N. Wang and B.-G. Wang, J. Nat. Prod., 2016, 79, 806–811, DOI:10.1021/acs.jnatprod.5b00893.
- X.-M. Hou, C.-Y. Wang, Y.-C. Gu and C.-L. Shao, Nat. Prod. Res., 2016, 30, 2274–2277, DOI:10.1080/14786419.2016.1163695.
- O. I. Zhuravleva, M. P. Sobolevskaya, V. A. Denisenko, N. N. Kirichuk, M. E. Zhidkov, S. P. Ermakova, N. Y. Kim, A. S. Antonov, E. V. Leshchenko and S. S. Afiyatullov, Nat. Prod. Commun., 2016, 11, 207–217 Search PubMed.
- M. Okabe, T. Sugita, K. Kinoshita and K. Koyama, J. Nat. Prod., 2016, 79, 1208–1212, DOI:10.1021/acs.jnatprod.6b00019.
- S. Chen, J. Wang, X. Lin, B. Zhao, X. Wei, G. Li, K. Kaliaperumal, S. Liao, B. Yang, X. Zhou, J. Liu, S. Xu and Y. Liu, Org. Lett., 2016, 18, 3650–3653, DOI:10.1021/acs.orglett.6b01699.
- L. Yi, C.-B. Cui, C.-W. Li, J.-X. Peng and Q.-Q. Gu, RSC Adv., 2016, 6, 43975–43979, 10.1039/c6ra06250d.
- N. Wang, C.-B. Cui and C.-W. Li, Arch. Pharmacal Res., 2016, 39, 762–770, DOI:10.1007/s12272-016-0751-7.
- C.-W. Li, M.-W. Xia, C.-B. Cui, J.-X. Peng and D.-H. Li, RSC Adv., 2016, 6, 82277–82281, 10.1039/c6ra17087k.
- C.-W. Li, C.-J. Wu, C.-B. Cui, L.-L. Xu, F. Cao and H.-J. Zhu, RSC Adv., 2016, 6, 73383–73387, 10.1039/c6ra14904a.
- H. Shin, G. Pil, S.-J. Heo, H.-S. Lee, J. Lee, Y.-J. Lee, J. Lee and H. Won, Mar. Drugs, 2016, 14, 14, DOI:10.3390/md14010014.
- M. P. Sobolevskaya, O. I. Zhuravleva, E. V. Leshchenko, A. M. Zakharenko, V. A. Denisenko, N. N. Kirichuk, R. S. Popov, D. V. Berdyshev, E. A. Pislyagin, M. V. Pivkin and S. S. Afiyatullov, Phytochem. Lett., 2016, 15, 7–12, DOI:10.1016/j.phytol.2015.10.016.
- M. P. Sobolevskaya, E. V. Leshchenko, T. P. T. Hoai, V. A. Denisenko, S. A. Dyshlovoy, N. N. Kirichuk, Y. V. Khudyakova, N. Y. Kim, D. V. Berdyshev, E. A. Pislyagin, A. S. Kuzmich, A. V. Gerasimenko, R. S. Popov, G. von Amsberg, A. S. Antonov and S. S. Afiyatullov, J. Nat. Prod., 2016, 79, 3031–3038, DOI:10.1021/acs.jnatprod.6b00624.
- Y.-Y. Bu, H. Yamazaki, O. Takahashi, R. Kirikoshi, K. Ukai and M. Namikoshi, J. Antibiot., 2016, 69, 57–61, DOI:10.1038/ja.2015.82.
- S. S. A. Murshid, J. M. Badr and D. T. A. Youssef, Rev. Bras. Farmacogn., 2016, 26, 29–33, DOI:10.1016/j.bjp.2015.09.007.
- E. Abdel-Sattar, M. Abdul-Aziz Al-Yahya, N. Nakamura and M. Hattori, Phytochemistry, 2001, 57, 1213–1217, DOI:10.1016/S0031-9422(01)00163-7.
- M. V. de Castro, L. P. Ióca, D. E. Williams, B. Z. Costa, C. M. Mizuno, M. F. C. Santos, K. de Jesus, É. L. F. Ferreira, M. H. R. Seleghim, L. D. Sette, E. R. Pereira Filho, A. G. Ferreira, N. S. Gonçalves, R. A. Santos, B. O. Patrick, R. J. Andersen and R. G. S. Berlinck, J. Nat. Prod., 2016, 79, 1668–1678, DOI:10.1021/acs.jnatprod.6b00295.
- A. N. Yurchenko, O. F. Smetanina, E. V. Ivanets, A. I. Kalinovsky, Y. V. Khudyakova, N. N. Kirichuk, R. S. Popov, C. Bokemeyer, G. von Amsberg, E. A. Chingizova, S. S. Afiyatullov and S. A. Dyshlovoy, Mar. Drugs, 2016, 14, 122, DOI:10.3390/md14070122.
- C. Pan, Y. Shi, B. Nazia Auckloo, X. Chen, C.-T. Arthur Chen, X. Tao and B. Wu, Mar. Drugs, 2016, 14, 156, DOI:10.3390/md14080156.
- Z.-F. Hu, L.-L. Qin, W.-J. Ding, Y. Liu and Z.-J. Ma, Nat. Prod. Res., 2016, 30, 2311–2315, DOI:10.1080/14786419.2016.1169414.
- L.-h. Zhang, Y. Long, X.-l. Lei, J.-y. Xu, Z.-j. Huang, Z.-g. She, Y.-c. Lin, J. Li and L. Liu, Phytochem. Lett., 2016, 18, 180–186, DOI:10.1016/j.phytol.2016.10.010.
- F. D. Kong, L. M. Zhou, Q. Y. Ma, S. Z. Huang, P. Wang, H.-F. Dai and Y.-X. Zhao, Phytochem. Lett., 2016, 59–63, DOI:10.1016/j.phytol.2016.07.014.
- S. S. A. Mourshid, J. M. Badr, A. L. Risinger, S. L. Mooberry and D. T. A. Youssef, Z. Naturforsch., C: J. Biosci., 2016, 71, 387–392, DOI:10.1515/znc-2015-0242.
- F. Song, H. He, R. Ma, X. Xiao, Q. Wei, Q. Wang, Z. Ji, H. Dai, L. Zhang and R. J. Capon, Tetrahedron Lett., 2016, 57, 3851–3852, DOI:10.1016/j.tetlet.2016.07.049.
- O. F. Smetanina, A. N. Yurchenko, E. V. Ivanets, N. N. Kirichuk, Y. V. Khudyakova, E. A. Yurchenko and S. S. Afiyatullov, Chem. Nat. Compd., 2016, 52, 111–112, DOI:10.1007/s10600-016-1560-4.
- Y. Yaoita and K. Machida, Nat. Prod. Commun., 2016, 11, 947–948 Search PubMed.
- Q. Xing, L.-S. Gan, X.-F. Mou, W. Wang, C.-Y. Wang, M.-Y. Wei and C.-L. Shao, RSC Adv., 2016, 6, 22653–22658, 10.1039/c6ra00374e.
- Y. Zhao, D. Liu, P. Proksch, S. Yu and W. Lin, Chem. Biodiversity, 2016, 13, 1186–1193, DOI:10.1002/cbdv.201600012.
- M. F. Elsebai and H. A. Ghabbour, Tetrahedron Lett., 2016, 57, 354–356, DOI:10.1016/j.tetlet.2015.12.024.
- E. La Kim, J. Lin Li, J. Hong, W. D. Yoon, H. S. Kim, Y. Liu, X. Wei and J. H. Jung, Tetrahedron Lett., 2016, 57, 2803–2806, DOI:10.1016/j.tetlet.2016.05.050.
- M.-S. Lee, S.-W. Wang, G.-J. Wang, K.-L. Pang, C.-K. Lee, Y.-H. Kuo, H.-J. Cha, R.-K. Lin and T.-H. Lee, J. Nat. Prod., 2016, 79, 2983–2990, DOI:10.1021/acs.jnatprod.6b00407.
- F. Cao, J.-K. Yang, Y.-F. Liu, H.-J. Zhu and C.-Y. Wang, Nat. Prod. Res., 2016, 30, 2448–2452, DOI:10.1080/14786419.2016.1198352.
- W.-J. Lan, K.-T. Wang, M.-Y. Xu, J.-J. Zhang, C.-K. Lam, G.-H. Zhong, J. Xu, D.-P. Yang, H.-J. Li and L.-Y. Wang, RSC Adv., 2016, 6, 76206–76213, 10.1039/c6ra06661e.
- K.-T. Wang, M.-Y. Xu, W. Liu, H.-J. Li, J. Xu, D.-P. Yang, W.-J. Lan and L.-Y. Wang, Molecules, 2016, 21, 442, DOI:10.3390/molecules21040442.
- K.-C. Hu, M.-Y. Xu, H.-J. Li, J. Yuan, G. Tang, J. Xu, D.-P. Yang and W.-J. Lan, RSC Adv., 2016, 6, 94763–94770, 10.1039/c6ra21142a.
- M. S. Elnaggar, S. S. Ebada, M. L. Ashour, W. Ebrahim, W. E. G. Müller, A. Mándi, T. Kurtán, A. Singab, W. Lin, Z. Liu and P. Proksch, Tetrahedron, 2016, 72, 2411–2419, DOI:10.1016/j.tet.2016.03.073.
- X. Liang, X.-Y. Zhang, X.-H. Nong, J. Wang, Z.-H. Huang and S.-H. Qi, Tetrahedron, 2016, 72, 3092–3097, DOI:10.1016/j.tet.2016.04.032.
- S. Niu, L. Si, D. Liu, A. Zhou, Z. Zhang, Z. Shao, S. Wang, L. Zhang, D. Zhou and W. Lin, Eur. J. Med. Chem., 2016, 108, 229–244, DOI:10.1016/j.ejmech.2015.09.037.
- S. Buttachon, W. May Zin, T. Dethoup, L. Gales, J. Pereira, A. Silva and A. Kijjoa, Planta Med., 2016, 82, 888–896, DOI:10.1055/s-0042-103687.
- R. Guo, Y. Zhang, D. Duan, Q. Fu, X. Zhang, X. Yu, S. Wang, B. Bao and W. Wu, Chin. J. Chem., 2016, 34, 1194–1198, DOI:10.1002/cjoc.201600623.
- J. W. Kim, S.-K. Ko, H.-M. Kim, G.-H. Kim, S. Son, G. S. Kim, G. J. Hwang, E. S. Jeon, K.-S. Shin, I.-J. Ryoo, Y.-S. Hong, H. Oh, K. H. Lee, N.-K. Soung, D. Hashizume, T. Nogawa, S. Takahashi, B. Y. Kim, H. Osada, J.-H. Jang and J. S. Ahn, J. Nat. Prod., 2016, 79, 2703–2708, DOI:10.1021/acs.jnatprod.6b00641.
- C. Almeida, F. El Maddah, S. Kehraus, G. Schnakenburg and G. M. König, Org. Lett., 2016, 18, 528–531, DOI:10.1021/acs.orglett.5b03553.
- F. El Maddah, S. Kehraus, M. Nazir, C. Almeida and G. M. König, J. Nat. Prod., 2016, 79, 2838–2845, DOI:10.1021/acs.jnatprod.6b00601.
- H.-L. Li, X.-M. Li, H. Liu, L.-H. Meng and B.-G. Wang, Mar. Drugs, 2016, 14, 223, DOI:10.3390/md14120223.
- A. L. Grunwald, F. Berrué, D. P. Overy and R. G. Kerr, Can. J. Chem., 2016, 94, 444–448, DOI:10.1139/cjc-2015-0439.
- Y.-L. Sun, X.-Y. Zhang, X.-H. Nong, X.-Y. Xu and S.-H. Qi, Tetrahedron Lett., 2016, 57, 366–370, DOI:10.1016/j.tetlet.2015.12.026.
- H. Yamazaki, H. Rotinsulu, O. Takahashi, R. Kirikoshi and M. Namikoshi, Tetrahedron Lett., 2016, 57, 5764–5767, DOI:10.1016/j.tetlet.2016.11.028.
- X.-R. Liang, F.-P. Miao, Y.-P. Song, Z.-Y. Guo and N.-Y. Ji, Nat. Prod. Res., 2016, 30, 1605–1610, DOI:10.1080/14786419.2015.1126264.
- X.-R. Liang, F.-P. Miao, Y.-P. Song, X.-H. Liu and N.-Y. Ji, Bioorg. Med. Chem. Lett., 2016, 26, 5029–5031, DOI:10.1016/j.bmcl.2016.08.093.
- M. Suzue, T. Kikuchi, R. Tanaka and T. Yamada, Tetrahedron Lett., 2016, 57, 5070–5073, DOI:10.1016/j.tetlet.2016.10.004.
- M. Mohamed-Benkada, Y. François Pouchus, P. Vérité, F. Pagniez, N. Caroff and N. Ruiz, Chem. Biodiversity, 2016, 13, 521–530, DOI:10.1002/cbdv.201500159.
- J. Arunpanichlert, V. Rukachaisirikul, S. Phongpaichit, O. Supaphon and J. Sakayaroj, Nat. Prod. Res., 2016, 30, 46–51, DOI:10.1080/14786419.2015.1032282.
- M. Ishino, H. Kamauchi, K. Takatori, K. Kinoshita, T. Sugita and K. Koyama, Tetrahedron Lett., 2016, 57, 4341–4344, DOI:10.1016/j.tetlet.2016.08.016.
- I. Kavianinia, L. Kunalingam, P. W. R. Harris, G. M. Cook and M. A. Brimble, Org. Lett., 2016, 18, 3878–3881, DOI:10.1021/acs.orglett.6b01886.
- Z. Ren, Y. Hao and X. Hu, Org. Lett., 2016, 18, 4958–4961, DOI:10.1021/acs.orglett.6b02424.
- S.-H. Hou, Y.-Q. Tu, S.-H. Wang, C.-C. Xi, F.-M. Zhang, S.-H. Wang, Y.-T. Li and L. Liu, Angew. Chem., Int. Ed., 2016, 55, 4456–4460, DOI:10.1002/anie.201600529.
- S. K. Chettu, R. B. Madhu, G. B. Raolji, K. R. Babu, N. S. Kameswara Rao, S. Gopalakrishnan, A. Ismail, G. Bhanuprakash Reddy and S. Shafi, RSC Adv., 2016, 6, 61555–61565, 10.1039/c6ra08861a.
- B. Zhang, W. Zheng, X. Wang, D. Sun and C. Li, Angew. Chem., Int. Ed., 2016, 55, 10435–10438, DOI:10.1002/anie.201604754.
- E. V. Mercado-Marin and R. Sarpong, Chem. Sci., 2015, 6, 5048–5052, 10.1039/c5sc01977j.
- B. Poornima, A. Venkanna, B. Swetha, K. R. Kamireddy, B. Siva, V. S. Phani Babu, R. Ummanni and K. Suresh Babu, Tetrahedron, 2016, 72, 4789–4797, DOI:10.1016/j.tet.2016.06.042.
- B. Seetharamsingh, P. V. Khairnar and D. Srinivasa Reddy, J. Org. Chem., 2016, 81, 290–296, DOI:10.1021/acs.joc.5b02318.
- M. C. Pirrung, F. Zhang, S. Ambadi and Y. Gangadhara Rao, Org. Biomol. Chem., 2016, 14, 8367–8375, 10.1039/c6ob01233g.
- S. Kr Ghosh and R. Nagarajan, Tetrahedron Lett., 2016, 57, 4277–4279, DOI:10.1016/j.tetlet.2016.08.018.
- X.-M. Hou, C.-Y. Wang, Z.-G. She, Y.-C. Gu and C.-L. Shao, Chem. Nat. Compd., 2016, 52, 478–479, DOI:10.1007/s10600-016-1677-5.
- D.-C. Kim, K.-H. Cho, W. Ko, C.-S. Yoon, J. Sohn, J. Yim, Y.-C. Kim and H. Oh, Int. J. Mol. Sci., 2016, 17, 529, DOI:10.3390/ijms17040529.
- J. Wiese, J. F. Imhoff, T. A. M. Gulder, A. Labes and R. Schmaljohann, Mar. Drugs, 2016, 14, 200, DOI:10.3390/md14110200.
- C. Roullier, S. Bertrand, E. Blanchet, M. Peigné, T. Robiou du Pont, Y. Guitton, Y. Pouchus and O. Grovel, Mar. Drugs, 2016, 14, 103, DOI:10.3390/md14050103.
- X. Zhang, H. He, Y. Yin, W. Zhou, M. Cai, X. Zhou and Y. Zhang, J. Biotechnol., 2016, 221, 34–42, DOI:10.1016/j.jbiotec.2016.01.021.
- L. Yu, W. Ding and Z. Ma, Nat. Prod. Res., 2016, 30, 1718–1723, DOI:10.1080/14786419.2015.1136910.
- Y. Liu, Y. Wu, R. Zhai, Z. Liu, X. Huang and Z. She, RSC Adv., 2016, 6, 72127–72132, 10.1039/c6ra16214b.
- C. Tan, Z. Liu, S. Chen, X. Huang, H. Cui, Y. Long, Y. Lu and Z. She, Sci. Rep., 2016, 6, 36609, DOI:10.1038/srep36609.
- Z. Liu, Y. Chen, S. Chen, Y. Liu, Y. Lu, D. Chen, Y. Lin, X. Huang and Z. She, Org. Lett., 2016, 18, 1406–1409, DOI:10.1021/acs.orglett.6b00336.
- Z. Xiao, S. Chen, R. Cai, S. Lin, K. Hong and Z. She, Beilstein J. Org. Chem., 2016, 12, 2077–2085, DOI:10.3762/bjoc.12.196.
- S. Chokpaiboon, P. Unagul, S. Kongthong, K. Danwisetkanjana, A. Pilantanapak, S. Suetrong and T. Bunyapaiboonsri, Tetrahedron Lett., 2016, 57, 1171–1173, DOI:10.1016/j.tetlet.2016.02.002.
- Z.-r. Ju, X. Qin, X.-p. Lin, J.-f. Wang, K. Kaliyaperumal, Y.-q. Tian, J. Liu, F. Liu, Z. Tu, S.-h. Xu and Y. Liu, Nat. Prod. Res., 2016, 30, 192–198, DOI:10.1080/14786419.2015.1050670.
- H. He, Z. Ma, Q. Wang, Y. Liu and H. Xu, Nat. Prod. Res., 2016, 30, 1526–1531, DOI:10.1080/14786419.2015.1116000.
- Y. Shiono, N. Miyazaki, T. Murayama, T. Koseki, Harizon, D. Gede Katja, U. Supratman, J. Nakata, Y. Kakihara, M. Saeki, J. Yoshida, S. Uesugi and K.-i. Kimura, Phytochem. Lett., 2016, 18, 122–127, DOI:10.1016/j.phytol.2016.09.007.
- L.-H. Meng, A. Mándi, X.-M. Li, Y. Liu, T. Kurtán and B.-G. Wang, Chirality, 2016, 28, 581–584, DOI:10.1002/chir.22613.
- J. Li, Y. Xue, J. Yuan, Y. Lu, X. Zhu, Y. Lin and L. Liu, Nat. Prod. Res., 2016, 30, 755–760, DOI:10.1080/14786419.2015.1062762.
- S. Chen, D. Chen, R. Cai, H. Cui, Y. Long, Y. Lu, C. Li and Z. She, J. Nat. Prod., 2016, 79, 2397–2402, DOI:10.1021/acs.jnatprod.6b00639.
- S.-S. Gao, X.-M. Li, K. Williams, P. Proksch, N.-Y. Ji and B.-G. Wang, J. Nat. Prod., 2016, 79, 2066–2074, DOI:10.1021/acs.jnatprod.6b00403.
- H. Cui, Y. Liu, Y. Nie, Z. Liu, S. Chen, Z. Zhang, Y. Lu, L. He, X. Huang and Z. She, Mar. Drugs, 2016, 14, 86, DOI:10.3390/md14050086.
- G. Yu, G. Zhou, M. Zhu, W. Wang, T. Zhu, Q. Gu and D. Li, Org. Lett., 2016, 18, 244–247, DOI:10.1021/acs.orglett.5b02964.
- L.-H. Meng, C.-Y. Wang, A. Mándi, X.-M. Li, X.-Y. Hu, M. U. Kassack, T. Kurtán and B.-G. Wang, Org. Lett., 2016, 18, 5304–5307, DOI:10.1021/acs.orglett.6b02620.
- S. Huang, H. Chen, W. Li, X. Zhu, W. Ding and C. Li, Mar. Drugs, 2016, 14, 172, DOI:10.3390/md14100172.
- C.-J. Zheng, G.-L. Huang, Y. Xu, X.-M. Song, J. Yao, H. Liu, R.-P. Wang and X.-P. Sun, Nat. Prod. Res., 2016, 30, 821–825, DOI:10.1080/14786419.2015.1072712.
- G.-L. Huang, X.-M. Zhou, M. Bai, Y.-X. Liu, Y.-L. Zhao, Y.-P. Luo, Y.-Y. Niu, C.-J. Zheng and G.-Y. Chen, Mar. Drugs, 2016, 14, 177, DOI:10.3390/md14100177.
- R. Xu, X.-M. Li and B.-G. Wang, Phytochem. Lett., 2016, 17, 114–118, DOI:10.1016/j.phytol.2016.07.003.
- T. Liu, S. Zhang, Z. Li, Y. Wang, Z. Chen, J. Bai, L. Tian, Y. Pei and H. Hua, Nat. Prod. Res., 2016, 30, 1025–1029, DOI:10.1080/14786419.2015.1101693.
- H. Liu, S. Chen, W. Liu, Y. Liu, X. Huang and Z. She, Mar. Drugs, 2016, 14, 217, DOI:10.3390/md14120217.
- S.-l. Zhou, M. Wang, H.-g. Zhao, Y.-h. Huang, Y.-y. Lin, G.-h. Tan and S.-l. Chen, Arch. Pharmacal Res., 2016, 39, 1621–1627, DOI:10.1007/s12272-016-0828-3.
- B. Ding, Z. Wang, X. Huang, Y. Liu, W. Chen and Z. She, Nat. Prod. Res., 2016, 30, 2805–2812, DOI:10.1080/14786419.2016.1164702.
- C. F. P. Hemphill, G. Daletos, Z. Liu, W. Lin and P. Proksch, Tetrahedron Lett., 2016, 57, 2078–2083, DOI:10.1016/j.tetlet.2016.03.101.
- S. Liu, H. Dai, G. Makhloufi, C. Heering, C. Janiak, R. Hartmann, A. Mándi, T. Kurtán, W. E. G. Müller, M. U. Kassack, W. Lin, Z. Liu and P. Proksch, J. Nat. Prod., 2016, 79, 2332–2340, DOI:10.1021/acs.jnatprod.6b00473.
- M. Huang, J. Li, L. Liu, S. Yin, J. Wang and Y. Lin, Mar. Drugs, 2016, 14, 215, DOI:10.3390/md14110215.
- Y.-F. Luo, M. Zhang, J.-G. Dai, P. Pedpradab, W.-J. Wang and J. Wu, Phytochem. Lett., 2016, 17, 162–166, DOI:10.1016/j.phytol.2016.07.027.
- M. Wibowo, V. Prachyawarakorn, T. Aree, C. Mahidol, S. Ruchirawat and P. Kittakoop, Phytochemistry, 2016, 122, 126–138, DOI:10.1016/j.phytochem.2015.11.016.
- S. Chokpaiboon, S. Choodej, N. Boonyuen, T. Teerawatananond and K. Pudhom, Phytochemistry, 2016, 122, 172–177, DOI:10.1016/j.phytochem.2015.12.010.
- M. Moussa, W. Ebrahim, M. El-Neketi, A. Mándi, T. Kurtán, R. Hartmann, W. Lin, Z. Liu and P. Proksch, Tetrahedron Lett., 2016, 57, 4074–4078, DOI:10.1016/j.tetlet.2016.07.091.
- S. Chen, Y. Liu, Z. Liu, R. Cai, Y. Lu, X. Huang and Z. She, RSC Adv., 2016, 6, 26412–26420, 10.1039/c6ra02566h.
- M. Zhang, J.-M. Liu, J.-L. Zhao, N. Li, R.-D. Chen, K.-B. Xie, W.-J. Zhang, K. -P. Feng, Z. Yan, N. Wang and J.-G. Dai, Chin. Chem. Lett., 2016, 27, 957–960, DOI:10.1016/j.cclet.2016.02.008.
- S. Choodej, T. Teerawatananond, T. Mitsunaga and K. Pudhom, Mar. Drugs, 2016, 14, 132, DOI:10.3390/md14070132.
- J. Wang and R. Tong, J. Org. Chem., 2016, 81, 4325–4339, DOI:10.1021/acs.joc.6b00788.
- Y.-Q. Yang, Synth. Commun., 2016, 46, 1263–1267, DOI:10.1080/00397911.2016.1198813.
- J. McNulty, D. McLeod and H. A. Jenkins, Eur. J. Org. Chem., 2016, 2016, 688–692, DOI:10.1002/ejoc.201501592.
- H. Chai, R. Yin, Y. Liu, H. Meng, X. Zhou, G. Zhou, X. Bi, X. Yang, T. Zhu, W. Zhu, Z. Deng and K. Hong, Sci. Rep., 2016, 6, 27181, DOI:10.1038/srep27181.
- H. Ogawa, A. Iwasaki, S. Sumimoto, Y. Kanamori, O. Ohno, M. Iwatsuki, A. Ishiyama, R. Hokari, K. Otoguro, S. Omura and K. Suenaga, J. Nat. Prod., 2016, 79, 1862–1866, DOI:10.1021/acs.jnatprod.6b00171.
- J. A. V. Lopez, S. S. Al-Lihaibi, W. M. Alarif, A. A. Lateff, Y. Nogata, K. Washio, M. Morikawa and T. Okino, J. Nat. Prod., 2016, 79, 1213–1218, DOI:10.1021/acs.jnatprod.6b00051.
- K. Sueyoshi, M. Kaneda, S. Sumimoto, S. Oishi, N. Fujii, K. Suenaga and T. Teruya, Tetrahedron, 2016, 72, 5472–5478, DOI:10.1016/j.tet.2016.07.031.
- R. A. Medina, D. E. Goeger, P. Hills, S. L. Mooberry, N. Huang, L. I. Romero, E. Ortega-Barria, W. H. Gerwick and K. L. McPhail, J. Am. Chem. Soc., 2008, 130, 6324, DOI:10.1021/ja801383f.
- G. Yao, Z. Pan, C. Wu, W. Wang, L. Fang and W. Su, J. Am. Chem. Soc., 2015, 137, 13488–13491, DOI:10.1021/jacs.5b09286.
- H. Luesch, W. Y. Yoshida, R. E. Moore, V. J. Paul and T. H. Corbett, J. Am. Chem. Soc., 2001, 123, 5418–5423, DOI:10.1021/ja010453j.
- J. D. Serrill, X. Wan, A. M. Hau, H. S. Jang, D. J. Coleman, A. K. Indra, A. W. G. Alani, K. L. McPhail and J. E. Ishmael, Invest. New Drugs, 2016, 34, 24–40, DOI:10.1007/s10637-015-0303-x.
- W. Cai, J. Matthews, V. Paul and H. Luesch, Planta Med., 2016, 82, 897–902, DOI:10.1055/s-0042-105157.
- M. J. Bertin, O. Demirkiran, G. Navarro, N. A. Moss, J. Lee, G. M. Goldgof, E. Vigil, E. A. Winzeler, F. A. Valeriote and W. H. Gerwick, Phytochemistry, 2016, 122, 113–118, DOI:10.1016/j.phytochem.2015.11.011.
- A. Awadhi, R. Ratnayake, V. J. Paul and H. Luesch, Bioorg. Med. Chem., 2016, 24, 3276–3282, DOI:10.1016/j.bmc.2016.04.062.
- D. Youssef, S. Ibrahim, L. Shaala, G. Mohamed and Z. Banjar, Molecules, 2016, 21, 324, DOI:10.3390/molecules21030324.
- S. Sumimoto, A. Iwasaki, O. Ohno, K. Sueyoshi, T. Teruya and K. Suenaga, Org. Lett., 2016, 18, 4884–4887, DOI:10.1021/acs.orglett.6b02364.
- L. Spoof, A. Błaszczyk, J. Meriluoto, M. Cegłowska and H. Mazur-Marzec, Mar. Drugs, 2016, 14, 8, DOI:10.3390/md14010008.
- T. B. Afonso, M. Sofia Costa, R. R. de Castro, S. Freitas, A. Silva, M. P. C. Schneider, R. Martins and P. N. Leão, J. Nat. Prod., 2016, 79, 2504–2513, DOI:10.1021/acs.jnatprod.6b00351.
- M. S. Costa, A. Rego, V. Ramos, T. B. Afonso, S. Freitas, M. Preto, V. Lopes, V. Vasconcelos, C. Magalhães and P. N. Leão, Sci. Rep., 2016, 6, 23436, DOI:10.1038/srep23436.
- Y. Kanamori, A. Iwasaki, S. Sumimoto and K. Suenaga, Tetrahedron Lett., 2016, 57, 4213–4216, DOI:10.1016/j.tetlet.2016.08.012.
- S. P. Gunasekera, L. Imperial, C. Garst, R. Ratnayake, L. H. Dang, V. J. Paul and H. Luesch, J. Nat. Prod., 2016, 79, 1867–1871, DOI:10.1021/acs.jnatprod.6b00203.
- S. P. Gunasekera, Y. Li, R. Ratnayake, D. Luo, J. Lo, J. H. Reibenspies, Z. Xu, M. J. Clare-Salzler, T. Ye, V. J. Paul and H. Luesch, Chem.–Eur. J., 2016, 22, 8158–8166, DOI:10.1002/chem.201600674.
- A. M. Fenner, N. Engene, C. Spadafora, W. H. Gerwick and M. J. Balunas, Org. Lett., 2016, 18, 352–355, DOI:10.1021/acs.orglett.5b03110.
- L. A. Salvador, V. J. Paul and H. Luesch, J. Nat. Prod., 2016, 79, 452, DOI:10.1021/acs.jnatprod.6b00120.
- E. Sato, Y. Tanabe, N. Nakajima, A. Ohkubo and K. Suenaga, Org. Lett., 2016, 18, 2047–2049, DOI:10.1021/acs.orglett.6b00660.
- S. Das, D. Paul and R. K. Goswami, Org. Lett., 2016, 18, 1908–1911, DOI:10.1021/acs.orglett.6b00713.
- B. Long, J. Zhang, X. Tang and Z. Wu, Org. Biomol. Chem., 2016, 14, 9712–9715, 10.1039/c6ob01783e.
- M. Kaneda, K. Sueyoshi, T. Teruya, H. Ohno, N. Fujii and S. Oishi, Org. Biomol. Chem., 2016, 14, 9093–9104, 10.1039/c6ob01583b.
- D. Luo, Q.-Y. Chen and H. Luesch, J. Org. Chem., 2016, 81, 532–544, DOI:10.1021/acs.joc.5b02386.
- S. Shinomiya, A. Iwasaki, O. Ohno and K. Suenaga, Phytochemistry, 2016, 132, 109–114, DOI:10.1016/j.phytochem.2016.10.005.
- A. Iwasaki, T. Teruya and K. Suenaga, Tetrahedron Lett., 2010, 51, 959–960, DOI:10.1016/j.tetlet.2009.12.041.
- K. Kunifuda, A. Iwasaki, M. Nagamoto and K. Suenaga, Tetrahedron Lett., 2016, 57, 3121–3123, DOI:10.1016/j.tetlet.2016.06.002.
- A. Gil, J. Lamariano-Merketegi, A. Lorente, F. Albericio and M. Álvarez, Chem.–Eur. J., 2016, 22, 7033–7035, DOI:10.1002/chem.201600770.
- A. Kolleth, J. Gebauer, A. ElMarrouni, R. Lebeuf, C. Prévost, E. Brohan, S. Arseniyadis and J. Cossy, Front. Chem., 2016, 4, 34, DOI:10.3389/fchem.2016.00034.
- D. N. Reddy, F. Ballante, T. Chuang, A. Pirolli, B. Marrocco and G. R. Marshall, J. Med. Chem., 2016, 59, 1613–1633, DOI:10.1021/acs.jmedchem.5b01632.
- J. Guo, I. N. Chaithanya Kiran, J. Gao, R. Santhosh Reddy and Y. He, Tetrahedron Lett., 2016, 57, 3481–3484, DOI:10.1016/j.tetlet.2016.06.091.
- M. Scherer, D. Bezold and K. Gademann, Angew. Chem., Int. Ed., 2016, 55, 9427–9431, DOI:10.1002/anie.201602755.
- K. Harju, H. Koskela, A. Kremp, S. Suikkanen, P. de la Iglesia, C. O. Miles, B. Krock and P. Vanninen, Toxicon, 2016, 112, 68–76, DOI:10.1016/j.toxicon.2016.01.064.
- M. Akakabe, K. Kumagai, M. Tsuda, Y. Konishi, A. Tominaga, D. Kaneno, E. Fukushi, J. Kawabata, A. Masuda and M. Tsuda, Chem. Pharm. Bull., 2016, 64, 1019–1023, DOI:10.1248/cpb.c16-00026.
- R. Irie, R. Suzuki, K. Tachibana, P. T. Holland, D. T. Harwood, F. Shi, P. McNabb, V. Beuzenberg, F. Hayashi, H. Zhang and M. Satake, Heterocycles, 2016, 92, 45–54, DOI:10.3987/COM-15-13332.
- P. Cai, S. He, C. Zhou, A. R. Place, S. Haq, L. Ding, H. Chen, Y. Jiang, C. Guo, Y. Xu, J. Zhang and X. Yan, Harmful Algae, 2016, 58, 66–73, DOI:10.1016/j.hal.2016.08.001.
- S. A. Rasmussen, S. Meier, N. G. Andersen, H. E. Blossom, J. Ø. Duus, K. F. Nielsen, P. J. Hansen and T. O. Larsen, J. Nat. Prod., 2016, 79, 2250–2256, DOI:10.1021/acs.jnatprod.6b00345.
- M. Kanto and M. Sasaki, Org. Lett., 2016, 18, 112–115, DOI:10.1021/acs.orglett.5b03346.
- M. Kanto, S. Sato, M. Tsuda and M. Sasaki, J. Org. Chem., 2016, 81, 9105–9121, DOI:10.1021/acs.joc.6b01700.
- H. Takamura, T. Fujiwara, Y. Kawakubo, I. Kadota and D. Uemura, Chem.–Eur. J., 2016, 22, 1979–1983, DOI:10.1002/chem.201503880.
- H. Takamura, T. Fujiwara, Y. Kawakubo, I. Kadota and D. Uemura, Chem.–Eur. J., 2016, 22, 1984–1996, DOI:10.1002/chem.201503881.
- M. Anttila, W. Strangman, R. York, C. Tomas and J. L. C. Wright, J. Nat. Prod., 2016, 79, 484–489, DOI:10.1021/acs.jnatprod.5b00869.
- T. Kubota, H. Sato, T. Iwai and J. Kobayashi, Chem. Pharm. Bull., 2016, 64, 979–981, DOI:10.1248/cpb.c16-00202.
- S. Tsuchiya, Y. Cho, K. Konoki, K. Nagasawa, Y. Oshima and M. Yotsu-Yamashita, Sci. Rep., 2016, 6, 20340, DOI:10.1038/srep20340.
- K.-C. Cheng, P.-C. Kuo, H.-Y. Hung, K.-H. Yu, T.-L. Hwang, P.-C. Shieh, J.-S. Chang and T.-S. Wu, Phytochem. Lett., 2016, 18, 113–116, DOI:10.1016/j.phytol.2016.09.008.
- X. Sun, H. Jin, L. Zhang, W. Hu, Y. Li and N. Xu, Chin. J. Oceanol. Limnol., 2016, 34, 781–788, DOI:10.1007/s00343-016-4383-z.
- Y.-y. Sun, H. Wang, G.-l. Guo, Y.-f. Pu, B.-l. Yan and C.-h. Wang, Environ. Sci. Pollut. Res., 2016, 23, 1449–1459, DOI:10.1007/s11356-015-5377-7.
- Z. Manzoor, J.-E. Koo, I. Ali, J.-E. Kim, S.-H. Byeon, E.-S. Yoo, H.-K. Kang, J.-W. Hyun, N.-H. Lee and Y.-S. Koh, Mar. Drugs, 2016, 14, 88, DOI:10.3390/md14050088.
- M. Kamio, M. Koyama, N. Hayashihara, K. Hiei, H. Uchida, R. Watanabe, T. Suzuki and H. Nagai, J. Chem. Ecol., 2016, 42, 452–460, DOI:10.1007/s10886-016-0703-1.
- T. Yan, Y. Zhao, X. Zhang and X. Lin, Mar. Drugs, 2016, 14, 56, DOI:10.3390/md14030056.
- C. Paliwal, T. Ghosh, B. George, I. Pancha, R. Maurya, K. Chokshi, A. Ghosh and S. Mishra, Algal Res., 2016, 15, 24–31, DOI:10.1016/j.algal.2016.01.017.
- S. Hielscher-Michael, C. Griehl, M. Buchholz, H.-U. Demuth, N. Arnold and L. A. Wessjohann, Mar. Drugs, 2016, 14, 203, DOI:10.3390/md14110203.
- C. de los Reyes, M. J. Ortega, H. Zbakh, V. Motilva and E. Zubía, J. Nat. Prod., 2016, 79, 395–405, DOI:10.1021/acs.jnatprod.5b01067.
- N.-Y. Ji, Y.-P. Song, F.-P. Miao and X.-R. Liang, Magn. Reson. Chem., 2016, 54, 88–90, DOI:10.1002/mrc.4319.
- A. Othmani, R. Bunet, J.-L. Bonnefont, J.-F. Briand and G. Culioli, J. Appl. Phycol., 2016, 28, 1975–1986, DOI:10.1007/s10811-015-0668-4.
- M. Dimou, E. Ioannou, M. G. Daskalaki, L. A. Tziveleka, S. C. Kampranis and V. Roussis, J. Nat. Prod., 2016, 79, 584–589, DOI:10.1021/acs.jnatprod.5b01031.
- C. Vieira, O. P. Thomas, G. Culioli, G. Genta-Jouve, F. Houlbreque, J. Gaubert, O. De Clerck and C. E. Payri, Sci. Rep., 2016, 6, 18637, DOI:10.1038/srep18637.
- D. C. Soares, M. M. Szlachta, V. L. Teixeira, A. R. Soares and E. M. Saraiva, Mar. Drugs, 2016, 14, 163, DOI:10.3390/md14090163.
- J.-A. Kim, F. Karadeniz, B.-N. Ahn, M. Sook Kwon, O.-J. Mun, M. Joo Bae, Y. Seo, M. Kim, S.-H. Lee, Y. Yong Kim, J. Mi-Soon and C.-S. Kong, J. Sci. Food Agric., 2016, 96, 783–790, DOI:10.1002/jsfa.7148.
- H. Zbakh, E. Talero, J. Avila, A. Alcaide, C. de los Reyes, E. Zubía and V. Motilva, Mar. Drugs, 2016, 14, 149, DOI:10.3390/md14080149.
- C. de Souza Barros, C. C. Cirne-Santos, V. Garrido, I. Barcelos, P. R. S. Stephens, V. Giongo, V. L. Teixeira and I. C. N. de Palmer Paixão, J. Appl. Phycol., 2016, 28, 2523–2527, DOI:10.1007/s10811-015-0776-1.
- L. R. Velatooru, C. B. Baggu and V. R. Janapala, Mol. Carcinog., 2016, 55, 2222–2235, DOI:10.1002/mc.22463.
- C. L. Hugelshofer and T. Magauer, J. Am. Chem. Soc., 2016, 138, 6420–6423, DOI:10.1021/jacs.6b03720.
- C. L. Hugelshofer and T. Magauer, Chem.–Eur. J., 2016, 22, 15125–15136, DOI:10.1002/chem.201603061.
- C. A. Incerti-Pradillos, M. A. Kabeshov, P. S. O'Hora, S. A. Shipilovskikh, A. E. Rubtsov, V. A. Drobkova, S. Y. Balandina and A. V. Malkov, Chem.–Eur. J., 2016, 22, 14390–14396, DOI:10.1002/chem.201602440.
- D. Zhao, L. Zheng, L. Qi, S. Wang, L. Guan, Y. Xia and J. Cai, Mar. Drugs, 2016, 14, 123, DOI:10.3390/md14070123.
- V. K. Sali, D. P. Mansingh and H. R. Vasanthi, MedChemComm, 2016, 7, 1429–1435, 10.1039/C6MD00178E.
- E. da Costa, J. Silva, S. Mendonça, M. Abreu and M. Domingues, Mar. Drugs, 2016, 14, 101, DOI:10.3390/md14050101.
- O. V. Tabakaeva and A. V. Tabakaev, Chem. Nat. Compd., 2016, 52, 777–781, DOI:10.1007/s10600-016-1776-3.
- H. Eo, T.-H. Kwon, G. Park, H. Song, Su-J. Lee, N.-H. Park and J. Jeong, Mar. Drugs, 2016, 14, 69, DOI:10.3390/md14040069.
- A.-R. Kim, B. Lee, E.-J. Joung, W.-G. Gwon, T. Utsuki, N.-G. Kim and H.-R. Kim, Immunopharmacol. Immunotoxicol., 2016, 38, 244–252, DOI:10.3109/08923973.2016.1173060.
- B.-N. Ahn, F. Karadeniz, C.-S. Kong, K.-H. Nam, M.-S. Jang, Y. Seo and H. S. Kim, Mar. Drugs, 2016, 14, 168, DOI:10.3390/md14090168.
- J.-J. Kim, Y.-J. Kang, S.-A. Shin, D.-H. Bak, J. Won Lee, K. Bok Lee, Y. Choon Yoo, D.-K. Kim, B. Ho Lee, D. Woon Kim, J. Lee, E.-K. Jo and J.-M. Yuk, PLoS One, 2016, 11, e0163433, DOI:10.1371/journal.pone.0163433.
- E.-A. Kim, S.-H. Lee, J.-H. Lee, N. Kang, J.-Y. Oh, S.-h. Seun-heui, G. Ahn, S. C. Ko, S. P. Fernando, S.-Y. Kim, S.-J. Park, Y.-T. Kim and Y.-J. Jeon, RSC Adv., 2016, 6, 78570–78575, 10.1039/C6RA12724J.
- S.-H. Cha, S.-J. Heo, Y.-J. Jeon and S. M. Park, RSC Adv., 2016, 6, 110040–110046, 10.1039/C6RA21697H.
- R. Wei, M.-S. Lee, B. Lee, C.-W. Oh, C.-G. Choi and H.-R. Kim, J. Appl. Phycol., 2016, 28, 3535–3545, DOI:10.1007/s10811-016-0847-y.
- S.-H. Lee, S.-H. Eom, N.-Y. Yoon, M.-M. Kim, Y.-X. Li, S. Keun Ha and S.-K. Kim, J. Chem., 2016, 2016, 6509212, DOI:10.1155/2016/6509212.
- G. Rajauria, B. Foley and N. Abu-Ghannam, Innovative Food Sci. Emerging Technol., 2016, 37, 261–268, DOI:10.1016/j.ifset.2016.02.005.
- S.-H. Lee, S.-C. Ko, M.-C. Kang, D. Ho Lee and Y.-J. Jeon, Food Chem. Toxicol., 2016, 91, 58–64, DOI:10.1016/j.fct.2016.02.022.
- R. Karthik, V. Manigandan, R. Sheeba, R. Saravanan and P. R. Rajesh, J. Appl. Phycol., 2016, 28, 3561–3573, DOI:10.1007/s10811-016-0851-2.
- J. Lin, L. Huang, J. Yu, S. Xiang, J. Wang, J. Zhang, X. Yan, W. Cui, S. He and Q. Wang, Mar. Drugs, 2016, 14, 67, DOI:10.3390/md14040067.
- Y. Yang, M. Bae, B. Kim, Y.-K. Park, S. I. Koo and J.-Y. Lee, J. Nutr. Biochem., 2016, 29, 21–26, DOI:10.1016/j.jnutbio.2015.11.005.
- Y. Sugiura, Y. Kinoshita, M. Usui, R. Tanaka, T. Matsushita and M. Miyata, Food Sci. Technol. Res., 2016, 22, 227–234, DOI:10.3136/fstr.22.227.
- Y. Liu, J. Zheng, Y. Zhang, Z. Wang, Y. Yang, M. Bai and Y. Dai, Neurochem. Res., 2016, 41, 2728–2751, DOI:10.1007/s11064-016-1989-7.
- C. Vizetto-Duarte, L. Custódio, K. N. Gangadhar, J. H. G. Lago, C. Dias, A. M. Matos, N. Neng, J. M. F. Nogueira, L. Barreira, F. Albericio, A. P. Rauter and J. Varela, Phytomedicine, 2016, 23, 550–557, DOI:10.1016/j.phymed.2016.02.008.
- H. Ah Jung, M. Yousof Ali, R. Joo Choi, H. Oh Jeong, H. Young Chung and J. S. Choi, Food Chem. Toxicol., 2016, 89, 104–111, DOI:10.1016/j.fct.2016.01.014.
- Y. Liu, M. Liu, X. Zhang, Q. Chen, H. Chen, L. Sun and G. Liu, J. Agric. Food Chem., 2016, 64, 416–424, DOI:10.1021/acs.jafc.5b05436.
- J.-I. Kang, E.-S. Yoo, J.-W. Hyun, Y.-S. Koh, N. Ho Lee, M.-H. Ko, C.-S. Ko and H.-K. Kang, Biol. Pharm. Bull., 2016, 39, 1273–1283, DOI:10.1248/bpb.b16-00024.
- J.-H. Jang, J.-H. Lee, H. S. Chand, J.-S. Lee, Y. Lin, N. Weathington, R. Mallampalli, Y.-J. Jeon and T. Nyunoya, Mar. Drugs, 2016, 14, 140, DOI:10.3390/md14070140.
- J.-H. Choi, N.-H. Kim, S.-J. Kim, H.-J. Lee and S. Kim, J. Biochem. Mol. Toxicol., 2016, 30, 111–119, DOI:10.1002/jbt.21769.
- A. Grasa-López, Á. Miliar-García, L. Quevedo-Corona, N. Paniagua-Castro, G. Escalona-Cardoso, E. Reyes-Maldonado and M.-E. Jaramillo-Flores, Mar. Drugs, 2016, 14, 148, DOI:10.3390/md14080148.
- A. A. Goda, K. M. Naguib, M. M. Mohamed, H. A. Amra, S. A. Nada, A.-R. B. Abdel-Ghaffar, C. R. Gissendanner and K. A. El Sayed, Mar. Drugs, 2016, 14, 208, DOI:10.3390/md14110208.
- T. D. Tran, N. B. Pham and R. J. Quinn, J. Nat. Prod., 2016, 79, 570–577, DOI:10.1021/acs.jnatprod.5b00989.
- A. Gutiérrez-Cepeda, J. J. Fernández, M. Norte, M. López-Rodríguez, I. Brito, C. D. Muller and M. L. Souto, J. Nat. Prod., 2016, 79, 1184–1188, DOI:10.1021/acs.jnatprod.5b01080.
- J. G. Hall and J. A. Reiss, Aust. J. Chem., 1986, 39, 1401–1409, DOI:10.1071/CH9861401.
- B. S. Dyson, J. W. Burton, T. Sohn, B. Kim, H. Bae and D. Kim, J. Am. Chem. Soc., 2012, 134, 11781–11790, DOI:10.1021/ja304554e.
- S. Urban, R. Brkljača, M. Hoshino, S. Lee and M. Fujita, Angew. Chem., Int. Ed., 2016, 55, 2678–2682, DOI:10.1002/anie.20150976.1.
- M. Suzuki, S. Nakano, Y. Takahashi, T. Abe and M. Masuda, Phytochemistry, 1999, 51, 657–662, DOI:10.1016/S0031-9422(99)00102-8.
- I. Shin, D. Lee and H. Kim, Org. Lett., 2016, 18, 4420–4423, DOI:10.1021/acs.orglett.6b02239.
- J.-Y. Chen, C.-Y. Huang, Y.-S. Lin, T.-L. Hwang, W.-L. Wang, S.-F. Chiou and J.-H. Sheu, J. Nat. Prod., 2016, 79, 2315–2323, DOI:10.1021/acs.jnatprod.6b00452.
- W. M. Alarif, K. O. Al-Footy, M. S. Zubair, M. Halid PH, M. A. Ghandourah, S. A. Basaif, S. S. Al-Lihaibi, S.-E. N. Ayyad and F. A. Badria, Nat. Prod. Res., 2016, 30, 1150–1155, DOI:10.1080/14786419.2015.1046378.
- N.-Y. Ji, X.-M. Li, L.-P. Ding and B.-G. Wang, Biochem. Syst. Ecol., 2016, 64, 1–5, DOI:10.1016/j.bse.2015.11.010.
- K. Chakraborty, D. Joseph, M. Joy and V. K. Raola, Food Chem., 2016, 212, 778–788, DOI:10.1016/j.foodchem.2016.06.039.
- V. H. Woolner, C. M. Jones, J. J. Field, N. H. Fadzilah, A. B. Munkacsi, J. H. Miller, R. A. Keyzers and P. T. Northcote, J. Nat. Prod., 2016, 79, 463–469, DOI:10.1021/acs.jnatprod.5b00831.
- M.-C. Li, W.-S. Sun, W. Cheng, D. Liu, H. Liang, Q.-Y. Zhang and W.-H. Lin, Bioorg. Med. Chem. Lett., 2016, 26, 3590–3593, DOI:10.1016/j.bmcl.2016.06.015.
- D. Mikami, H. Kurihara, M. Ono, S. Moo Kim and K. Takahashi, Fitoterapia, 2016, 108, 20–25, DOI:10.1016/j.fitote.2015.11.002.
- A. G. Gonzalez and J. Darias, Tetrahedron Lett., 1976, 3051–3054, DOI:10.1016/0040-4039(76)80067-6.
- O. Salvador-Neto, S. Gomes, A. Soares, F. Machado, R. Samuels, R. Nunes da Fonseca, J. Souza-Menezes, J. Moraes, E. Campos, F. Mury and J. Silva, Mar. Drugs, 2016, 14, 20, DOI:10.3390/md14020020.
- H. Kurihara, T. Mitani, J. Kawabata and K. Takahashi, J. Nat. Prod., 1999, 62, 882–884, DOI:10.1021/np980324p.
- F. Xu, F. Wang, Z. Wang, W. Lv, W. Wang and Y. Wang, PLoS One, 2016, 11, e0147748, DOI:10.1371/journal.pone.0147748.
- K. Becker, A. Hartmann, M. Ganzera, D. Fuchs and J. M. Gostner, Mar. Drugs, 2016, 14, 119, DOI:10.3390/md14060119.
- R. E. Moore and G. Bartolini, J. Am. Chem. Soc., 1981, 103, 2491–2494, DOI:10.1021/ja00399a093.
- S. Mori, K. Sugahara, M. Maeda, K. Nomoto, T. Iwashita and T. Yamagaki, Tetrahedron Lett., 2016, 57, 3612–3617, DOI:10.1016/j.tetlet.2016.06.108.
- J. J. Sims, G. H. Y. Lin and R. M. Wing, Tetrahedron Lett., 1974, 3487–3490, DOI:10.1016/S0040-4039(01)91944-6.
- D. B. Sudatti, M. T. Fujii, S. V. Rodrigues, A. Turra, H. M. Duarte, A. R. Soares and R. C. Pereira, Biochem. Syst. Ecol., 2016, 64, 131–135, DOI:10.1016/j.bse.2015.12.001.
- F. L. S. Machado, H. M. Duarte, L. M. S. Gestinari, V. Cassano, C. R. Kaiser and A. R. Soares, Chem. Biodiversity, 2016, 13, 845–851, DOI:10.1002/cbdv.201500246.
- F. L. Calegario, PLoS One, 2016, 11, e0165954, DOI:10.1371/journal.pone.0165954.
- Y. Andriani, D. Fitrya Syamsumir, T. Ching Yee, F. Shaharom Harisson, G. Ming Herng and S. A. Abdullah, Nat. Prod. Commun., 2016, 11, 1117–1120 Search PubMed.
- W. Xu, X. Liao, S. Xu, J. Diao, B. Du, X. Zhou and S. Pan, Org. Lett., 2008, 10, 4569–4572, DOI:10.1021/ol801799d.
- J. Lunagariya, S. Zhong, J. Chen, D. Bai, P. Bhadja, W. Long, X. Liao, X. Tang and S. Xu, Mar. Drugs, 2016, 14, 161, DOI:10.3390/md14090161.
- S. Ankisetty, S. Nandiraju, H. Win, Y. C. Park, C. D. Amsler, J. B. McClintock, J. A. Baker and T. K. Diyabalanage, J. Nat. Prod., 2004, 67, 1295–1302, DOI:10.1021/np049965c.
- F. J. Seidl and N. Z. Burns, Beilstein J. Org. Chem., 2016, 12, 1361–1365, DOI:10.3762/bjoc.12.129.
- J. Clarke, K. J. Bonney, M. Yaqoob, S. Solanki, H. S. Rzepa, A. J. P. White, D. S. Millan and D. Christopher Braddock, J. Org. Chem., 2016, 81, 9539–9552, DOI:10.1021/acs.joc.6b02008.
- A. Gutierrez-Cepeda, J. J. Fernandez, L. V. Gil, M. Lopez-Rodriguez, M. Norte and M. L. Souto, J. Nat. Prod., 2011, 74, 441–448, DOI:10.1021/np100866g.
- A. G. Cepeda, J. J. Fernandez, M. Norte and M. L. Souto, Org. Lett., 2011, 13, 2690–2693, DOI:10.1021/ol200792v.
- D. R. Abou-Hussein and D. T. A. Youssef, Mar. Drugs, 2016, 14, 155, DOI:10.3390/md14080155.
- H. Gaspar, A. Cutignano, L. Grauso, N. Neng, V. Cachatra, A. Fontana, J. Xavier, M. Cerejo, H. Vieira and S. Santos, Mar. Drugs, 2016, 14, 179, DOI:10.3390/md14100179.
- N. M. Carballeira, N. Montano, L. A. Amador, A. D. Rodríguez, M. Y. Golovko, S. A. Golovko, R. M. Reguera, R. Álvarez-Velilla and R. Balaña-Fouce, Lipids, 2016, 51, 245–256, DOI:10.1007/s11745-015-4114-9.
- K. Takada, Y. Imae, Y. Ise, S. Ohtsuka, A. Ito, S. Okada, M. Yoshida and S. Matsunaga, J. Nat. Prod., 2016, 79, 2384–2390, DOI:10.1021/acs.jnatprod.6b00588.
- E. Einarsdottir, H.-B. Liu, J. Freysdottir, C. Gotfredsen and S. Omarsdottir, Planta Med., 2016, 82, 903–909, DOI:10.1055/s-0042-105877.
- A. G. Guzii, T. N. Makarieva, V. A. Denisenko, P. S. Dmitrenok, A. S. Kuzmich, S. A. Dyshlovoy, G. von Amsberg, V. B. Krasokhin and V. A. Stonik, Org. Lett., 2016, 18, 3478–3481, DOI:10.1021/acs.orglett.6b01678.
- D. B. Abdjul, H. Yamazaki, O. Takahashi, R. Kirikoshi, K. Ukai and M. Namikoshi, Chem. Pharm. Bull., 2016, 64, 733–736, DOI:10.1248/cpb.c16-00061.
- M. T. Jamison, D. S. Dalisay and T. F. Molinski, J. Nat. Prod., 2016, 79, 555–563, DOI:10.1021/acs.jnatprod.5b00951.
- G. Chianese, H.-B. Yu, F. Yang, C. Sirignano, P. Luciano, B.-N. Han, S. Khan, H.-W. Lin and O. Taglialatela-Scafati, J. Org. Chem., 2016, 81, 5135–5143, DOI:10.1021/acs.joc.6b00695.
- S. Khokhar, G. K. Pierens, J. N. A. Hooper, M. G. Ekins, Y. Feng and R. A. Davis, J. Nat. Prod., 2016, 79, 946–953, DOI:10.1021/acs.jnatprod.5b01029.
- N. Tanokashira, S. Kukita, H. Kato, T. Nehira, E. D. Angkouw, R. E. P. Mangindaan, N. J. de Voogd and S. Tsukamoto, Tetrahedron, 2016, 72, 5530–5540, DOI:10.1016/j.tet.2016.07.045.
- G. Esposito, G. Della Sala, R. Teta, A. Caso, M.-L. Bourguet-Kondracki, J. R. Pawlik, A. Mangoni and V. Costantino, Eur. J. Org. Chem., 2016, 2016, 2871–2875, DOI:10.1002/ejoc.201600370.
- K. Ishida and M. Murakami, J. Org. Chem., 2000, 65, 5898–5900, DOI:10.1021/jo991918f.
- Y. Nakashima, Y. Egami, M. Kimura, T. Wakimoto and I. Abe, PLoS One, 2016, 11, e0164468, DOI:10.1371/journal.pone.0164468.
- K. Fukuhara, K. Takada, S. Okada and S. Matsunaga, J. Nat. Prod., 2016, 79, 1694–1697, DOI:10.1021/acs.jnatprod.6b00261.
- A. H. Afifi, A. H. El-Desoky, H. Kato, R. E. P. Mangindaan, N. J. de Voogd, N. M. Ammar, M. S. Hifnawy and S. Tsukamoto, Tetrahedron Lett., 2016, 57, 1285–1288, DOI:10.1016/j.tetlet.2016.02.031.
- J. Sun, W. Cheng, N. J. de Voogd, P. Proksch and W. Lin, Tetrahedron Lett., 2016, 57, 4288–4292, DOI:10.1016/j.tetlet.2016.08.024.
- C. Urda, M. Pérez, J. Rodríguez, C. Jiménez, C. Cuevas and R. Fernández, Tetrahedron Lett., 2016, 57, 3239–3242, DOI:10.1016/j.tetlet.2016.05.054.
- L. A. Shaala, D. T. A. Youssef, S. R. M. Ibrahim and G. A. Mohamed, Nat. Prod. Res., 2016, 30, 2783–2790, DOI:10.1080/14786419.2016.1155577.
- M. Stierhof, K. Ø. Hansen, M. Sharma, K. Feussner, K. Subko, F. F. Díaz-Rullo, J. Isaksson, I. Pérez-Victoria, D. Clarke, E. Hansen, M. Jaspars and J. N. Tabudravu, Tetrahedron, 2016, 72, 6929–6934, DOI:10.1016/j.tet.2016.09.016.
- M. Su, H. Li, H. Wang, E. L. Kim, H. S. Kim, E.-H. Kim, J. Lee and J. H. Jung, Bioorg. Med. Chem., 2016, 24, 2979–2987, DOI:10.1016/j.bmc.2016.05.006.
- D. Hahn, H. Kim, I. Yang, J. Chin, H. Hwang, D. Hwan Won, B. Lee, S.-J. Nam, M. Ekins, H. Choi and H. Kang, J. Nat. Prod., 2016, 79, 499–506, DOI:10.1021/acs.jnatprod.5b00871.
- K. C. Tan, T. Wakimoto and I. Abe, Org. Lett., 2014, 16, 3256–3259, DOI:10.1021/ol501271v.
- K. Co Tan, T. Wakimoto and I. Abe, J. Nat. Prod., 2016, 79, 2418–2422, DOI:10.1021/acs.jnatprod.6b00586.
- M. T. Jamison and T. F. Molinski, J. Nat. Prod., 2016, 79, 2243–2249, DOI:10.1021/acs.jnatprod.6b00336.
- B. Yang, J. Huang, X. Lin, Y. Zhang, H. Tao and Y. Liu, Rec. Nat. Prod., 2016, 10, 117–121 CAS.
- C.-K. Kim, J.-K. Woo, Y.-J. Lee, H.-S. Lee, C. J. Sim, D.-C. Oh, K.-B. Oh and J. Shin, J. Nat. Prod., 2016, 79, 1179–1183, DOI:10.1021/acs.jnatprod.5b01078.
- D. B. Abdjul, H. Yamazaki, S.-i. Kanno, O. Takahashi, R. Kirikoshi, K. Ukai and M. Namikoshi, J. Nat. Prod., 2016, 79, 1149–1154, DOI:10.1021/acs.jnatprod.6b00095.
- B. Harrison, S. Talapatra, E. Lobkovsky, J. Clardy and P. Crews, Tetrahedron Lett., 1996, 37, 9151–9154, DOI:10.1016/S0040-4039(96)02165-X.
- G. Esposito, M.-L. Bourguet-Kondracki, L. H. Mai, A. Longeon, R. Teta, L. Meijer, R. Van Soest, A. Mangoni and V. Costantino, J. Nat. Prod., 2016, 79, 2953–2960, DOI:10.1021/acs.jnatprod.6b00939.
- M. Arai, K. Kamiya, D. Shin, H. Matsumoto, T. Hisa, A. Setiawan, N. Kotoku and M. Kobayashi, Chem. Pharm. Bull., 2016, 64, 766–771, DOI:10.1248/cpb.c16-00118.
- H. Zhang, S. T. Loveridge, K. Tenney and P. Crews, Nat. Prod. Res., 2016, 30, 1262–1265, DOI:10.1080/14786419.2015.1054826.
- B. P. Bashyal, G. W. J. Fleet, M. J. Gough and P. W. Smith, Tetrahedron, 1987, 43, 3083–3093, DOI:10.1016/S0040-4020(01)86850-2.
- S. M. Al-Massarani, A. A. El-Gamal, M. S. Al-Said, M. S. Abdel-Kader, A. E. Ashour, A. Kumar, W. M. Abdel-Mageed, A. J. Al-Rehaily, H. A. Ghabbour and H.-K. Fun, Pharmacogn. Mag., 2016, 12, 114–119, DOI:10.4103/0973-1296.177906.
- E. K. Olsen, E. Hansen, L. W. K. Moodie, J. Isaksson, K. Sepčić, M. Cergolj, J. Svenson and J. H. Andersen, Org. Biomol. Chem., 2016, 14, 1629–1640, 10.1039/c5ob02416a.
- Y. Hitora, K. Takada, Y. Ise, S. Okada and S. Matsunaga, J. Nat. Prod., 2016, 79, 2973–2976, DOI:10.1021/acs.jnatprod.6b00710.
- T. Kubota, K. Nakamura, K. Sakai, J. Fromont, T. Gonoi and J. Kobayashi, Chem. Pharm. Bull., 2016, 64, 975–978, DOI:10.1248/cpb.c16-00201.
- S. R. M. Ibrahim and G. A. Mohamed, Phytochem. Lett., 2016, 18, 168–171, DOI:10.1016/j.phytol.2016.10.014.
- N. K. Utkina and V. A. Denisenko, Nat. Prod. Commun., 2016, 11, 1259–1260 Search PubMed.
- R.-Y. Huang, W.-T. Chen, T. Kurtán, A. Mándi, J. Ding, J. Li, X.-W. Li and Y.-W. Guo, Future Med. Chem., 2016, 8, 17–27, DOI:10.4155/fmc.15.169.
- A. El-Demerdash, C. Moriou, M. Thérèse Martin, A. d. S. R. Stien, S. Petek, M. Demoy-Schneider, K. Hall, J. N. A. Hooper, C. Debitus and A. Al-Mourabit, J. Nat. Prod., 2016, 79, 1929–1937, DOI:10.1021/acs.jnatprod.6b00168.
- K. M. Tabakmakher, T. N. Makarieva, L. K. Shubina, V. A. Denisenko, R. S. Popov, A. S. Kuzmich, H.-S. Lee, Y.-J. Lee and V. A. Stonik, Nat. Prod. Commun., 2016, 11, 1817–1820 Search PubMed.
- Y. Zhu, Y. Wang, B.-B. Gu, F. Yang, W.-H. Jiao, G.-H. Hu, H.-B. Yu, B.-N. Han, W. Zhang, Y. Shen and H.-W. Lin, Tetrahedron, 2016, 72, 2964–2971, DOI:10.1016/j.tet.2016.04.020.
- K.-K. Gong, X.-L. Tang, Y.-S. Liu, P.-L. Li and G.-Q. Li, Molecules, 2016, 21, 150, DOI:10.3390/molecules21020150.
- A. Mokhlesi, R. Hartmann, E. Achten, Chaidir, T. Hartmann, W. Lin, G. Daletos and P. Proksch, Eur. J. Org. Chem., 2016, 2016, 639–643, DOI:10.1002/ejoc.201501250.
- J. Muñoz and M. Köck, J. Nat. Prod., 2016, 79, 434–437, DOI:10.1021/acs.jnatprod.5b00265.
- J. Dai, S. M. Parrish, W. Y. Yoshida, M. L. Richard Yip, J. Turkson, M. Kelly and P. Williams, Bioorg. Med. Chem. Lett., 2016, 26, 499–504, DOI:10.1016/j.bmcl.2015.11.086.
- N. Sirimangkalakitti, M. Yokoya, S. Chamni, P. Chanvorachote, A. Plubrukrn, N. Saito and K. Suwanborirux, Chem. Pharm. Bull., 2016, 64, 258–262, DOI:10.1248/cpb.c15-00901.
- J. Rodríguez, C. Jiménez, M. Blanco, G. Tarazona, R. Fernández and C. Cuevas, Org. Lett., 2016, 18, 5832–5835, DOI:10.1021/acs.orglett.6b02832.
- B. C. Hinshaw, J. F. Gerster, R. K. Robins and L. B. Townsend, J. Org. Chem., 1970, 35, 236–241, DOI:10.1021/jo00826a049.
- D. Wang, Y. Feng, M. Murtaza, S. Wood, G. Mellick, J. N. A. Hooper and R. J. Quinn, J. Nat. Prod., 2016, 79, 353–361, DOI:10.1021/acs.jnatprod.5b00987.
- H. M. Nguyen, T. Ito, N. N. Win, T. Kodama, V. Q. Hung, H. T. Nguyen and H. Morita, Phytochem. Lett., 2016, 17, 288–292, DOI:10.1016/j.phytol.2016.08.012.
- D. B. Abdjul, H. Yamazaki, O. Takahashi, R. Kirikoshi, K. Ukai and M. Namikoshi, J. Nat. Prod., 2016, 79, 1842–1847, DOI:10.1021/acs.jnatprod.6b00367.
- X. Zhang, H.-Y. Xu, A.-M. Huang, L. Wang, Q. Wang, P.-Y. Cao and P.-M. Yang, Chem. Pharm. Bull., 2016, 64, 1036–1042, DOI:10.1248/cpb.c16-00183.
- W.-H. Jiao, G.-H. Shi, T.-T. Xu, G.-D. Chen, B.-B. Gu, Z. Wang, S. Peng, S.-P. Wang, J. Li, B.-N. Han, W. Zhang and H.-W. Lin, J. Nat. Prod., 2016, 79, 406–411, DOI:10.1021/acs.jnatprod.5b01079.
- T. Li, B. Wang, N. J. de Voogd, X.-L. Tang, Q. Wang, M.-J. Chu, P.-L. Li and G.-Q. Li, Chin. Chem. Lett., 2016, 27, 1048–1051, DOI:10.1016/j.cclet.2016.05.017.
- H.-L. Liu, D.-Q. Xue, S.-H. Chen, X.-W. Li and Y.-W. Guo, Helv. Chim. Acta, 2016, 99, 650–653, DOI:10.1002/hlca.201600077.
- H. Prawat, C. Mahidol, W. Kaweetripob, V. Prachyawarakorn, P. Tuntiwachwuttikul and S. Ruchirawat, Tetrahedron, 2016, 72, 4222–4229, DOI:10.1016/j.tet.2016.05.060.
- P. Van Kiem, N. Xuan Nhiem, B. Huu Tai, H. Le Tuan Anh, D. Thi Thuy Hang, N. Thi Cuc, L. Thi Huyen, N. Hoai Nam, P. Hai Yen, D. Cong Thung and C. Van Minh, Nat. Prod. Commun., 2016, 11, 439–480 Search PubMed.
- A. Srikrishna and S. Gharpure, Perkin 1, 2000, 3191–3193, 10.1039/b005943i.
- E. K. Olsen, K. L. Søderholm, J. Isaksson, J. H. Andersen and E. Hansen, J. Nat. Prod., 2016, 79, 1285–1291, DOI:10.1021/acs.jnatprod.5b00966.
- K. Ota, Y. Hamamoto, W. Eda, K. Tamura, A. Sawada, A. Hoshino, H. Mitome, K. Kamaike and H. Miyaoka, J. Nat. Prod., 2016, 79, 996–1004, DOI:10.1021/acs.jnatprod.5b01069.
- M. Shingaki, T. Wauke, P. Ahmadi and J. Tanaka, Chem. Pharm. Bull., 2016, 64, 272–275, DOI:10.1248/cpb.c15-00726.
- A. H. El-Desoky, H. Kato, E. D. Angkouw, R. E. P. Mangindaan, N. J. de Voogd and S. Tsukamoto, J. Nat. Prod., 2016, 79, 1922–1928, DOI:10.1021/acs.jnatprod.6b00158.
- J. L. von Salm, C. G. Witowski, R. M. Fleeman, J. B. McClintock, C. D. Amsler, L. N. Shaw and B. J. Baker, Org. Lett., 2016, 18, 2596–2599, DOI:10.1021/acs.orglett.6b00979.
- C.-c. Su, H.-j. Su, K.-j. Liang, S.-j. Tsaif and J.-h. Su, Nat. Prod. Commun., 2016, 11, 445–451 Search PubMed.
- F. Yang, R.-P. Wang, B. Xu, H.-B. Yu, G.-Y. Ma, G.-F. Wang, S.-W. Dai, W. Zhang, W.-H. Jiao, S.-J. Song and H.-W. Lin, Bioorg. Med. Chem. Lett., 2016, 26, 2084–2087, DOI:10.1016/j.bmcl.2016.02.070.
- S. S. Elhady, A. M. Al-Abd, A. M. El-Halawany, A. M. Alahdal, H. A. Hassanean and S. A. Ahmed, Mar. Drugs, 2016, 14, 130, DOI:10.3390/md14070130.
- S. Elhady, A. El-Halawany, A. Alahdal, H. Hassanean and S. Ahmed, Molecules, 2016, 21, 82, DOI:10.3390/molecules21010082.
- K.-H. Lai, Y.-C. Liu, J.-H. Su, M. El-Shazly, C.-F. Wu, Y.-C. Du, Y.-M. Hsu, J.-C. Yang, M.-K. Weng, C.-H. Chou, G.-Y. Chen, Y.-C. Chen and M.-C. Lu, Sci. Rep., 2016, 6, 36170, DOI:10.1038/srep36170.
- M. Wang, I. Tietjen, M. Chen, D. E. Williams, J. Daoust, M. A. Brockman and R. J. Andersen, J. Org. Chem., 2016, 81, 11324–11334, DOI:10.1021/acs.joc.6b02312.
- J. Dai, W. Y. Yoshida, M. Kelly and P. Williams, J. Nat. Prod., 2016, 79, 1464–1467, DOI:10.1021/acs.jnatprod.6b00042.
- W.-F. He, D.-Q. Xue, L.-G. Yao, J. Li, H.-L. Liu and Y.-W. Guo, J. Asian Nat. Prod. Res., 2016, 18, 195–199, DOI:10.1080/10286020.2015.1056521.
- Z. Cheng, D. Liu, N. J. de Voogd, P. Proksch and W. Lin, Helv. Chim. Acta, 2016, 99, 588–596, DOI:10.1002/hlca.201600021.
- M. Chen, X.-D. Wu, Q. Zhao and C.-Y. Wang, Mar. Drugs, 2016, 14, 146, DOI:10.3390/md14080146.
- A. El-Gamal, S. Al-Massarani, L. Shaala, A. Alahdald, M. Al-Said, A. Ashour, A. Kumar, M. Abdel-Kader, W. Abdel-Mageed and D. Youssef, Mar. Drugs, 2016, 14, 82, DOI:10.3390/md14050082.
- H. Mitome, H. Miyaoka, M. Nakano and Y. Yamada, Tetrahedron Lett., 1995, 36, 8231–8234, DOI:10.1016/0040-4039(95)01772-A.
- C. Djerassi and R. W. Lang, Tetrahedron Lett., 1982, 23, 2063–2066, DOI:10.1016/S0040-4039(00)87261-5.
- W.-G. Xu, J. Wang, G.-S. Xing, J.-J. Xu, W. Qiao, C. Zhao and S.-A. Tang, Z. Naturforsch., C: J. Biosci., 2016, 71, 111–114, DOI:10.1515/znc-2016-0054.
- T.-Y. Eom, Y.-J. Lee and H.-S. Lee, Ocean Polar Res., 2016, 38, 287–294, DOI:10.4217/OPR.2016.38.4.287.
- A. Zampella, A. Randazzo, N. Borbone, S. Luciani, L. Trevisi, C. Debitus and M. V. D'Auria, Tetrahedron Lett., 2002, 43, 6163–6166, DOI:10.1016/S0040-4039(02)01334-5.
- M. V. D'Auria, V. Sepe, R. D'Orsi, F. Bellotta, C. Debitus and A. Zampella, Tetrahedron, 2007, 63, 131–140, DOI:10.1016/j.tet.2006.10.032.
- M. Kikuchi and H. Konno, Biosci., Biotechnol., Biochem., 2016, 80, 1066–1069, DOI:10.1080/09168451.2016.1148581.
- I. Paterson, S. M. Dalby, J. C. Roberts, G. J. Naylor, E. A. Guzman, R. Isbrucker, T. P. Pitts, P. Linley, D. Divlianska, J. K. Reed and A. E. Wright, Angew. Chem., Int. Ed., 2011, 50, 3219–3223, DOI:10.1002/anie.201007719.
- E. A. Guzmán, Q. Xu, T. P. Pitts, K. O. Mitsuhashi, C. Baker, P. A. Linley, J. Oestreicher, K. Tendyke, P. L. Winder, E. M. Suh and A. E. Wright, Int. J. Cancer, 2016, 139, 2116–2126, DOI:10.1002/ijc.30253.
- P. Ciminiello, C. Dell'Aversano, E. Fattorusso, S. Magno and M. Pansini, J. Nat. Prod., 2000, 63, 263–266, DOI:10.1021/np990343e.
- L. W. K. Moodie, M. C. Žužek, R. Frangež, J. H. Andersen, E. Hansen, E. K. Olsen, M. Cergolj, K. Sepčić, K. Ø. Hansen and J. Svenson, Org. Biomol. Chem., 2016, 14, 11220–11229, 10.1039/C6OB02120D.
- N. B. Perry, J. W. Blunt and M. H. G. Munro, Tetrahedron, 1988, 44, 1727–1734, DOI:10.1016/S0040-4020(01)86737-5.
- D. C. Radisky, E. S. Radisky, L. R. Barrows, B. R. Copp, R. A. Kramer and C. M. Ireland, J. Am. Chem. Soc., 1993, 115, 1632–1638, DOI:10.1021/ja00058a003.
- A. K. L. Goey, C. H. Chau, T. M. Sissung, K. M. Cook, D. J. Venzon, A. Castro, T. R. Ransom, C. J. Henrich, T. C. McKee, J. B. McMahon, T. Grkovic, M. M. Cadelis, B. R. Copp, K. R. Gustafson and W. D. Figg, J. Nat. Prod., 2016, 79, 1267–1275, DOI:10.1021/acs.jnatprod.5b00846.
- E. W. Schmidt, M. K. Harper and D. J. Faulkner, J. Nat. Prod., 1995, 58, 1861–1867, DOI:10.1021/np50126a008.
- E. Alonso, R. Alvariño, M. Leirós, J. N. Tabudravu, K. Feussner, M. A. Dam, M. E. Rateb, M. Jaspars and L. M. Botana, Mar. Drugs, 2016, 14, 197, DOI:10.3390/md14110197.
- S. Forenza, L. Minale, R. Riccio and E. Fattorusso, J. Chem. Soc., Chem. Commun., 1971, 1129–1130, 10.1039/c29710001129.
- R. P. Walker, D. J. Faulkner, D. Van Engen and J. Clardy, J. Am. Chem. Soc., 1981, 103, 6772–6773, DOI:10.1021/ja00412a052.
- R. J. Melander, H.-B. Liu, M. D. Stephens, C. A. Bewley and C. Melander, Bioorg. Med. Chem. Lett., 2016, 26, 5863–5866, DOI:10.1016/j.bmcl.2016.11.018.
- A. O'Rourke, S. Kremb, T. Bader, M. Helfer, P. Schmitt-Kopplin, W. Gerwick, R. Brack-Werner and C. Voolstra, Mar. Drugs, 2016, 14, 28, DOI:10.3390/md14020028.
- G. Cimino, S. De Rosa, S. De Stefano, R. Self and G. Sodano, Tetrahedron Lett., 1983, 24, 3029–3032, DOI:10.1016/S0040-4039(00)88087-9.
- C. Florean, M. Schnekenburger, J.-Y. Lee, K. Rok Kim, A. Mazumder, S. Song, J.-M. Kim, C. Grandjenette, J.-G. Kim, A.-Y. Yoon, M. Dicato, K.-W. Kim, C. Christov, B.-W. Han, P. Proksch and M. Diederich, Oncotarget, 2016, 7, 24027–24049, DOI:10.18632/oncotarget.8210.
- https://openinnovation.lilly.com/, accessed 4 September 2017.
- V. Anjaneyulu, M. M. Kirshna and P. Radhika, Acta Cienc. Indica, Chem., 1996, 22, 59–60 CAS.
- S. Kohmoto, O. J. McConnell, A. Wright, F. Koehn, W. Thompson, M. Lui and K. M. Snader, J. Nat. Prod., 1987, 50, 336, DOI:10.1021/np50050a064.
- H. Ebrahim and K. El Sayed, Mar. Drugs, 2016, 14, 57, DOI:10.3390/md14030057.
- L. Minale, R. Riccio and G. Sodano, Tetrahedron Lett., 1974, 3401–3404, DOI:10.1016/S0040-4039(01)91918-5.
- K. A. Alvi, M. C. Diaz, P. Crews, D. L. Slate, R. H. Lee and R. Moretti, J. Org. Chem., 1992, 57, 6604–6607, DOI:10.1021/jo00050a043.
- R. T. Luibrand, T. R. Erdman, J. J. Vollmer, P. J. Scheuer, J. Finer and J. Clardy, Tetrahedron, 1979, 35, 609–612, DOI:10.1016/0040-4020(79)87004-0.
- R. Kazlauskas, P. T. Murphy, R. G. Warren, R. J. Wells and J. F. Blount, Aust. J. Chem., 1978, 31, 2685–2697, DOI:10.1071/CH9782685.
- E. L. Chaikina, N. K. Utkina and M. M. Anisimov, Nat. Prod. Commun., 2016, 11, 11–13 Search PubMed.
- B. W. Sullivan, D. J. Faulkner, K. T. Okamoto, M. H. M. Chen and J. Clardy, J. Org. Chem., 1986, 51, 5134–5136, DOI:10.1021/jo00376a014.
- D. B. Abdjul, S.-I. Kanno, H. Yamazaki, K. Ukai and M. Namikoshi, Bioorg. Med. Chem. Lett., 2016, 26, 315–317, DOI:10.1016/j.bmcl.2015.12.022.
- L. Mayol, V. Piccialli and D. Sica, Tetrahedron Lett., 1985, 26, 1357–1360, DOI:10.1016/S0040-4039(00)94893-7.
- A. Poiner and W. C. Taylor, Aust. J. Chem., 1990, 43, 1713–1727, DOI:10.1071/CH9901713.
- A. Rueda, A. Losada, R. Fernandez, C. Cabanas, L. F. Garcia-Fernandez, F. Reyes and C. Cuevas, Lett. Drug Des. Discovery, 2006, 3, 753–760, DOI:10.2174/157018006778631875.
- M. E. Rateb, W. E. Houssen, M. Schumacher, W. T. A. Harrison, M. Diederich, R. Ebel and M. Jaspars, J. Nat. Prod., 2009, 72, 1471–1476, DOI:10.1021/np900233c.
- P. Karuso, P. R. Bergquist, R. C. Cambie, J. S. Buckleton, G. R. Clark and C. E. F. Rickard, Aust. J. Chem., 1986, 39, 1643–1653, DOI:10.1071/CH9861643.
- J. A. Sánchez, A. Alfonso, M. Leirós, E. Alonso, M. E. Rateb, M. Jaspars, W. E. Houssen, R. Ebel, J. Tabudravu and L. M. Botana, Pharmacol. Res., 2016, 107, 407–414, DOI:10.1016/j.phrs.2016.03.029.
- J. A. Sánchez, A. Alfonso, I. Rodriguez, E. Alonso, J. M. Cifuentes, R. Bermudez, M. E. Rateb, M. Jaspars, W. E. Houssen, R. Ebel, J. Tabudravu and L. M. Botana, Front. Immunol., 2016, 7, 452, DOI:10.3389/fimmu.2016.00452.
- J. Rae Rho, H.-S. Lee, C. J. Sim and J. Shin, Tetrahedron, 2002, 58, 9585–9591, DOI:10.1016/S0040-4020(02)01257-7.
- H. Y. Lee, E. J. Jang, S. Y. Bae, J.-e. Jeon, H. J. Park, J. Shin and S. K. Lee, Mar. Drugs, 2016, 14, 212, DOI:10.3390/md14110212.
- V. Costantino, E. Fattorusso, C. Imperatore and A. Mangoni, J. Org. Chem., 2008, 73, 6158–6165, DOI:10.1021/jo800837k.
- P.-C. Gao, S.-Y. Zhu, H. Cao and J.-S. Yang, J. Am. Chem. Soc., 2016, 138, 1684–1688, DOI:10.1021/jacs.5b12589.
- K. Watanabe, Y. Tsuda, Y. Yamane, K. Takahashi, K. Iguchi, H. Naoki and T. Fujita, Tennen Yuki Kagobutsu Toronkai Koen Yoshishu, 2000, 42, 367–372 Search PubMed.
- F. Liu, J. Zhong, S. Li, M. Li, L. Wu, Q. Wang, J. Mao, S. Liu, B. Zheng, M. Wang and Q. Bian, J. Nat. Prod., 2016, 79, 244–247, DOI:10.1021/acs.jnatprod.5b00713.
- S. Aoki, K. Matsui, H. Wei, N. Murakami and M. Kobayashi, Tetrahedron, 2002, 58, 5417–5422, DOI:10.1016/S0040-4020(02)00519-7.
- V. A. D'yakonov, L. U. Dzhemileva, A. A. Makarov, E. N. Andreev and U. M. Dzhemilev, Russ. J. Org. Chem., 2016, 52, 1844–1846, DOI:10.1134/S107042801612023X.
- J.-X. Gong, H.-Y. Wang, L.-G. Yao, X.-W. Li and Y.-W. Guo, Synlett, 2016, 27, 391–394, DOI:10.1055/s-0035-1560807.
- K. Horikawa, T. Yagyu, Y. Yoshioka, T. Fujiwara, A. Kanamoto, T. Okamoto and M. Ojika, Tetrahedron, 2013, 69, 101–106, DOI:10.1016/j.tet.2012.10.063.
- P. Gangadhar, A. Sathish Reddy and P. Srihari, Tetrahedron, 2016, 72, 5807–5817, DOI:10.1016/j.tet.2016.08.009.
- L. Carletti, G. Massiot and C. Debitus, WO20110513880 A1, 2011.
- C. I. MacGregor, B. Y. Han, J. M. Goodman and I. Paterson, Chem. Commun., 2016, 52, 4632–4635, 10.1039/c6cc01074a.
- C. Festa, G. Lauro, S. De Marino, M. V. D'Auria, M. C. Monti, A. Casapullo, C. D'Amore Barbara Renga, A. Mencarelli, S. Petek, G. Bifulco, S. Fiorucci and A. Zampella, J. Med. Chem., 2012, 55, 8303–8317, DOI:10.1021/jm300911g.
- D. B. Stierle and D. J. Faulkner, J. Org. Chem., 1980, 45, 3396–3401, DOI:10.1021/jo01305a005.
- M. D. Norris and M. V. Perkins, J. Org. Chem., 2016, 81, 6848–6854, DOI:10.1021/acs.joc.6b01196.
- C. Festa, S. De Marino, M. V. D'Auria, E. Deharo, G. Gonzalez, C. Deyssard, S. Petek, G. Bifulco and A. Zampella, Tetrahedron, 2012, 68, 10157–10163, DOI:10.1016/j.tet.2012.09.106.
- S. A. Ruider and E. M. Carreira, Org. Lett., 2016, 18, 220–223, DOI:10.1021/acs.orglett.5b03356.
- J. K. Woo, J. E. Jeon, C. K. Kim, C. J. Sim, D. C. Oh, K. B. Oh and J. Shin, J. Nat. Prod., 2013, 76, 1380–1383, DOI:10.1021/np4003367.
- P. M. Garcia-Barrantes and C. W. Lindsley, Org. Lett., 2016, 18, 3810–3813, DOI:10.1021/acs.orglett.6b01825.
- M. R. Vippila, S. Nikhar, A. P. Gracia and G. D. Cuny, Org. Lett., 2016, 18, 4726–4729, DOI:10.1021/acs.orglett.6b02379.
- X. Wang, B. I. Morinaka and T. F. Molinski, J. Nat. Prod., 2014, 77, 625–630, DOI:10.1021/np400891s.
- R. Dahiya, S. Singh, A. Sharma, S. V. Chennupati and S. Maharaj, Mar. Drugs, 2016, 14, 228, DOI:10.3390/md14120228.
- H. J. Zhang, Y. H. Yi, G. J. Yang, M. Y. Hu, G. D. Cao, F. Yang and H. W. Lin, J. Nat. Prod., 2010, 73, 650–655, DOI:10.1021/np9008267.
- F. Shaheen, M. Asad Ziaee, S. A. Ali, S. U. Simjee, A. Ahmed and M. Iqbal Choudhary, Rec. Nat. Prod., 2016, 10, 397–406 CAS.
- C. Festa, S. De Marino, V. Sepe, M. V. D'Auria, G. Bifulco, C. Debitus, M. Bucci, V. Vellecco and A. Zampella, Org. Lett., 2011, 13, 1532–1535, DOI:10.1021/ol200221n.
- K. Kashinath, G. R. Jachak, P. R. Athawale, U. K. Marelli, R. G. Gonnade and D. Srinivasa Reddy, Org. Lett., 2016, 18, 3178–3181, DOI:10.1021/acs.orglett.6b01395.
- X. Zhang, M. R. Jacob, R. Ranga Rao, Y.-H. Wang, A. K. Agarwal, D. J. Newman, I. A. Khan, A. M. Clark and X.-C. Li, Res. Rep. Med. Chem., 2012, 2, 7–14, DOI:10.2147/RRMC.S30895.
- E. Y. Melikhova, R. D. C. Pullin, C. Winter and T. J. Donohoe, Angew. Chem., Int. Ed., 2016, 55, 9753–9757, DOI:10.1002/anie.201604764.
- C.-D. Pham, R. Hartmann, P. Böhler, B. Stork, S. Wesselborg, W. Lin, D. Lai and P. Proksch, Org. Lett., 2014, 16, 266–269, DOI:10.1021/ol403241v.
- J. Zhou, B. Gao, Z. Xu and T. Ye, J. Am. Chem. Soc., 2016, 138, 6948–6951, DOI:10.1021/jacs.6b03533.
- A. K. Ghosh and L. A. Kassekert, Org. Lett., 2016, 18, 3274–3277, DOI:10.1021/acs.orglett.6b01523.
- A. K. Ghosh, L. A. Kassekert and J. D. Bungard, Org. Biomol. Chem., 2016, 14, 11357–11370, 10.1039/c6ob02051h.
- A. S. Ratnayake, R. A. Davis, M. K. Harper, C. A. Veltri, C. D. Andjelic, L. R. Barrows and C. M. Ireland, J. Nat. Prod., 2005, 68, 104–107, DOI:10.1021/np049721s.
- M. Petermichl, S. Loscher and R. Schobert, Angew. Chem., Int. Ed., 2016, 55, 10122–10125, DOI:10.1002/anie.201604912.
- K. H. Jang, G. W. Kang, J. E. Jeon, C. Lim, H. S. Lee, C. J. Sim, K. B. Oh and J. Shin, Org. Lett., 2009, 11, 1713–1716, DOI:10.1021/ol900282m.
- L.-D. Guo, X.-Z. Huang, S.-P. Luo, W.-S. Cao, Y.-P. Ruan, J.-L. Ye and P.-Q. Huang, Angew. Chem., Int. Ed., 2016, 55, 4064–4068, DOI:10.1002/anie.201512005.
- N. Daikuhara, Y. Tada, S. Yamaki, K. Charupant, S. Amnuoypol, K. Suwanborirux and N. Saito, Tetrahedron Lett., 2009, 50, 4276–4278, DOI:10.1016/j.tetlet.2009.05.014.
- M. Yokoya, R. Toyoshima, T. Suzuki, V. H. Le, R. M. Williams and N. Saito, J. Org. Chem., 2016, 81, 4039–4047, DOI:10.1021/acs.joc.6b00327.
- J. Jia, R. Chen, H. Liu, X. Li, Y. Jia and X. Chen, Org. Biomol. Chem., 2016, 14, 7334–7344, 10.1039/c6ob01064d.
- Y. H. Chang, D. Shin, Z. Na, H. S. Lee, D. D. Kim, K. B. Oh and J. Shin, J. Nat. Prod., 2008, 71, 779–783, DOI:10.1021/np078015z.
- S. Yu, F. Li, H. Jeon, S. Lee, J. Shin and S. Kim, Org. Lett., 2016, 18, 2986–2989, DOI:10.1021/acs.orglett.6b01336.
- Y. J. Lee, D. G. Lee, H. S. Rho, V. B. Krasokhin, H. J. Shin, J. S. Lee and H. S. Lee, J. Heterocycl. Chem., 2013, 50, 1400–1404, DOI:10.1002/jhet.1599.
- N. H. Ansari and B. C. G. Söderberg, Tetrahedron, 2016, 72, 4214–4221, DOI:10.1016/j.tet.2016.05.057.
- H. Zhang, M. M. Conte, Z. Khalil, X. C. Huang and R. J. Capon, RSC Adv., 2012, 2, 4209–4214, 10.1039/c2ra20322g.
- W. Zhang and J. M. Ready, J. Am. Chem. Soc., 2016, 138, 10684–10692, DOI:10.1021/jacs.6b06460.
- E. A. Jares-Erijman, A. A. Ingrum, F. Sun and K. L. Rinehart, J. Nat. Prod., 1993, 56, 2186–2188, DOI:10.1021/np50102a025.
- A. Nakazaki, Y. Nakane, Y. Ishikawa, M. Yotsu-Yamashita and T. Nishikawa, Org. Biomol. Chem., 2016, 14, 5304–5309, 10.1039/c6ob00914j.
- M. S. Buchanan, A. R. Carroll, D. Wessling, M. Jobling, V. M. Avery, R. A. Davis, Y. Feng, J. N. A. Hooper and R. J. Quinn, J. Nat. Prod., 2009, 72, 973–975, DOI:10.1021/np8008013.
- M. P. Badart, C. M. L. Squires, S. K. Baird and B. C. Hawkins, Tetrahedron Lett., 2016, 57, 5108–5111, DOI:10.1016/j.tetlet.2016.10.019.
- N. K. Utkina, V. A. Denisenko, O. V. Scholokova and M. V. Virovaya, Tetrahedron Lett., 2003, 44, 101–102, DOI:10.1016/S0040-4039(02)02497-8.
- K. Speck, R. Wildermuth and T. Magauer, Angew. Chem., Int. Ed., 2016, 55, 14131–14135, DOI:10.1002/anie.201608040.
- J. C. Braekman, D. Daloze, G. Hulot, B. Tursch, J. P. Declercq, G. Germain and M. van Meerssche, Bull. Soc. Chim. Belg., 1978, 87, 917–926, DOI:10.1002/bscb.19780871114.
- J. Salva and D. J. Faulkner, J. Org. Chem., 1990, 55, 1941–1943, DOI:10.1021/jo00293a047.
- Y. C. Shen, M. C. Hung, C. V. S. Prakash and J. J. Wang, J. Chin. Chem. Soc., 2000, 47, 567–570, DOI:10.1002/jccs.200000076.
- W. Yu, P. Hjerrild, J. Overgaard and T. B. Poulsen, Angew. Chem., Int. Ed., 2016, 55, 8294–8298, DOI:10.1002/anie.201602476.
- P. A. Horton and P. Crews, J. Nat. Prod., 1995, 58, 44–50, DOI:10.1021/np50115a005.
- A. Torres, P. Gutierrez, R. Alvarez-Manzaneda, R. Chahboun and E. Alvarez-Manzaneda, Org. Biomol. Chem., 2016, 14, 9836–9845, 10.1039/c6ob01640e.
- S. C. Bobzin and D. J. Faulkner, J. Org. Chem., 1989, 54, 3902–3907, DOI:10.1021/jo00277a029.
- Z. Wang, Z. Xing, L. Liu, H. Zhang, Z. Zhong, X. Xie and X. She, ChemistrySelect, 2016, 1, 2225–2227, DOI:10.1002/slct.201600635.
- K. D. Wellington, R. C. Cambie, P. S. Rutledge and P. R. Bergquist, J. Nat. Prod., 2000, 63, 79–85, DOI:10.1021/np9903494.
- A. J. Singh, J. D. Dattelbaum, J. J. Field, Z. Smart, E. F. Woolly, J. M. Barber, R. Heathcott, J. H. Miller and P. T. Northcote, Org. Biomol. Chem., 2013, 11, 8041–8051, 10.1039/c3ob41305e.
- J. D. Dattelbaum, A. Jonathan Singh, J. J. Field, J. H. Miller and P. T. Northcote, J. Org. Chem., 2015, 80, 304–312, DOI:10.1021/jo502370b.
- X. Li, D. Xue, C. Wang and S. Gao, Angew. Chem., Int. Ed., 2016, 55, 9942–9946, DOI:10.1002/anie.201604070.
- M. S. Butler and R. J. Capon, Aust. J. Chem., 1992, 45, 1705–1743, DOI:10.1071/CH9921705.
- M. Fujii, Y. Morimoto, M. Ono and H. Akita, J. Mol. Catal. B: Enzym., 2016, 123, 160–166, DOI:10.1016/j.molcatb.2015.11.019.
- G. Cimino, S. De Stefano, L. Minale and E. Fattorusso, Tetrahedron, 1971, 27, 4673–4679, DOI:10.1016/S0040-4020(01)98174-8.
- M. Kobayashi, R. Chavakula, O. Murata and N. S. Sarma, J. Chem. Res., Synop., 1992, 366–367 CAS.
- G. Cimino, S. De Stefano, L. Minale and E. Fattorusso, Tetrahedron, 1972, 28, 267–273, DOI:10.1016/0040-4020(72)80132-7.
- D.-X. Tan, Z.-J. Xu, H.-J. Chen, Y. Wu and J. You, Eur. J. Org. Chem., 2016, 2016, 946–957, DOI:10.1002/ejoc.201501489.
- A. Rudi, T. Yosief, S. Loya, A. Hizi, M. Schleyer and Y. Kashman, J. Nat. Prod., 2001, 64, 1451–1453, DOI:10.1021/np010121s.
- T. Zhou, F. Feng, Y. Shi and W.-S. Tian, Org. Lett., 2016, 18, 2308–2311, DOI:10.1021/acs.orglett.6b01029.
- T. Hamada, S. Matsunaga, G. Yano and N. Fusetani, J. Am. Chem. Soc., 2005, 127, 110–118, DOI:10.1021/ja045749e.
- A. Parent, A. Guillot, A. Benjdia, G. Chartier, J. Leprince and O. Berteau, J. Am. Chem. Soc., 2016, 138, 15515–15518, DOI:10.1021/jacs.6b06697.
- M. Litaudon and M. Guyot, Tetrahedron Lett., 1986, 27, 4455–4456, DOI:10.1016/S0040-4039(00)84977-1.
- K. Ø. Hanssen, G. Cervin, R. Trepos, J. Petitbois, T. Haug, E. Hansen, J. H. Andersen, H. Pavia, C. Hellio and J. Svenson, Mar. Biotechnol., 2014, 16, 684–694, DOI:10.1007/s10126-014-9583-y.
- P. Cárdenas, J. Chem. Ecol., 2016, 42, 339–347, DOI:10.1007/s10886-016-0693-z.
- M. Reverter, T. Perez, A. V. Ereskovsky and B. Banaigs, J. Chem. Ecol., 2016, 42, 60–70, DOI:10.1007/s10886-015-0664-9.
- B. W. Sullivan, D. J. Faulkner, G. K. Matsumoto, H. Cun-heng and J. Clardy, J. Org. Chem., 1986, 51, 4568–4573, DOI:10.1021/jo00374a015.
- J. A. Chan, A. J. Freyer, B. K. Carte, M. E. Hemling, G. A. Hofmann, M. R. Mattern, M. A. Mentzer and J. W. Westley, J. Nat. Prod., 1994, 57, 1543–1548, DOI:10.1021/np50113a011.
- A. Grube, M. Assmann, E. Lichte, F. Sasse, J. R. Pawlik and M. Kock, J. Nat. Prod., 2007, 70, 504–509, DOI:10.1021/np0603018.
- A. W. Markwell-Heys and J. H. George, Org. Biomol. Chem., 2016, 14, 5546–5549, 10.1039/C6OB00171H.
- D. A. Gold, J. Grabenstatter, A. de Mendoza, A. Riesgo, I. Ruiz-Trillo and R. E. Summons, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2684–2689, DOI:10.1073/pnas.1512614113.
- A. F. Abdel-Razik, M. I. Nassar, A. I. Elshamy, T. M. Kubacy, M.-E. F. Hegazy, N. Ibrahim, A.-C. Le Lamer and A.-R. H. Farrag, Arabian J. Chem., 2016, 9, 649–655, DOI:10.1016/j.arabjc.2014.11.055.
- A. G. Guzii, T. N. Makarieva, S. N. Fedorov, V. A. Denisenko, P. S. Dmitrenok, A. S. Kuzmich, V. B. Krasokhin, H.-S. Lee, Y.-J. Lee and V. A. Stonik, Nat. Prod. Commun., 2016, 11, 1263–1265 Search PubMed.
- Y.-M. Hsu, F.-R. Chang, I.-W. Lo, K.-H. Lai, M. El-Shazly, T.-Y. Wu, Y.-C. Du, T.-L. Hwang, Y.-B. Cheng and Y.-C. Wu, J. Nat. Prod., 2016, 79, 2674–2680, DOI:10.1021/acs.jnatprod.6b00625.
- G. Nuzzo, B. A. Gomes, E. Luongo, M. C. M. Torres, E. A. Santos, A. Cutignano, O. D. L. Pessoa, L. V. Costa-Lotufo and A. Fontana, J. Nat. Prod., 2016, 79, 1881–1885, DOI:10.1021/acs.jnatprod.6b00259.
- S.-Y. Cheng, S.-K. Wang, Y.-H. Ou and C.-Y. Duh, Bioorg. Med. Chem. Lett., 2016, 26, 879–881, DOI:10.1016/j.bmcl.2015.12.076.
- T. Ishii, C.-S. Phan, T. Kamada and C. S. Vairappan, Nat. Prod. Commun., 2016, 11, 1065–1066 Search PubMed.
- Y.-S. Lin, J.-H. Su, C.-L. Lo, C.-Y. Huang and J.-H. Sheu, Nat. Prod. Commun., 2016, 11, 577–585 Search PubMed.
- P. K. Roy, R. Ashimine, H. Miyazato, J. Taira and K. Ueda, Arch. Pharmacal Res., 2016, 39, 778–784, DOI:10.1007/s12272-016-0759-z.
- W. Yuan, S. Cheng, W. Fu, M. Zhao, X. Li, Y. Cai, J. Dong, K. Huang, K. R. Gustafson and P. Yan, J. Nat. Prod., 2016, 79, 1124–1131, DOI:10.1021/acs.jnatprod.6b00031.
- P. Georgantea, E. Ioannou, E. Evain-Bana, D. Bagrel, N. Martinet, C. Vagias and V. Roussis, Tetrahedron, 2016, 72, 3262–3269, DOI:10.1016/j.tet.2016.04.059.
- Q. Peng, F. Liu, H. Sun, X.-J. Liao, M.-R. Feng, T.-T. Liu, Z.-X. Xu, J. Zhang and S.-H. Xu, Nat. Prod. Res., 2016, 30, 2299–2304, DOI:10.1080/14786419.2016.1166496.
- M.-W. Xu and Y.-T. Hao, Nat. Prod. Res., 2016, 30, 2402–2406, DOI:10.1080/14786419.2016.1190720.
- E. G. Lyakhova, C. N. Diep, D. V. Berdyshev, S. A. Kolesnikova, A. I. Kalinovsky, P. S. Dmitrenok, V. A. Tu, N. X. Cuong, N. Van Thanh, N. H. Nam, P. Van Kiem, V. A. Stonik and C. Van Minh, Nat. Prod. Commun., 2016, 11, 913–916 Search PubMed.
- M. P. Rahelivao, M. Gruner, T. Lübken, D. Islamov, O. Kataeva, H. Andriamanantoanina, I. Bauer and H.-J. Knölker, Org. Biomol. Chem., 2016, 14, 989–1001, 10.1039/C5OB02280K.
- Y. Trifman, M. Aknin, A. Gauvin-Bialecki, Y. Benayahu, S. Carmeli and Y. Kashman, Mar. Drugs, 2016, 14, 41, DOI:10.3390/md14020041.
- C.-S. Phan, S.-Y. Ng, T. Kamada and C. S. Vairappan, Nat. Prod. Commun., 2016, 11, 899–900 Search PubMed.
- C. Angulo-Preckler, G. Genta-Jouve, N. Mahajan, M. de la Cruz, N. de Pedro, F. Reyes, K. Iken, C. Avila and O. P. Thomas, J. Nat. Prod., 2016, 79, 1132–1136, DOI:10.1021/acs.jnatprod.6b00040.
- T. Ishii, T. Kamada and C. S. Vairappan, J. Asian Nat. Prod. Res., 2016, 18, 415–422, DOI:10.1080/10286020.2016.1145670.
- M. Shaaban, M. A. Ghani and K. A. Shaaban, Z. Naturforsch., B: J. Chem. Sci., 2016, 71, 1211–1217, DOI:10.1515/znb-2016-0144.
- T. Kamada, C.-S. Phan, H.-S. Tin and C. S. Vairappan, Nat. Prod. Commun., 2016, 11, 1077–1078 Search PubMed.
- K.-M. Liu, Y.-H. Lan, C.-C. Su and P.-J. Sung, Nat. Prod. Commun., 2016, 11, 21–23 Search PubMed.
- G.-H. Tang, Z.-H. Sun, Y.-H. Zou and S. Yin, Molecules, 2016, 21, 587, DOI:10.3390/molecules21050587.
- C.-Y. Huang, Y.-J. Tseng, U. Chokkalingam, T.-L. Hwang, C.-H. Hsu, C.-F. Dai, P.-J. Sung and J.-H. Sheu, J. Nat. Prod., 2016, 79, 1339–1346, DOI:10.1021/acs.jnatprod.5b01142.
- M.-E. Hegazy, T. Mohamed, A. Elshamy, M. Al-Hammady, S. Ohta and P. Paré, Molecules, 2016, 21, 308, DOI:10.3390/molecules21030308.
- D. Torres-Mendoza, Y. González, J. Gómez-Reyes, H. Guzmán, J. López-Perez, W. Gerwick, P. Fernandez and M. Gutiérrez, Molecules, 2016, 21, 819, DOI:10.3390/molecules21060819.
- C.-H. Chao, C.-Y. Wu, C.-Y. Huang, H.-C. Wang, C.-F. Dai, Y.-C. Wu and J.-H. Sheu, Mar. Drugs, 2016, 14, 150, DOI:10.3390/md14080150.
- M. Zhao, S. Cheng, W. Yuan, Y. Xi, X. Li, J. Dong, K. Huang, K. R. Gustafson and P. Yan, Mar. Drugs, 2016, 14, 111, DOI:10.3390/md14060111.
- P. Roy, R. Ashimine, H. Miyazato, J. Taira and K. Ueda, Molecules, 2016, 21, 679, DOI:10.3390/molecules21050679.
- B. R. Chitturi, V. B. Tatipamula, C. B. Dokuburra, U. K. Mangamuri, V. R. Tuniki, S. V. Kalivendi, R. A. Bunce and V. Yenamandra, Tetrahedron, 2016, 72, 1933–1940, DOI:10.1016/j.tet.2016.02.056.
- M. Zubair, W. Alarif, K. Al-Footy, M. Ph, M. Ali, S. Basaif, S. Al-Lihaibi and S.-E. Ayyad, Turk. J. Chem., 2016, 40, 385–392, DOI:10.3906/kim-1502-82.
- P. Sun, Q. Yu, J. Li, R. Riccio, G. Lauro, G. Bifulco, T. Kurtán, A. Mándi, H. Tang, T.-J. Li, C.-L. Zhuang, W. H. Gerwick and W. Zhang, J. Nat. Prod., 2016, 79, 2552–2558, DOI:10.1021/acs.jnatprod.6b00453.
- Y.-D. Su, Z.-H. Wen, Y.-C. Wu, L.-S. Fang, Y.-H. Chen, Y.-C. Chang, J.-H. Sheu and P.-J. Sung, Tetrahedron, 2016, 72, 944–951, DOI:10.1016/j.tet.2015.12.058.
- Y.-D. Su, C.-S. Sung, Z.-H. Wen, Y.-H. Chen, Y.-C. Chang, J.-J. Chen, L.-S. Fang, Y.-C. Wu, J.-H. Sheu and P.-J. Sung, Int. J. Mol. Sci., 2016, 17, 79, DOI:10.3390/ijms17010079.
- C. Li, M.-P. La, H. Tang, P. Sun, B.-S. Liu, C.-L. Zhuang, Y.-H. Yi and W. Zhang, Mar. Drugs, 2016, 14, 201, DOI:10.3390/md14110201.
- F.-Y. Chang, U. Chokkalingam, C.-J. Tai, C.-Y. Huang, W.-C. Wei, N.-S. Yang, J.-H. Su, P.-J. Sung and J.-H. Sheu, Tetrahedron, 2016, 72, 192–198, DOI:10.1016/j.tet.2015.11.025.
- K.-Y. Peng, N.-F. Chen, Z.-C. Chen, K.-H. Tsui, Z.-H. Wen, Y.-D. Su, Y.-C. Chang, Y.-H. Chen, M.-C. Lu, L.-S. Fang, J.-J. Chen, T.-Y. Wu, Y.-C. Wu and P.-J. Sung, Tetrahedron Lett., 2016, 57, 4239–4242, DOI:10.1016/j.tetlet.2016.08.028.
- Y.-C. Chang, L.-M. Kuo, J.-H. Su, T.-L. Hwang, Y.-H. Kuo, C.-S. Lin, Y.-C. Wu, J.-H. Sheu and P.-J. Sung, Tetrahedron, 2016, 72, 999–1004, DOI:10.1016/j.tet.2015.12.072.
- Y.-C. Chang, T.-L. Hwang, J.-H. Sheu, Y.-C. Wu and P.-J. Sung, Mar. Drugs, 2016, 14, 218, DOI:10.3390/md14120218.
- Y.-C. Chang, L.-M. Kuo, T.-L. Hwang, J. Yeh, Z.-H. Wen, L.-S. Fang, Y.-C. Wu, C.-S. Lin, J.-H. Sheu and P.-J. Sung, Mar. Drugs, 2016, 14, 12, DOI:10.3390/md14010012.
- T. Inuzuka, Y. Kawazoe, S. Kobayashi, R. Matsumoto, J. Yabe, S. Ohmura and D. Uemura, Chem. Lett., 2016, 45, 81–82, DOI:10.1246/cl.150938.
- Y.-D. Su, C.-H. Cheng, Z.-H. Wen, Y.-C. Wu and P.-J. Sung, Bioorg. Med. Chem. Lett., 2016, 26, 3060–3063, DOI:10.1016/j.bmcl.2016.05.015.
- X.-W. Zhang, X.-L. Tang, B.-S. Liu, P.-L. Li and G.-Q. Li, Chem. Biodiversity, 2016, 13, 233–237, DOI:10.1002/cbdv.201500093.
- W. Cheng, J. Ren, Q. Huang, H. Long, H. Jin, L. Zhang, H. Liu, L. van Ofwegen and W. Lin, Steroids, 2016, 108, 99–104, DOI:10.1016/j.steroids.2016.02.003.
- C.-Y. Huang, C.-W. Chang, Y.-J. Tseng, J. Lee, P.-J. Sung, J.-H. Su, T.-L. Hwang, C.-F. Dai, H.-C. Wang and J.-H. Sheu, Mar. Drugs, 2016, 14, 180, DOI:10.3390/md14100180.
- Y.-B. Cheng, J.-C. Lee, I.-W. Lo, S.-R. Chen, H.-C. Hu, Y.-H. Wu, Y.-C. Wu and F.-R. Chang, Bioorg. Med. Chem. Lett., 2016, 26, 2344–2348, DOI:10.1016/j.bmcl.2016.03.029.
- J.-C. Lee, F.-R. Chang, S.-R. Chen, Y.-H. Wu, H.-C. Hu, Y.-C. Wu, A. Backlund and Y.-B. Cheng, Mar. Drugs, 2016, 14, 151, DOI:10.3390/md14080151.
- M.-E. F. Hegazy, A. M. Gamal-Eldeen, T. A. Mohamed, M. A. Alhammady, A. A. Hassanien, M. A. Shreadah, I. I. Abdelgawad, E. M. Elkady and P. W. Paré, Nat. Prod. Res., 2016, 30, 1266–1272, DOI:10.1080/14786419.2015.1055266.
- M. Chen, L. Han, X.-L. Zhang and C.-Y. Wang, Nat. Prod. Res., 2016, 30, 1431–1435, DOI:10.1080/14786419.2015.1063055.
- Y.-C. Chang, N.-F. Chen, T.-L. Hwang, C.-C. Tseng, T.-Y. Wu, B.-R. Peng, Z.-H. Wen, L.-S. Fang, Y.-C. Wu, J.-H. Sheu and P.-J. Sung, Steroids, 2016, 115, 123–129, DOI:10.1016/j.steroids.2016.08.018.
- M.-E. F. Hegazy, T. A. Mohamed, A. I. Elshamy, A. A. Hassanien, N. S. Abdel-Azim, M. A. Shreadah, I. I. Abdelgawad, E. M. Elkady and P. W. Paré, Nat. Prod. Res., 2016, 30, 340–344, DOI:10.1080/14786419.2015.1046871.
- W.-R. Tseng, C.-Y. Huang, Y.-Y. Tsai, Y.-S. Lin, T.-L. Hwang, J.-H. Su, P.-J. Sung, C.-F. Dai and J.-H. Sheu, Bioorg. Med. Chem. Lett., 2016, 26, 3253–3257, DOI:10.1016/j.bmcl.2016.05.060.
- N. Van Thanh, N. Thi Ngoc, H. Le Tuan Anh, D. Cong Thung, D. Thi Thao, N. Xuan Cuong, N. Hoai Nam, P. Van Kiem and C. Van Minh, J. Asian Nat. Prod. Res., 2016, 18, 938–944, DOI:10.1080/10286020.2016.1173676.
- M. Z. Končić, E. Ioannou, W. Richard Sawadogo, A. F. Abdel-Razik, C. Vagias, M. Diederich and V. Roussis, Steroids, 2016, 115, 130–135, DOI:10.1016/j.steroids.2016.08.017.
- F. Cardoso-Martínez, J. M. de la Rosa, A. R. Díaz-Marrero, J. Darias, L. D'Croz, M. D. Jiménez-Antón, M. Jesús Corral, R. García, J. M. Alunda and M. Cueto, RSC Adv., 2016, 6, 38579–38591, 10.1039/c6ra04521a.
- T.-Y. Whuang, W.-C. Tsai, N.-F. Chen, Z.-C. Chen, K.-H. Tsui, Z.-H. Wen, Y.-D. Su, Y.-C. Chang, Y.-H. Chen, M.-C. Lu, L.-S. Fang, J.-J. Chen, T.-Y. Wu, Y.-C. Wu and P.-J. Sung, Bioorg. Med. Chem. Lett., 2016, 26, 4966–4969, DOI:10.1016/j.bmcl.2016.09.007.
- H. Sun, F. Liu, M.-R. Feng, Q. Peng, X.-J. Liao, T.-T. Liu, J. Zhang and S.-H. Xu, Nat. Prod. Res., 2016, 30, 2819–2824, DOI:10.1080/14786419.2016.1166495.
- T.-C. Tsai, Y.-T. Huang, S.-K. Chou, M.-C. Shih, C.-Y. Chiang and J.-H. Su, Chem. Pharm. Bull., 2016, 64, 1519–1522, DOI:10.1248/cpb.c16-00426.
- N. T. Ngoc, P. T. M. Huong, N. Van Thanh, N. X. Cuong, N. H. Nam, D. C. Thung, P. Van Kiem and C. Van Minh, Chem. Pharm. Bull., 2016, 64, 1417–1419, DOI:10.1248/cpb.c16-00385.
- Y.-N. Lu, P. Cui, X.-Q. Tian, L.-G. Lou and C.-Q. Fan, Planta Med., 2016, 82, 882–887, DOI:10.1055/s-0042-106168.
- H. Takamura, T. Kikuchi, N. Endo, Y. Fukuda and I. Kadota, Org. Lett., 2016, 18, 2110–2113, DOI:10.1021/acs.orglett.6b00737.
- M. Kousara, F. Bideau, R. Ibrahim, A. Ferry, P.-E. Venot, C. Dejean, J. Raingeaud, J. Dubois, P. Retailleau and F. Dumas, Synthesis, 2016, 48, 1637–1646, DOI:10.1055/s-0035-1561430.
- Y. Ren, M. Presset, J. Godemert, N. Vanthuyne, J.-V. Naubron, M. Giorgi, J. Rodriguez and Y. Coquerel, Chem. Commun., 2016, 52, 6565–6568, 10.1039/c6cc01689h.
- E. J. Chamgordani, J. Paulsen and L.-L. Gundersen, Tetrahedron Lett., 2016, 57, 4926–4929, DOI:10.1016/j.tetlet.2016.09.078.
- M. Yang, X. Yang, H. Sun and A. Li, Angew. Chem., Int. Ed., 2016, 55, 2851–2855, DOI:10.1002/anie.201510568.
- H. Miyazato, J. Taira and K. Ueda, Bioorg. Med. Chem. Lett., 2016, 26, 4641–4644, DOI:10.1016/j.bmcl.2016.08.057.
- S. Caplan, B. Zheng, K. Dawson-Scully, C. White and L. West, Mar. Drugs, 2016, 14, 55, DOI:10.3390/md14030055.
- D. Gao, Y. Wang, W. Xie, T. Yang, Y. Jiang, Y. Guo, J. Guan and H. Liu, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2016, 1014, 17–23, DOI:10.1016/j.jchromb.2016.01.003.
- Y.-J. Wu, B.-S. Wong, S.-H. Yea, C.-I. Lu and S.-H. Weng, Mar. Drugs, 2016, 14, 142, DOI:10.3390/md14080142.
- S.-P. Tseng, W.-C. Hung, C.-Y. Huang, Y.-S. Lin, M.-Y. Chan, P.-L. Lu, L. Lin and J.-H. Sheu, Mar. Drugs, 2016, 14, 143, DOI:10.3390/md14080143.
- C.-H. Chen, N.-F. Chen, C.-W. Feng, S.-Y. Cheng, H.-C. Hung, K.-H. Tsui, C.-H. Hsu, P.-J. Sung, W.-F. Chen and Z.-H. Wen, Mar. Drugs, 2016, 14, 160, DOI:10.3390/md14090160.
- C.-W. Feng, H.-C. Hung, S.-Y. Huang, C.-H. Chen, Y.-R. Chen, C.-Y. Chen, S.-N. Yang, H.-M. David Wang, P.-J. Sung, J.-H. Sheu, K.-H. Tsui, W.-F. Chen and Z.-H. Wen, Mar. Drugs, 2016, 14, 187, DOI:10.3390/md14100187.
- J.-J. Lin, R. Wang, J.-C. Chen, C.-C. Chiu, M.-H. Liao and Y.-J. Wu, Int. J. Mol. Sci., 2016, 17, 1787, DOI:10.3390/ijms17111787.
- R. Vaikundamoorthy, R. Sundaramoorthy, V. Krishnamoorthy, R. Vilwanathan and R. Rajendran, Tumor Biol., 2016, 37, 10517–10531, DOI:10.1007/s13277-016-4947-8.
- C. Ishikawa, T. Jomori, J. Tanaka, M. Senba and N. Mori, Int. J. Oncol., 2016, 49, 1713–1721, DOI:10.3892/ijo.2016.3648.
- S. Aratake, Y. Taira, T. Fujii, M. C. Roy, J. D. Reimer, T. Yamazaki and H. Jenke-Kodama, Toxicon, 2016, 111, 86–90, DOI:10.1016/j.toxicon.2015.12.004.
- A. T. Wang, M. R. Prinsep and R. D. Martinus, SpringerPlus, 2016, 5, 742, DOI:10.1186/s40064-016-2397-9.
- J. Shimokawa, K. Chiyoda, H. Umihara and T. Fukuyama, Chem. Pharm. Bull., 2016, 64, 1239–1241, DOI:10.1248/cpb.c16-00256.
- B. D. Morris and M. R. Prinsep, J. Nat. Prod., 1999, 62, 688–693, DOI:10.1021/np980410p.
- D. Staveness, R. Abdelnabi, A. J. Schrier, B. A. Loy, V. A. Verma, B. A. DeChristopher, K. E. Near, J. Neyts, L. Delang, P. Leyssen and P. A. Wender, J. Nat. Prod., 2016, 79, 675–679, DOI:10.1021/acs.jnatprod.5b01016.
- D. Staveness, R. Abdelnabi, K. E. Near, Y. Nakagawa, J. Neyts, L. Delang, P. Leyssen and P. A. Wender, J. Nat. Prod., 2016, 79, 680–684, DOI:10.1021/acs.jnatprod.5b01017.
- G. Nuzzo, A. Cutignano, J. Moles, C. Avila and A. Fontana, Tetrahedron Lett., 2016, 57, 71–74, DOI:10.1016/j.tetlet.2015.11.067.
- M. Letizia Ciavatta, P. Devi, M. Carbone, V. Mathieu, R. Kiss, A. Casapullo and M. Gavagnin, Tetrahedron, 2016, 72, 625–631, DOI:10.1016/j.tet.2015.12.003.
- A. M. White, G. K. Pierens, L. C. Forster, A. E. Winters, K. L. Cheney and M. J. Garson, J. Nat. Prod., 2016, 79, 477–483, DOI:10.1021/acs.jnatprod.5b00866.
- Y. Hirayama, P. L. Katavic, A. M. White, G. K. Pierens, L. K. Lambert, A. E. Winters, H. Kigoshi, M. Kita and M. J. Garson, Aust. J. Chem., 2016, 69, 136–144, DOI:10.1071/ch15203.
- A. Bogdanov, C. Hertzer, S. Kehraus, S. Nietzer, S. Rohde, P. J. Schupp, H. Wägele and G. M. König, J. Nat. Prod., 2016, 79, 611–615, DOI:10.1021/acs.jnatprod.5b00860.
- M. Kita, A. Kawamura and H. Kigoshi, Tetrahedron Lett., 2016, 57, 858–860, DOI:10.1016/j.tetlet.2016.01.028.
- A. Karagiannis, N. Diddi and D. E. Ward, Org. Lett., 2016, 18, 3794–3797, DOI:10.1021/acs.orglett.6b01798.
- N. Alnafta, J. P. Schmidt, C. L. Nesbitt and C. S. P. McErlean, Org. Lett., 2016, 18, 6520–6522, DOI:10.1021/acs.orglett.6b03219.
- A. J. Burckle, V. H. Vasilev and N. Z. Burns, Angew. Chem., Int. Ed., 2016, 55, 11476–11479, DOI:10.1002/anie.201605722.
- R. Meier and D. Trauner, Angew. Chem., Int. Ed., 2016, 55, 11251–11255, DOI:10.1002/anie.201604102.
- R. K. Quinn, Z. A. Könst, S. E. Michalak, Y. Schmidt, A. R. Szklarski, A. R. Flores, S. Nam, D. A. Horne, C. D. Vanderwal and E. J. Alexanian, J. Am. Chem. Soc., 2016, 138, 696–702, DOI:10.1021/jacs.5b12308.
- D. J. Tao, Y. Slutskyy and L. E. Overman, J. Am. Chem. Soc., 2016, 138, 2186–2189, DOI:10.1021/jacs.6b00541.
- T. Maoka, A. Nishino, H. Yasui, Y. Yamano and A. Wada, Mar. Drugs, 2016, 14, 93, DOI:10.3390/md14050093.
- Y. Ashida, R. C. Yanagita, C. Takahashi, Y. Kawanami and K. Irie, Bioorg. Med. Chem., 2016, 24, 4218–4227, DOI:10.1016/j.bmc.2016.07.011.
- Y. Hirayama, K. Yamagishi, T. Suzuki, H. Kawagishi, M. Kita and H. Kigoshi, Bioorg. Med. Chem., 2016, 24, 2809–2814, DOI:10.1016/j.bmc.2016.04.049.
- K. L. Cheney, A. White, I. Wayan Mudianta, A. E. Winters, M. Quezada, R. J. Capon, E. Mollo and M. J. Garson, PLoS One, 2016, 11, e0145134, DOI:10.1371/journal.pone.0145134.
- J. Bernáldez, S. Jiménez, L. González, J. Ferro, E. Soto, E. Salceda, D. Chávez, M. Aguilar and A. Licea-Navarro, Toxins, 2016, 8, 39, DOI:10.3390/toxins8020039.
- S. Yu, T. Du, Z. Liu, Q. Wu, G. Feng, M. Dong, X. Zhou, L. Jiang and Q. Dai, Peptides, 2016, 81, 15–20, DOI:10.1016/j.peptides.2016.04.004.
- E. K. M. Lebbe, M. G. K. Ghequire, S. Peigneur, B. G. Mille, P. Devi, S. Ravichandran, E. Waelkens, L. D'Souza, R. De Mot and J. Tytgat, Mar. Drugs, 2016, 14, 199, DOI:10.3390/md14110199.
- A. Brust, D. E. Croker, B. Colless, L. Ragnarsson, Å. Andersson, K. Jain, S. Garcia-Caraballo, J. Castro, S. M. Brierley, P. F. Alewood and R. J. Lewis, J. Med. Chem., 2016, 59, 2381–2395, DOI:10.1021/acs.jmedchem.5b00911.
- X. Zhu, J. Bi, J. Yu, X. Li, Y. Zhang, D. Zhangsun and S. Luo, Mar. Drugs, 2016, 14, 11, DOI:10.3390/md14010011.
- J. R. Prashanth, S. Dutertre, A. H. Jin, V. Lavergne, B. Hamilton, F. C. Cardoso, J. Griffin, D. J. Venter, P. F. Alewood and R. J. Lewis, Mol. Ecol., 2016, 25, 598–615, DOI:10.1111/mec.13504.
- C. Imperatore, P. Luciano, A. Aiello, R. Vitalone, C. Irace, R. Santamaria, J. Li, Y.-W. Guo and M. Menna, J. Nat. Prod., 2016, 79, 1144–1148, DOI:10.1021/acs.jnatprod.6b00063.
- M. Nazari, J. D. Serrill, J. Sikorska, T. Ye, J. E. Ishmael and K. L. McPhail, Org. Lett., 2016, 18, 1374–1377, DOI:10.1021/acs.orglett.6b00308.
- J. Wang, A. Norrie Pearce, S. T. S. Chan, R. B. Taylor, M. J. Page, A. Valentin, M.-L. Bourguet-Kondracki, J. P. Dalton, S. Wiles and B. R. Copp, J. Nat. Prod., 2016, 79, 607–610, DOI:10.1021/acs.jnatprod.5b00770.
- H. Uchimasu, K. Matsumura, M. Tsuda, K. Kumagai, M. Akakabe, M. J. Fujita and R. Sakai, Tetrahedron, 2016, 72, 7185–7193, DOI:10.1016/j.tet.2016.09.051.
- S. T. S. Chan, R. R. Nani, E. A. Schauer, G. E. Martin, R. Thomas Williamson, J. Saurí, A. V. Buevich, W. A. Schafer, L. A. Joyce, A. K. L. Goey, W. D. Figg, T. T. Ransom, C. J. Henrich, T. C. McKee, A. Moser, S. A. MacDonald, S. Khan, J. B. McMahon, M. J. Schnermann and K. R. Gustafson, J. Org. Chem., 2016, 81, 10631–10640, DOI:10.1021/acs.joc.6b02380.
- C. Imperatore, M. Senese, A. Aiello, P. Luciano, S. Fiorucci, C. D'Amore, A. Carino and M. Menna, Mar. Drugs, 2016, 14, 117, DOI:10.3390/md14060117.
- K. M. Snyder, J. Sikorska, T. Ye, L. Fang, W. Su, R. G. Carter, K. L. McPhail and P. H.-Y. Cheong, Org. Biomol. Chem., 2016, 14, 5826–5831, 10.1039/C6OB00707D.
- L. Filippova, S. Antonsen, Y. Stenstrøm and T. V. Hansen, Tetrahedron, 2016, 72, 6572–6577, DOI:10.1016/j.tet.2016.08.070.
- S. Flock, S. Antonsen, H. Gallantree-Smith, A. Marie Langseter, L. Skattebøl and Y. Stenstrøm, Tetrahedron, 2016, 72, 4518–4522, DOI:10.1016/j.tet.2016.06.009.
- E. K. Davison and J. Sperry, Org. Chem. Front., 2016, 3, 38–42, 10.1039/C5QO00367A.
- A. C. Lindsay, I. K. H. Leung and J. Sperry, Org. Lett., 2016, 18, 5404–5407, DOI:10.1021/acs.orglett.6b02798.
- D. C. Medellin, Q. Zhou, R. Scott, R. Matthew Hill, S. K. Frail, R. Dasari, S. J. Ontiveros, S. C. Pelly, W. A. L. van Otterlo, T. Betancourt, C. B. Shuster, E. Hamel, R. Bai, D. V. LaBarbera, S. Rogelj, L. V. Frolova and A. Kornienko, J. Med. Chem., 2016, 59, 480–485, DOI:10.1021/acs.jmedchem.5b01426.
- T. Vijai Kumar Reddy, A. Jyotsna, B. L. A. Prabhavathi Devi, R. B. N. Prasad, Y. Poornachandra and C. Ganesh Kumar, Eur. J. Med. Chem., 2016, 120, 86–96, DOI:10.1016/j.ejmech.2016.04.073.
- M. H. Nguyen, M. Imanishi, T. Kurogi and A. B. Smith, J. Am. Chem. Soc., 2016, 138, 3675–3678, DOI:10.1021/jacs.6b01731.
- N. Veerasamy, A. Ghosh, J. Li, K. Watanabe, J. D. Serrill, J. E. Ishmael, K. L. McPhail and R. G. Carter, J. Am. Chem. Soc., 2016, 138, 770–773, DOI:10.1021/jacs.5b12318.
- T. M. Brütsch, P. Bucher and K.-H. Altmann, Chem.–Eur. J., 2016, 22, 1292–1300, DOI:10.1002/chem.201504230.
- R. Toyoshima, N. Mori, T. Suzuki, W. Lowtangkitcharoen, K. Suwanborirux and N. Saito, Chem. Pharm. Bull., 2016, 64, 966–969, DOI:10.1248/cpb.c16-00192.
- P. Guo, Z. Wang, G. Li, Y. Liu, Y. Xie and Q. Wang, J. Agric. Food Chem., 2016, 64, 4264–4272, DOI:10.1021/acs.jafc.6b01415.
- P. Comba, A. Eisenschmidt, N. Kipper and J. Schießl, J. Inorg. Biochem., 2016, 159, 70–75, DOI:10.1016/j.jinorgbio.2016.02.014.
- A. Asano, K. Minoura, T. Yamada and M. Doi, J. Pept. Sci., 2016, 22, 156–165, DOI:10.1002/psc.2853.
- Y. Ota, T. Chinen, K. Yoshida, S. Kudo, Y. Nagumo, Y. Shiwa, R. Yamada, H. Umihara, K. Iwasaki, H. Masumoto, S. Yokoshima, H. Yoshikawa, T. Fukuyama, J. Kobayashi and T. Usui, ChemBioChem, 2016, 17, 1616–1620, DOI:10.1002/cbic.201600075.
- J. M. Giulietti, P. M. Tate, A. Cai, B. Cho and S. P. Mulcahy, Bioorg. Med. Chem. Lett., 2016, 26, 4705–4708, DOI:10.1016/j.bmcl.2016.08.047.
- E. A. Vasileva, N. P. Mishchenko, P. A. Zadorozhny and S. A. Fedoreyev, Nat. Prod. Commun., 2016, 11, 821–825 Search PubMed.
- R. Ueoka, Y. Hitora, A. Ito, M. Yoshida, S. Okada, K. Takada and S. Matsunaga, J. Nat. Prod., 2016, 79, 2754–2757, DOI:10.1021/acs.jnatprod.6b00701.
- W. H. Gerwick, P. J. Proteau, D. G. Nagle, E. Hamel, A. Blokhin and D. L. Slate, J. Org. Chem., 1994, 59, 1243–1245, DOI:10.1021/jo00085a006.
- L. Thi Vien, T. Thi Hong Hanh, P. Thi Thanh Huong, N. Hai Dang, N. Van Thanh, E. Lyakhova, N. Xuan Cuong, N. Hoai Nam, P. Van Kiem, A. Kicha and C. Van Minh, Chem. Pharm. Bull., 2016, 64, 1654–1657, DOI:10.1248/cpb.c16-00585.
- T. T. H. Hanh, L. T. Vien, L. B. Vinh, N. Van Thanh, N. X. Cuong, N. H. Nam, D. C. Thung, P. Van Kiem and C. Van Minh, Chem. Pharm. Bull., 2016, 64, 1523–1527, DOI:10.1248/cpb.c16-00445.
- T. V. Malyarenko, A. A. Kicha, A. I. Kalinovsky, N. V. Ivanchina, R. S. Popov, E. A. Pislyagin, E. S. Menchinskaya, K. Pillai Padmakumar and V. A. Stonik, Chem. Biodiversity, 2016, 13, 998–1007, DOI:10.1002/cbdv.201500336.
- T. V. Malyarenko, S. D. Kharchenko, A. A. Kicha, N. V. Ivanchina, P. S. Dmitrenok, E. A. Chingizova, E. A. Pislyagin, E. V. Evtushenko, T. I. Antokhina, C. Van Minh and V. A. Stonik, J. Nat. Prod., 2016, 79, 3047–3056, DOI:10.1021/acs.jnatprod.6b00667.
- L. T. Vien, T. T. H. Hanh, P. T. T. Huong, V. A. Tu, N. V. Thanh, E. G. Lyakhova, N. X. Cuong, N. H. Nam, P. V. Kiem, C. V. Minh, A. A. Kicha and V. A. Stonik, Chem. Nat. Compd., 2016, 52, 1056–1060, DOI:10.1007/s10600-016-1860-8.
- A. Kicha, N. Ivanchina, T. Malyarenko, A. Kalinovsky, P. Dimitrenok, E. Pislyagin and E. Yurchenko, Nat. Prod. Commun., 2016, 11, 1243–1246 Search PubMed.
- J.-X. Kang, Y.-F. Kang and H. Han, Mar. Drugs, 2016, 14, 189, DOI:10.3390/md14100189.
- A. S. Silchenko, A. I. Kalinovsky, S. A. Avilov, P. V. Andryjaschenko, P. S. Dmitrenok, E. A. Yurchenko, I. Y. Dolmatov, S. S. Dautov, V. A. Stonik and V. I. Kalinin, Nat. Prod. Commun., 2016, 11, 741–747 Search PubMed.
- R. Popov, N. Ivanchina, A. Kalinovsky, S. Kharchenko, A. Kicha, T. Malyarenko, S. Ermakova and P. Dmitrenok, Nat. Prod. Commun., 2016, 11, 1247–1250 Search PubMed.
- A. S. Silchenko, A. I. Kalinovsky, S. A. Avilov, P. V. Andryjaschenko, P. S. Dmitrenok, V. I. Kalinin, E. A. Yurchenko and I. Y. Dolmatov, Nat. Prod. Commun., 2016, 11, 381–388 Search PubMed.
- A. Silchenko, A. Kalinovsky, S. Avilov, P. Andryjaschenko, P. Dmitrenok, V. Kalinin, E. Chingizova, K. Minin and V. Stonik, Molecules, 2016, 21, 939, DOI:10.3390/molecules21070939.
- A. Silchenko, A. Kalinovsky, S. Avilov, P. Andryjaschenko, P. Dimitrenok, V. Kalinin, E. Martyyas and K. Minin, Nat. Prod. Commun., 2016, 11, 939–945 Search PubMed.
- M. J. Graniel-Sabido, G. Mirón-López, L. V. León-Deniz, R. E. Moo-Puc, C. J. Quintal-Novelo, L. Quijano and G. J. Mena-Rejón, Tetrahedron Lett., 2016, 57, 4375–4378, DOI:10.1016/j.tetlet.2016.08.051.
- G. I. Melman, K. L. Borisova, N. D. Pokhilo, V. V. Makhankov and V. P. Anufriev, Tetrahedron Lett., 2016, 57, 736–738, DOI:10.1016/j.tetlet.2016.01.004.
- N. P. Mishchenko, E. A. Vasileva and S. A. Fedoreyev, Tetrahedron Lett., 2014, 55, 5967–5969, DOI:10.1016/j.tetlet.2014.09.018.
- D. Pelageev and V. Anufriev, Synthesis, 2016, 48, 761–764, DOI:10.1055/s-0035-1560389.
- R. Higuchi, S. Inoue, K. Inagaki, M. Sakai, T. Miyamoto, T. Komori, M. Inagaki and R. Isobe, Chem. Pharm. Bull., 2006, 54, 287–291, DOI:10.1248/cpb.54.287.
- K. Goto, M. Sawa, H. Tamai, A. Imamura, H. Ando, H. Ishida and M. Kiso, Chem.–Eur. J., 2016, 22, 8323–8331, DOI:10.1002/chem.201600970.
- M. Girard, J. Belanger, J. W. ApSimon, F. X. Garneau, C. Harvey and J. R. Brisson, Can. J. Chem., 1990, 68, 11–18, DOI:10.1139/v90-003.
- J. Al Shemaili, K. A. Parekh, R. A. Newman, B. Hellman, C. Woodward, A. Adem, P. Collin and T. E. Adrian, Mar. Drugs, 2016, 14, 115, DOI:10.3390/md14060115.
- S. A. Dyshlovoy, E. S. Menchinskaya, S. Venz, S. Rast, K. Amann, J. Hauschild, K. Otte, V. I. Kalinin, A. S. Silchenko, S. A. Avilov, W. Alsdorf, R. Madanchi, C. Bokemeyer, U. Schumacher, R. Walther, D. L. Aminin, S. N. Fedorov, L. K. Shubina, V. A. Stonik, S. Balabanov, F. Honecker and G. von Amsberg, Int. J. Cancer, 2016, 138, 2450–2465, DOI:10.1002/ijc.29977.
- I. Kitagawa, M. Kobayashi, T. Inamoto, M. Fuchida and Y. Kyogoku, Chem. Pharm. Bull., 1985, 33, 5214–5224, DOI:10.1248/cpb.33.5214.
- M. Wen, X. Fu, X. Han, X. Hu, P. Dong, J. Xu, Y. Xue, J. Wang, C. Xue and Y. Wang, J. Nutr. Sci. Vitaminol., 2016, 62, 170–177, DOI:10.3177/jnsv.62.170.
- I. Kitagawa, T. Inamoto, M. Fuchida, S. Okada, M. Kobayashi, T. Nishino and Y. Kyoboku, Chem. Pharm. Bull., 1980, 28, 1651–1653, DOI:10.1248/cpb.28.1651.
- I. I. Maltsev, V. A. Stonik, A. I. Kalinovsky and G. B. Elyakov, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 1984, 78, 421–426, DOI:10.1016/0305-0491(84)90052-X.
- S. Li, Y. Wang, T. Jiang, H. Wang, S. Yang and Z. Lv, Mar. Drugs, 2016, 14, 114, DOI:10.3390/md14060114.
- S. A. Avilov, Khim. Prir. Soedin., 1984, 6, 799–800 Search PubMed.
- D. Aminin, E. Pislyagin, M. Astashev, A. Es'kov, V. Kozhemyako, S. Avilov, E. Zelepuga, E. Yurchenko, L. Kaluzhskiy, E. Kozlovskaya, A. Ivanov and V. Stonik, Sci. Rep., 2016, 6, 39683, DOI:10.1038/srep39683.
- S. P. R. Thota, N. S. Sarma, Y. L. N. Murthy, V. S. S. N. Kantamreddi and C. W. Wright, Indian J. Chem., Sect. B: Org. Chem. Incl. Med. Chem., 2016, 55, 123–127 Search PubMed.
- Y. Chen, W.-J. Wang and J. Wu, J. Asian Nat. Prod. Res., 2016, 18, 41–45, DOI:10.1080/10286020.2015.1121998.
- K. Niu, L. Shen and J. Wu, J. Asian Nat. Prod. Res., 2016, 18, 36–40, DOI:10.1080/10286020.2015.1075006.
- Z.-F. Zhou, T. Kurtán, A. Mándi, Y.-C. Gu, L.-G. Yao, G.-R. Xin, X.-W. Li and Y.-W. Guo, Sci. Rep., 2016, 6, 33908, DOI:10.1038/srep33908.
- Y.-G. Dai, W.-S. Li, P. Pedpradab, J.-J. Liu, J. Wu and L. Shen, RSC Adv., 2016, 6, 85978–85984, 10.1039/C6RA14721F.
- S. A. Viczek, K. B. Jensen and K. A. Francesconi, Angew. Chem., Int. Ed., 2016, 55, 5259–5262, DOI:10.1002/anie.201512031.
- M. Feher and J. M. Schmidt, J. Chem. Inf. Comput. Sci., 2003, 43, 218–227, DOI:10.1021/ci0200467.
- J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. G. Munro and M. R. Prinsep, Nat. Prod. Rep., 2015, 32, 116–211, 10.1039/c4np00144c.
- P. A. Novick, O. F. Ortiz, J. Poelman, A. Y. Abdulhay and V. S. Pande, PLoS One, 2013, 8, e79568, DOI:10.1371/journal.pone.0079568.
- T. Sander, J. Freyss, M. von Korff and C. Rufener, J. Chem. Inf. Model., 2015, 55, 460–473, DOI:10.1021/ci500588j.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7np00052a |
| This journal is © The Royal Society of Chemistry 2018 |