Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Crosstalk of the structural and zinc buffering properties of mammalian metallothionein-2†
Agnieszka
Drozd‡§
a,
Dominika
Wojewska‡¶
 a,
Manuel David
Peris-Díaz
a,
Manuel David
Peris-Díaz
 a,
Piotr
Jakimowicz
b and
Artur
Krężel
a,
Piotr
Jakimowicz
b and
Artur
Krężel
 *a
*a
aDepartment of Chemical Biology, Faculty of Biotechnology, University of Wrocław, F. Joliot-Curie 14a, 50-383 Wrocław, Poland. E-mail: artur.krezel@uwr.edu.pl
bDepartment of Protein Biotechnology, Faculty of Biotechnology, University of Wrocław, F. Joliot-Curie 14a, 50-383 Wrocław, Poland
First published on 21st March 2018
Abstract
Metallothioneins (MTs), small cysteine-rich proteins, present in four major isoforms, are key proteins involved in zinc and copper homeostasis in mammals. To date, only one X-ray crystal structure of a MT has been solved. It demonstrates seven bivalent metal ions bound in two structurally independent domains with M4S11 (α) and M3S9 (β) clusters. Recent discoveries indicate that Zn(II) ions are bound with MT2 with the range from nano- to picomolar affinity, which determines its cellular zinc buffering properties that are demonstrated by the presence of partially Zn(II)-depleted MT2 species. These forms serve as Zn(II) donors or acceptors and are formed under varying cellular free Zn(II) concentrations. Due to the lack of appropriate methods, knowledge regarding the structure of partially-depleted metallothionein is lacking. Here, we describe the Zn(II) binding mechanism in human MT2 with high resolution with respect to particular Zn(II) binding sites, and provide structural insights into Zn(II)-depleted MT species. The results were obtained by the labelling of metal-free cysteine residues with iodoacetamide and subsequent top-down electrospray ionization analysis, MALDI MS, bottom-up nanoLC-MALDI-MS/MS approaches and molecular dynamics (MD) simulations. The results show that the α-domain is formed sequentially in the first stages, followed by the formation of the β-domain, although both processes overlap, which is in contrast to the widely investigated cadmium MT. Independent ZnS4 cores are characteristic for early stages of domain formation and are clustered in later stages. However, Zn–S network rearrangement in the β-domain upon applying the seventh Zn(II) ion explains its lower affinity. Detailed analysis showed that the weakest Zn(II) ion associates with the β-domain by coordination to Cys21, which was also found to dissociate first in the presence of the apo-form of sorbitol dehydrogenase. We found that Zn(II) binding to the isolated β-domain differs significantly from the whole protein, which explains its previously observed different Zn(II)-binding properties. MD results obtained for Zn(II) binding to the whole protein and isolated β-domain are highly convergent with mass spectrometry data. This study provides a comprehensive overview of the crosstalk of structural and zinc buffering related-to-thermodynamics properties of partially metal-saturated mammalian MT2 and sheds more light on other MT proteins and zinc homeostasis.
Significance to metallomicsThe growing interest in metallomics within metal-dependent biological systems provides an opportunity to explore the structural and buffering properties of metallothioneins (MTs). The wide relevance of MTs is obvious due to their multiple biological functions such as participation in zinc and copper homeostasis. However, the MT metalloproteome is still not fully explored and is rather complicated in vivo or in vitro because of its highly dynamic structure, which lacks secondary structural elements and metamorphic behaviour. We have undertaken comprehensive research into the Zn(II) ion binding process and localization, providing structural insights into partially Zn(II)-depleted species of MT2 through a combination of mass spectrometry strategies and molecular dynamics simulations. We attempted to decipher the Zn(II) binding process and its implications for reactivity and metal buffering. |
Introduction
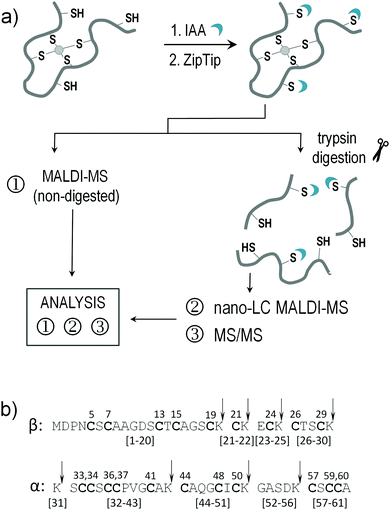
Mammalian metallothionein (MT) is a small ∼7 kDa cellular protein with a high (∼30%) content of cysteine residues. It was discovered 60 years ago as a Cd(II)-containing protein in horse kidneys.1 However, further studies showed that Zn(II) and Cu(I) ions are the most relevant biologically. Mammalian metallothionein is present in four major isoforms, MT1–MT4, which are tissue-specific proteins. MT1 (with 9 subisoforms) and MT2 are present in all kinds of tissues in various amounts and ratios, while MT3 and MT4 are mostly present in the brain and squamous epithelia.2,3 Structural studies of unsaturated MT have proven to be a challenge mainly due to its probably unfolded and highly dynamic structure. To date, only one X-ray structure of metallothionein has been solved: hepatic rat MT2.4 However, NMR structures of individual (isolated) domains of human, rat, rabbit and mouse MT1–MT3 were also determined.5–8 Mammalian metallothionein folds into two separate domains with two Zn–S clusters, one with 3 Zn(II) and 9 Cys (β-cluster) and the other with 4 Zn(II) and 11 Cys (α-cluster) (Fig. 1a). In both domains metal ions are present in tetrahedral geometry and tetrathiolate environments, where sulfur donors are bound to one or two (bridging donors) metal ions. It is worth noting that the X-ray structure is based on Cd(II)-induced in vivo protein,4 which has one Cd(II) ion in the β-domain at position IV and four ions in all positions (I, V, VI and VII) in the α-domain (Fig. 1b). MT reconstituted in vitro also binds seven Zn(II) or seven Cd(II) ions. Most of the current knowledge regarding the structural, biophysical and even biochemical properties of MTs is based on cadmium MTs, which are more convenient to study due to their better spectroscopic properties, less dynamic nature or higher protein affinity, in comparison with a zinc counterpart. Zinc metallothioneins are much less explored mostly due to the lack of appropriate methods to study the structural or biophysical features of their complexes.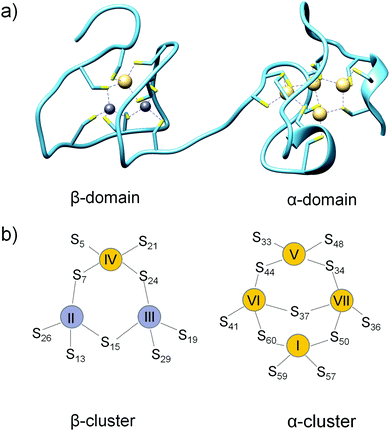 | ||
| Fig. 1 Crystal structure and cluster organization in hepatic rat MT2. (a) Crystal structure of Zn2Cd5MT isolated from rat liver (PDB ID: 4MT2).4 (b) Schematic representations of β- and α-metallothionein clusters in MT2. Yellow and grey circles refer to Cd(II) and Zn(II), respectively, found in the crystal structure. Metal ion numbering corresponds to the order of 113Cd-NMR chemical shifts of Cd7MT2. | ||
In early studies it was shown that seven Zn(II) ions bind to the apo-form of metallothionein (thionein) with the same, undistinguishable affinity which varies from ∼10−12 to 10−13 M in terms of average dissociation constant, depending on the literature source.9,10 In these approaches Zn(II)-to-protein affinity was determined by competition with either protons or chelating agents assuming either identical pKa of all cysteine thiols or equal contribution of ligand-to-metal charge transfer (LMCT) of Zn–S bonds in observable absorbances in the UV range. Our previous study performed on highly sensitive zinc fluorescent probes, FluoZin-3 and RhodZin-3, showed that Zn(II) ions bind to human MT2 with three ranges of affinity.11 Four Zn(II) ions are bound tightly with an average dissociation constant of ∼10−12 M as previously determined for all metal ions. Another two Zn(II) ions bind moderately with a dissociation constant of 10−11 to 10−10 M. The last and the weakest Zn(II) is bound with nanomolar affinity (Kd ∼ 10−8 M). The differentiation of Zn(II) stability constants has been recently shown for MT3 by ITC experiments with EDTA as a competitor, showing that differentiation of Zn(II)-to-protein affinity is common across mammalian metallothionein isoforms.12 In consequence, Zn(II) binding or dissociation occurs sequentially and intermediates such as Zn4MT2, Zn5MT2 and Zn6MT2 are formed, which has been shown in several examples in ESI-MS titrations.13,14 It is worth noting that these species play an important role in free Zn(II) buffering due to their unsaturated nature and various Zn(II) affinities. They can serve as a Zn(II) donor and acceptor at the same time and buffer Zn(II) from pico- to nanomolar concentration. Considering cellular free Zn(II) concentration and its natural fluctuations in the range from 10−11 to 10−9 M, it is apparent that partially Zn(II)-depleted metallothionein species should be present as stable MT2 forms under cellular conditions. Studies performed on HT-29 cells have shown that cells contain in the cytosol fraction a surplus (∼10%) of unoccupied tightly Zn(II) binding proteins. Differential chemical fluorescent modification of metallothionein indicated that a major fraction of this surplus belongs to partially Zn(II)-saturated metallothionein.15–17 Moreover, a number of studies have already shown that various ratios of thionein (T) to total MT (T + MT) control the activity of zinc enzymes or Zn(II)-modulated enzymes, zinc finger folding and saturation of other structural zinc sites.18–21
In this study we aim to connect the current knowledge regarding the structural properties of mammalian metallothionein and its Zn(II) buffering properties. Assuming that metallothionein is always naturally present as Zn7MT species is not valid, and Zn(II)-to-protein content depends on many factors such as oxidative or nitrosative state, thionein induction level and cellular or local free Zn(II) concentration.22,23 The way how Zn(II) is buffered and muffled is still not fully understood. One way to explore this issue is to understand the structural and biophysical features of highly dynamic Zn(II)-depleted MTs species, especially those with five (Zn5MT) and six (Zn6MT) Zn(II) ions, which seem to be the most relevant for the cellular zinc buffering process. Although knowledge regarding the physicochemical properties of Zn(II)-depleted species is growing, currently little is known about the structure and exact Zn(II) distribution among those species of various MT isoforms.11,24 Moreover, the molecular bases that differentiate Zn(II)-to-protein affinity in both domains of metallothionein are also not fully understood.25 In this study we combined classical chemical labelling of cysteine residues by iodoacetamide (IAA) with mass spectrometry approaches. Although metallothionein labelling with IAA has been applied in the past for Cd(II) binding to MT, the lack of high resolution techniques made it impossible to determine the distribution of metal ions accurately and assign them to particular cysteines.26,27 Since spectroscopic methodologies rely on measuring an average structure of multiple species and due to the lack of secondary structural elements and aromatic amino acids, limited information about partially depleted MT could be obtained. In contrast, as demonstrated in early studies, mass spectrometry is a useful and unique technique to identify particular saturated species in MT.28 Recently, the usefulness of mass spectrometry with ion mobility detection for studying conformational changes in partially Cd(II)-depleted MT2 species has been confirmed.29 However, Cd(II) coordination positions across cysteine residues were not assessed. It is worth noting that besides simple ESI-MS studies, none systematic research on zinc metallothionein with bottom-up and top-down MALDI-MS has been performed. Therefore, here we aimed to explore the crosstalk of structural and zinc buffering properties of human metallothionein-2 to shed more light on the properties and importance of Zn(II)-depleted species which are critical for understanding the basis of the cellular zinc buffering process and role of metallothionein in zinc homeostasis.
Experimental
Materials
All reagents used in these studies were of high purity and purchased from Sigma-Aldrich, Acros Organic, Roth, BioShop, VWR International, Avantor, and Iris Biotech. All pH buffers prepared with Milli-Q water were incubated with Chelex 100 resin to eliminate trace metal ion contamination. More detailed information about the used materials is listed in the ESI.†Expression and purification of MT2
The cDNA encoding metallothionien-2 (MT2) was synthesized (GenScript, USA) and cloned into the pTYB21 vector using SapI and EcoRV (Thermo Scientific, USA) restriction sites. The primer pair sequence used in the PCR reaction is provided in the ESI.† The pTYB21 vector encoding MT2 – deposited in the Addgene plasmid repository (https://www.addgene.org), plasmid ID 105693 (MT2a) – was transformed into BL21(DE3)pLysS E. coli cells. The culture medium (1.1% tryptone, 2.2% yeast extract, 0.45% glycerol, 1.3% K2HPO4, 0.38% KH2PO4) was prepared as described previously.30,31 Transformed bacteria (4 l) were cultured at 37 °C until OD600 reached 0.5, then induced with 0.1 mM IPTG and incubated overnight at 20 °C with vigorous shaking. All subsequent steps of purification were conducted at 4 °C. Cells were collected by centrifugation at 4000 × g for 10 min, resuspended in 50 ml of cold buffer A (20 mM HEPES, pH 8.0, 500 mM NaCl, 1 mM EDTA, 1 mM TCEP) and sonicated for 30 min (1 min “on”, 1 min “off”) followed by centrifugation at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min. The supernatant was incubated with 20 ml of chitin resin in buffer A and kept overnight with mild shaking. After the incubation resin was washed 4–5 times with 50 ml of buffer A, to induce the cleavage reaction 25 ml of buffer B (20 mM HEPES, pH 8.0, 500 mM NaCl, 1 mM EDTA, 100 mM DTT) was added to the resin and the mixture was incubated for 36–48 h at room temperature on a rocking bed. The reaction was monitored by SDS-PAGE. Briefly, the centrifuged solution containing protein was acidified to pH ∼ 2.5 with 7% HCl and subsequently concentrated to a small volume using Amicon Ultra-4 Centrifugal Filter Units with a membrane NMWL of 3 kDa (Merck Millipore, USA) and purified on an SEC-70 gel filtration column (Bio-Rad, USA) equilibrated with 5 mM HCl. The identity of apo-MT2 protein (thionein, T) was confirmed by ESI-MS, using an API 2000 instrument (Applied Biosystems, USA). The average molecular weight (MW) calculated was 6042.0/6042.2 Da, respectively. Purified apo-MT2 was used for experiments immediately after purification due to its instability under aerobic conditions. The yield of pure MT2 production was 3.5 mg per liter of bacterial culture.
000 × g for 15 min. The supernatant was incubated with 20 ml of chitin resin in buffer A and kept overnight with mild shaking. After the incubation resin was washed 4–5 times with 50 ml of buffer A, to induce the cleavage reaction 25 ml of buffer B (20 mM HEPES, pH 8.0, 500 mM NaCl, 1 mM EDTA, 100 mM DTT) was added to the resin and the mixture was incubated for 36–48 h at room temperature on a rocking bed. The reaction was monitored by SDS-PAGE. Briefly, the centrifuged solution containing protein was acidified to pH ∼ 2.5 with 7% HCl and subsequently concentrated to a small volume using Amicon Ultra-4 Centrifugal Filter Units with a membrane NMWL of 3 kDa (Merck Millipore, USA) and purified on an SEC-70 gel filtration column (Bio-Rad, USA) equilibrated with 5 mM HCl. The identity of apo-MT2 protein (thionein, T) was confirmed by ESI-MS, using an API 2000 instrument (Applied Biosystems, USA). The average molecular weight (MW) calculated was 6042.0/6042.2 Da, respectively. Purified apo-MT2 was used for experiments immediately after purification due to its instability under aerobic conditions. The yield of pure MT2 production was 3.5 mg per liter of bacterial culture.
Synthesis of β-domain of MT2
The N-terminal fragment of MT2 (Met1-Ser32) corresponding to the β-domain (βMT2) was synthesized on TentaGel R RAM resin (loading 0.19 mmol g−1, Rapp Polymere GmbH) using a Liberty 1 microwave-assisted synthesizer (CEM, USA) according to the standard Fmoc strategy described in detail before.32,33 The resin-attached peptide was cleaved from the resin by 2 h incubation with a mixture of TFA/thioanisole/EDT/anisole (90/5/3/2, v/v/v/v), followed by precipitation in cold (−20 °C) Et2O. The crude peptide was purified by HPLC (Dionex Ultimate 3000 system) on a Phenomenex C18 column (Gemini-NX 5 μm, 110 Å) using 0.1% TFA with a 0–45% ACN gradient in 30 min and then lyophilized. The identity of the obtained pure peptide was confirmed by ESI-MS (API 2000 Applied Biosystems, USA). The average mass calculated for the β-domain was 3252.3/3252.8 Da.Reconstitution of MT2 and the β-domain with Zn(II)
Aliquots of apo-MT2 and apo-βMT2 in diluted HCl (pH 2.5 with 1 mM TCEP) were mixed with zinc sulfate at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 or 1
9 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 under a nitrogen blanket, and the pH adjusted to 8.6 with 1 M solution of Tris base.34 The samples were concentrated with Amicon Ultra-4 Centrifugal Filter Units with a membrane NMWL of 3 kDa, and purified on an SEC-70 column (Bio-Rad, USA) equilibrated with 20 mM Tris–HCl buffer, pH 8.6. The concentration of the purified protein was obtained spectrophotometrically, via DTNB and PAR assays regarding thiol and Zn(II) concentration, respectively.35,36 Additionally, samples were analysed by ICP (ICP-AES iCAP 7400, Thermo Scientific) to confirm the spectrophotometric results.
4 under a nitrogen blanket, and the pH adjusted to 8.6 with 1 M solution of Tris base.34 The samples were concentrated with Amicon Ultra-4 Centrifugal Filter Units with a membrane NMWL of 3 kDa, and purified on an SEC-70 column (Bio-Rad, USA) equilibrated with 20 mM Tris–HCl buffer, pH 8.6. The concentration of the purified protein was obtained spectrophotometrically, via DTNB and PAR assays regarding thiol and Zn(II) concentration, respectively.35,36 Additionally, samples were analysed by ICP (ICP-AES iCAP 7400, Thermo Scientific) to confirm the spectrophotometric results.
ESI-MS-monitored titration of apo-MT2 with Zn(II)
The Zn(II) titration study was performed in degassed 10 mM (NH4)2CO3/ACN (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) buffer solution, pH 7.6. A solution of 25 μM apo-MT2 was mixed with 0–7 molar equivalents of zinc acetate (500 μM), and buffer solution was added to a final volume of 200 μl. The samples were incubated for 30 s and injected by a syringe pump (40 μl min−1 flow rate) into an ESI mass spectrometer (API 2000 Applied Biosystems, USA). MS spectra were recorded in positive ion mode during 5 min in the 1000–1800 m/z range.
1, v/v) buffer solution, pH 7.6. A solution of 25 μM apo-MT2 was mixed with 0–7 molar equivalents of zinc acetate (500 μM), and buffer solution was added to a final volume of 200 μl. The samples were incubated for 30 s and injected by a syringe pump (40 μl min−1 flow rate) into an ESI mass spectrometer (API 2000 Applied Biosystems, USA). MS spectra were recorded in positive ion mode during 5 min in the 1000–1800 m/z range.
Alkylation of MT2 for MALDI-MS studies
All solutions were degassed by purging with nitrogen just before use. To an Eppendorf plastic tube (1.5 ml) filled with nitrogen the following were added sequentially: 1 nmol of apo-MT2 in 10 mM HCl, 20 μl of 100 mM (NH4)2CO3, pH ∼ 8, and an appropriate amount of 500 μM ZnSO4 (1, 2, 3, 4, 5, 6 and 7 molar equivalents over apo-MT2, respectively). A control experiment with no Zn(II) ions added was also performed. The final volume was adjusted to 55 μl with Milli-Q water. The pH of the final solution was 7.5. After gently mixing, 3 μl of 10 mM iodoacetamide (IAA) (30 nmol, 1.5 molar equivalents over one Cys) was added. Alkylation was carried out under a nitrogen blanket in darkness for 15 min, followed by freezing the samples on dry ice to prevent further reaction. Each sample was desalted while removing excess IAA using ZipTip μ-C18 (Merck Millipore, USA), and eluted with 10 μl of Milli-Q water/ACN solution (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v). MALDI spectra were recorded for each sample (1 μl of sample diluted 12 times with CHCA matrix in ACN) on an ABI 4800 MALDI TOF/TOF mass spectrometer (Applied Biosystems, USA).
50, v/v). MALDI spectra were recorded for each sample (1 μl of sample diluted 12 times with CHCA matrix in ACN) on an ABI 4800 MALDI TOF/TOF mass spectrometer (Applied Biosystems, USA).
Trypsin digestion of alkylated MT2
4.5 μl of a desalted solution of partially alkylated MT2 was pipetted into a 250 μl PCR tube. Acetonitrile was removed before digestion with trypsin on a Maxi dry plus speed vacuum system (Heto Lab Equipment, UK) for approximately 10 minutes, followed by adding 1.5 μl of diluted trypsin (0.5 μg ml−1 in 50 mM AcOH) and 3.5 μl of 100 mM (NH4)2CO3 to each sample. Digestion was carried out for 10 minutes at 37 °C followed by freezing the sample on dry ice to prevent further digestion. All samples were lyophilized and kept at −20 °C before analysis.Nano-LC separation and MS measurements of digested samples
Lyophilized samples were dissolved in 30 μl of 0.1% TFA in LC-MS quality water and transferred into low protein binding autosampler vials. Nano-LC separations were carried out on an Easy nano-LC II instrument (Bruker, Germany) using an Acclaim PepMap C18 3 μm column (75 μm i.d. × 15 cm, 100 Å) with a gradient of 0–60% B in A (A: 0.1% TFA/H2O, B: 0.1% TFA/ACN) during 24 minutes. Collected fractions were pooled every 15 s and spotted on a MALDI plate pre-spotted with HCCA matrix (PAC II 384, Bruker, Germany) as a single spot (96 spots in total for each sample). MS spectra were recorded off-line using an Ultraflextreme MALDI spectrometer (Bruker, Germany), in the 700–3500 m/z range, calibrated before each of four adjacent spots with Peptide Calibration Standard PACII (Bruker, Germany). The laser intensity was 29% for calibration and 31% for measurement. MS/MS spectra were recorded in LIFT mode, in the 800–3000 m/z range, with the laser intensity adjusted manually.Preparation of metal-depleted SDH
An apo-form of sorbitol dehydrogenase (SDH) was prepared according to a previously established procedure by the treatment of commercial SDH (Roche) with dipicolinic acid and subsequent purification.18,37 Concentration of purified apo-SDH was examined by its titration with standard Zn(II) ions followed by enzyme activity determination. The activity was determined spectrophotometrically as time-course measurements at 340 nm as a result of NADH oxidation to NAD+ upon conversion of D-fructose to D-sorbitol. For that purpose protein samples were incubated in 50 mM HEPES buffer, pH 7.4, with 100 μM TCEP, 0.1 M D-fructose and 0.125 mM NADH.Zn(II) transfer from MT2 to metal-depleted SDH (apo-SDH)
The rate and efficiency of Zn(II) transfer from MT2 to apo-SDH were monitored enzymatically using the assay described above. Metal-depleted SDH was mixed in a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with freshly prepared 1 μM MT2 and incubated in 50 mM HEPES buffer, pH 7.4, in the presence of 100 μM TCEP. Protein aliquots were taken after certain periods of time (0–120 min) and assayed for the recovery of enzymatic activity. Full recovery of the SDH was reached by apo-SDH saturation with Zn(II) ions under the same conditions. Samples for mass spectrometry were prepared in a different way. For that purpose 1 nmol of MT2 and apo-SDH were mixed with 20 μl of 100 mM (NH4)2CO3 and incubated for 4, 30, 60 or 120 min at room temperature. After that time the sample was modified with IAA according to the procedure described above and finally measured using an ABI 4800 MALDI TOF-TOF mass spectrometer (Applied Biosystems) and subjected to nano-LC separation and a further procedure as described above.
1 with freshly prepared 1 μM MT2 and incubated in 50 mM HEPES buffer, pH 7.4, in the presence of 100 μM TCEP. Protein aliquots were taken after certain periods of time (0–120 min) and assayed for the recovery of enzymatic activity. Full recovery of the SDH was reached by apo-SDH saturation with Zn(II) ions under the same conditions. Samples for mass spectrometry were prepared in a different way. For that purpose 1 nmol of MT2 and apo-SDH were mixed with 20 μl of 100 mM (NH4)2CO3 and incubated for 4, 30, 60 or 120 min at room temperature. After that time the sample was modified with IAA according to the procedure described above and finally measured using an ABI 4800 MALDI TOF-TOF mass spectrometer (Applied Biosystems) and subjected to nano-LC separation and a further procedure as described above.
Mass spectrometry data analysis
Obtained mass spectrometry data were analysed and processed using Data Explorer Software (Version 4.9, Applied Biosystems), flexAnalysis (Version 3.4, Bruker Daltonik) or Data Analyst (Version 1.4.2, Applied Biosystems) according to the equipment used.Circular dichroism
Circular dichroism (CD) spectra of zinc metallothionein were recorded using a J-1500 Jasco spectropolarimeter (JASCO) at 25 °C in a 2 mm quartz cuvette, under a constant nitrogen flow over the range of 196–270 nm with a 100 nm min−1 speed scan. Final spectra were averaged from three independent scans. 10 μM thionein (apo-MT2) was incubated with 6 and 7 eq. of ZnSO4 in 20 mM Tris–HCl buffer (100 mM NaCl, 200 μM TCEP pH 7.4) and recorded. Independently, reconstituted 10 μM MT2 was incubated with 0–100 μM of EGTA or EDTA and spectra were recorded after 10 and 30 minutes.Molecular dynamics
Initial coordinates were taken from the crystal structure of rat Zn2Cd5MT2 (PDB ID: 4MT2). All molecular dynamics simulations were performed using an AMBER99SD force field for protein atoms and a semi-bonded approach for ZnxSy clusters.38,39 Hydrogen bonds were added to the metalloprotein using the LEAP module of the AMBER program. The structure was solvated in a box of TIP3P water molecules with a density of 0.813 g cc−1 and counter-ions were added to neutralize the system. After solvation and neutralization, periodic bound conditions were set up using the particle mesh Ewald method with a cut-off of 12 Å. The system was firstly energy minimized for 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps by means of a conjugate gradient algorithm followed by a steep descent algorithm, setting up the switching cut-off from 10 to 12 Å for computing electrostatics and Lennard-Jones energies. The LINCS constraint was applied to all bonds involving hydrogens. MD simulations were performed in two steps. The system was equilibrated using the NVT ensemble with leapfrog Verlet integrator (leap) and Hoover thermostat (303.15 K and 1 atm) for 1 ns with a timestep of 0.002 (2 fs) and NVT production for 4 ns with a 2 fs timestep. Molecular dynamics simulations were carried out with the module LEAP from AMBER and with the GROMACS software.40 The PROPKA 2.0 method was used for the prediction of the pKa values of ionizable residues.41
000 steps by means of a conjugate gradient algorithm followed by a steep descent algorithm, setting up the switching cut-off from 10 to 12 Å for computing electrostatics and Lennard-Jones energies. The LINCS constraint was applied to all bonds involving hydrogens. MD simulations were performed in two steps. The system was equilibrated using the NVT ensemble with leapfrog Verlet integrator (leap) and Hoover thermostat (303.15 K and 1 atm) for 1 ns with a timestep of 0.002 (2 fs) and NVT production for 4 ns with a 2 fs timestep. Molecular dynamics simulations were carried out with the module LEAP from AMBER and with the GROMACS software.40 The PROPKA 2.0 method was used for the prediction of the pKa values of ionizable residues.41
Results
In order to gain detailed structural insights into the Zn(II) complexation pathway in mammalian MT2, we combined differential alkylation of free cysteine thiols by iodoacetamide (IAA) in metal-free and partially Zn(II)-depleted protein with mass spectrometry techniques. To do so, samples of human MT2 were subjected to alkylation with IAA to “fix” the coordination pattern in the desired structural state, and subsequently analysed by (i) MALDI-MS, (ii) nano-LC-MALDI-MS for the partially alkylated tryptic digest and finally by (iii) MS/MS for the nano-LC separated tryptic peptides (Fig. 2). The same approach was used to study the complexation process of the isolated β-domain of MT2 (βMT2) and to map free cysteine residues formed upon Zn(II) transfer from fully Zn(II)-loaded MT2 to metal-depleted sorbitol dehydrogenase (apo-SDH). We supported our study with direct titration of apo-MT2 with consecutive Zn(II) molar equivalents, ESI-MS measurements and molecular dynamics, which were used to examine structure conformational changes upon Zn(II) binding. In the first step of the study, MALDI spectrum of metal-free or partially Zn(II)-loaded MT2 modified by IAA was analysed and compared with the signal profile of unmodified protein obtained by ESI-MS. Then, detailed analysis of tryptic digest MS and MS/MS spectra were carried out and taken into account to propose the stoichiometry of the formed MT2 species and coordination mode. Finally, experimental results were compared with molecular dynamics simulations.Alkylation of metal-free metallothionein-2
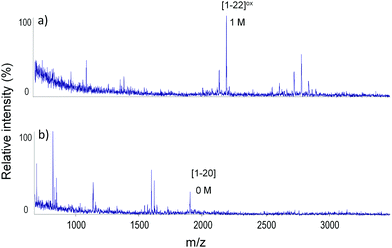
The MALDI spectrum recorded for unmodified MT2 demonstrates one major signal of 6043.7 m/z (Table S1 and Fig. S1a, ESI†), which is expected for a pure apo-form. However, alkylated apo-MT2 indicates signal distribution with pairwise intensities with clearly visible signals with 12, 14, 16, 18 and 20 modifications (Fig. 3a, Fig. S1b and Table S1, ESI†), which is contradictory to the expected single signal of an entirely alkylated protein (20 acetamide moieties). The observed pattern did not change when either extending the reaction time or applying higher IAA concentrations. This effect was largely suppressed when the protein was treated with 6 M urea (Fig. S1c, ESI†). In this case the most abundant signal has 20 modifications, but peaks with 17, 18 and 19 modifications are still observed (Fig. S1c, ESI†). The tryptic digest revealed significantly incomplete alkylation in regions [1–20] and [32–43], but incomplete modification was also observed in other peptides (Table 1, Tables S2 and S3, ESI†). The pKa values of cysteine thiols predicted by PROPKA method revealed an acidity shift of Cys residues, which ranges from 6.9 to 11.9.41 The protonation state of Cys affects solvent exposure and accessibility for Cys to be modified by IAA. Calculations of absolute surface area (ASA) of each residue in apo-MT2 obtained by MD after 4 ns showed cysteine residues from Cys6 up to Cys20 and from Cys34 to Cys42 to be less accessible for solvent (Table S4, ESI†). These results are in agreement with those obtained from tryptic digestion and match fragments [1–20] and [32–43] in which alkylation was not completed, possibly due to the lower solvent accessibility.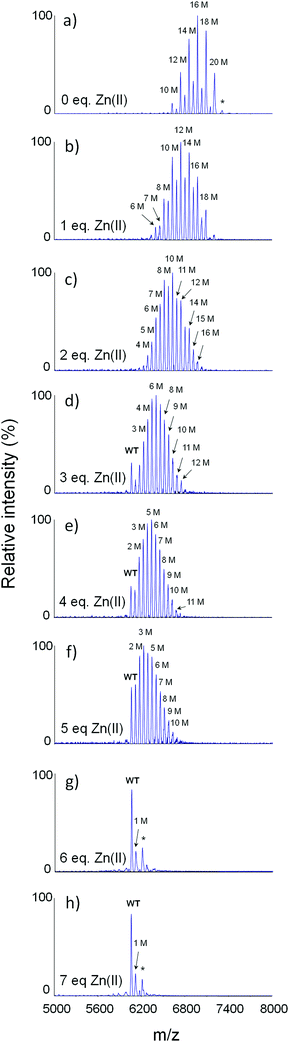 | ||
| Fig. 3 Comparison of MALDI-MS spectra of IAA-modified apo-MT2 treated with 0–7 molar equivalents of Zn(II). Found and calculated m/z are listed in Table S1 (ESI†). M denotes cysteine modification. | ||
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ) modified by IAA are indicated by n M (where n indicates the number). n.d. – not detected. All identified tryptic fragments can be found in Table S3 (ESI). The full list of calculated and found m/z for all tryptic fragments can be found in Table S4 (ESI)
) modified by IAA are indicated by n M (where n indicates the number). n.d. – not detected. All identified tryptic fragments can be found in Table S3 (ESI). The full list of calculated and found m/z for all tryptic fragments can be found in Table S4 (ESI)
| Zn(II) eq. | MDPN![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) S S![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) AAGDS AAGDS![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) T T![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) AGS AGS![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) K [1–20] MDPN K [1–20] MDPN![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) S S![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) AAGDS AAGDS![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) T T![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) AGS AGS![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) KCK [1–22] KCK [1–22] |
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) TS TS![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) KK [26–31] KK [26–31] |
S![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) S S![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) PVG PVG![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) AK [32–43] AK [32–43] |
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) AQG AQG![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) I I![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) K [44–51] K [44–51] ![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) AQG AQG![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) I I![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) K GASDK K GASDK![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) S S![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) A [44–61] A [44–61] |
GASDK![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) S S![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) A [52–61] A [52–61] |
|---|---|---|---|---|---|
| 0 | 3–5 M | 1 M | 3–5 M | 2 M | 0 M, 2 M |
| 1 | 3–5 M | 2 M | 2–5 M | 2 M | 2 M |
| 2 | 2–5 M | 1 M | 1–5 M | 1–2 M | 1–2 M |
| 3 | 5 M | 1 M | 1–5 M | 1–2 M | 0–2 M |
| 4 | 3 M | n.d. | 0–5 M | 0–2 M | 0 M |
| 5 | 0–2 M | 1 M | 0–2 M | 0–1 M | 0 M |
| 6 | 0 M | 0 M | 0 M | 0 M | n.d. |
| 7 | 0 M | n.d. | 0 M | 0 M | n.d. |
| 7 | 1 M | n.d. | 0 M | 0 M | n.d. |
Alkylation of partially Zn(II)-depleted metallothionein-2
Alkylation of apo-MT2 was performed in such a way as to determine the positions of modified and protected Cys residues upon addition of each molar equivalent of Zn(II) from 1 up to 7 in order to map the Zn(II)-bound position with the highest possible resolution. The MALDI spectrum of apo-MT2 with one Zn(II) equivalent shows a maximum of 18 modifications, which is a sign of two cysteine residues protected from alkylation (Fig. 3b). The most abundant signal corresponds to 12 modified residues, whereas in the spectra for metal free MT it corresponded to 16 modifications. Therefore, these four cysteine residues not observed in MS spectra should be involved in metal binding when one Zn(II) equivalent is applied. The ESI spectrum of the same Zn(II)-to-protein ratio reveals the co-existence of three species with 0–2 bound Zn(II) ions, Zn0–2MT2 (Fig. S2b and Table S5, ESI†). Additionally, MS/MS spectra show that the [32–43] region is more resistant to alkylation since it showed a tryptic fragment with fewer modifications (2 M) compared to the metal-free protein sample (3 M) (Table 1). MS/MS spectra of selected tryptic fragments revealed Cys36, Cys48 and Cys60 residues as highly protected, so more likely involved in Zn(II) ion coordination (Fig. S3, ESI†). Because Cys41 is modified, and Zn(II) coordinates tetrahedrally, one of Cys33 or Cys34 and Cys37 may be involved in coordination of the first Zn(II) ion, although these occur in both modified and unmodified forms (Fig. 4a). Molecular dynamics results indicated that Cys37 is coordinated to Zn(II), whereas Cys33 and Cys34 remain far from the metal ion coordination sphere (Fig. S4a, ESI†). A stabilized structure by interaction between Lys44 and Cys37 was found, which would increase Zn(II)-bound thiolate reactivity towards the Zn(II) ion. Positively charged lysine will lower the pKa value of cysteine thiol and thus promote more nucleophilic thiolate formation.42,43 The observed peaks indicate labile coordination of the first Zn(II) ion under the applied conditions. Surprisingly, the residues responsible for Zn(II) binding do not correlate with any positions observed in the crystal structure of fully saturated MT2 (Fig. 1).4,5,7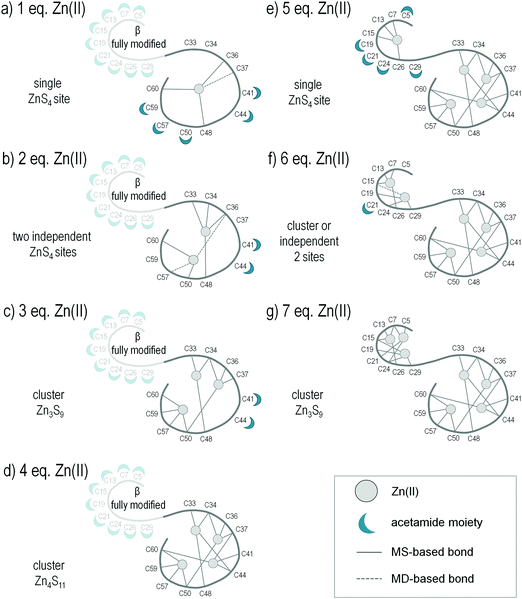 | ||
| Fig. 4 Scheme of the most prevalent sequential Zn(II) binding pathway in MT2 based on MS/MS analysis of tryptic peptides differently modified by IAA and MD calculations. | ||
The addition of two Zn(II) equivalents to apo-MT2 and subsequent alkylation results in 4 up to 16 cysteines modified observable in the MALDI spectrum, indicating a minimum of four highly protected cysteine residues, which is two modifications less in comparison with results obtained for a sample with one Zn(II) equivalent (Fig. 3b and c). Characteristic pairwise peak distribution is no longer visible. Interestingly, a singly modified cysteine residue (1 M) in the tryptic fragment [32–43] emerged; hence if only one cysteine could be modified, the other four Cys residues might be protected in this region (Table 1). In the tryptic fragment [44–51], the signal intensity in MALDI-MS spectra suggests the prevalence of double modification, which would lead to one protected cysteine (Fig. S5, ESI†). MS/MS data revealed that both Cys48 and Cys50 are protected from alkylation, wherein Cys50 is less resistant to those two (Fig. S6a, ESI†). Tryptic fragment [52–61] occurs in singly and doubly modified form, which corresponds to two and/or one protected cysteine (Table 1 and Table S2, ESI†). Because Cys60 was alkylation protected for the sample with one Zn(II) equivalent, similarity with the two Zn(II) equivalent sample is highly possible. The tetrahedral ZnS4 coordination sphere is probably filled up by either Cys57 or Cys59 (Fig. 4b). Therefore, from the results obtained we may conclude that Cys33, 34, 50 and 59 or 57 residues were available to coordinate Zn(II), besides those reported previously (Cys36, 48 and 60). Regarding molecular dynamics simulations, Cys33, 34, 36 and 48 coordinate one Zn(II), whereas Cys36, 50, 57 and 60 coordinate a second Zn(II) (Fig. S4b, ESI†).
The MALDI spectrum of undigested apo-MT2 with three Zn(II) equivalents shows a maximum of 12 modifications, strongly suggesting at least eight protected Cys residues, which is four less than in the previous step (Fig. 3d). Strikingly, a small fraction of unmodified protein (indicated as WT, which corresponds to Zn7MT2) is observed. ESI titration results are consistent with this finding, as the observed peaks correspond to co-existing forms from Zn3MT2 up to Zn7MT2 (Fig. S2d, ESI†). The presence of the unmodified tryptic fragment [52–61] showed that Cys57, 59 and 60 are protected against alkylation to some extent (Table 1), but singly and doubly modified [52–61] forms are still visible (Fig. S7, ESI†). Fragment [32–43] exhibits an almost unchanged modification mode as compared to the protein with two Zn(II) equivalents, so Cys33, 34, 36 and 37 are visible as modified and unmodified (Table 1). MS/MS data for singly and doubly modified tryptic fragments [44–51] show that one or two cysteines are involved in Zn(II) binding, i.e. Cys48 and/or Cys50 may possibly be involved in cluster formation (Fig. S8, ESI†). Based on the above results, the existence of a Zn3S8 or Zn3S9 cluster is inferred (Fig. 4c). Molecular dynamics results indicated that both Cys48 and 50 are involved in Zn3S9 cluster formation (Fig. S4c, ESI†).
Iodoacetamide modification of apo-MT2 incubated with four Zn(II) equivalents results in 0 and up to 11 modifications of cysteines observed in the MALDI spectrum, which can be attributed to maximally 9 alkylated cysteines in the β-domain and two alkylated residues in the α-domain (Fig. 3e). The most abundant signals correspond to five and four modified cysteines, which translates to 15 or 16 cysteine residues involved in Zn(II) binding. This can be attributed to the most present Zn4MT2 and Zn5MT2 forms. The ESI-MS of Zn(II) titration study indicates species diversity and the presence of Zn4MT2, Zn5MT2, Zn6MT2 and Zn7MT2 (Fig. S2e, ESI†), but one should note that the presence of particular Zn(II)-depleted species must be taken qualitatively not quantitatively. Tryptic fragment [1–20] is triply modified, which means two cysteine residues resistant to alkylation reside in the β-domain and probably coordinate to the same Zn(II) ion. Tryptic fragments [32–43], [44–51] and [52–61] exist in non-alkylated forms (Table 1), which proves full saturation of the Zn4αMT cluster (Fig. 4d). Molecular dynamics simulation showed Zn(II) distribution along the Zn4αMT domain (Fig. S4d, ESI†). Whilst fragment [52–61] is observed in unmodified form only, both [32–43] and [44–51] exist in partially modified forms. For fragment [44–51] a fully modified form was not detected (Fig. S9, ESI†).
The most abundant signal in the MALDI profile for apo-MT2 with five Zn(II) equivalents conforms to three modified cysteine residues (Fig. 3f); hence the dominant form should have 17 cysteine residues involved in the coordination, which is shifted to two modifications less than in apo-MT2 with four Zn(II) equivalents. This relates to the Zn4αZn2βMT2 form (Zn4S11 in α- and Zn2S6 in β-domain). Moreover, the maximum number of modifications observed is 10, so the rest would be involved in Zn(II) coordination forming α-cluster Zn3S9–10 with one labile cysteine (Fig. 3f). Furthermore, increased abundance of unmodified protein is observed (e.g. Zn7MT2 WT form) in MALDI-MS spectra. ESI-MS titration shows that the forms Zn6MT2 and Zn7MT2 predominate, although minor intensities from Zn3MT2, Zn4MT2 and Zn5MT2 are still observed (Fig. S2f, ESI†). Tryptic fragments [32–43] and [44–51] show decreased heterogeneity regarding the number of cysteine modifications in the α-cluster (with a range of 0 to 2 and 0 to 1 modified cysteines, respectively) (Table 1). Interestingly, a new pattern is observed when adding 5 Zn(II) equivalents compared to previous titrations regarding tryptic fragment [1–20] since 0 to 2 cysteine residues are modified (0–2 M) (Table 1). So, a minimum of 3 and a maximum of 5 cysteine residues would be involved in Zn(II) coordination. Tryptic fragment [26–31] with two cysteines is observed as singly modified, so the other cysteine might be involved in metal binding to fill up Zn(II) coordination in the β-domain. MD simulations present a structure with Zn4S11 and ZnS4 clusters in α- and β-domains, respectively (Fig. 5a and Fig. 4e). As observed in Fig. S12a (ESI†), the structure obtained by molecular dynamics showed a comparable α-domain to the X-ray MT2 structure (PDB ID: 4MT2).4
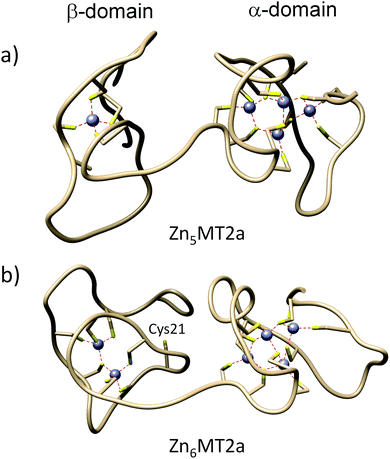 | ||
| Fig. 5 Simulated metallothionein-2 structures obtained from molecular dynamics. (a) Molecular structure of Zn5MT2; (b) molecular structure for Zn6MT2. Zn(II) ions are represented by grey colour. | ||
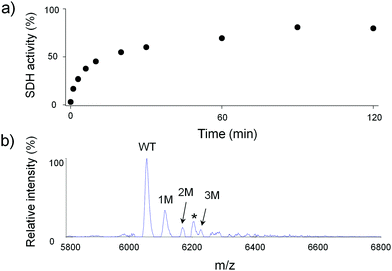
Apo-MT2 with six Zn(II) equivalents shows in the MALDI spectrum only two peaks, with one and without any modification (Fig. 3g). This is consistent with ESI-MS titration (Fig. S2g, ESI†), where in the case of six Zn(II) equivalents only Zn7MT2 and Zn6MT2 species are observed. All tryptic fragments are found to be unmodified, except for the region 21–25, which was not detected (Table 1). MS/MS spectra for tryptic peptides confirm the lack of modification in region 31–61 (α-domain) (Fig. S10, ESI†). Because there is one single modification, and it is proved not to be found in this region (31–61), there is strong evidence that either Cys21 or Cys24 should be modified by IAA. MD results showed a possible structure where Zn(II) binds tetrahedrally to Cys24, whereas Cys21 is not coordinated (Fig. 5b). Regarding whether Cys21 or Cys24 is modified, our MD results indicate H-bonding interaction from Ser6 to Cys21 stabilizing the latter, and thus Cys21 is modified (Fig. 4f). On the other hand, Cys5 is found to not be involved in coordination. Fig. S12b (ESI†) showed overlapping structures between the X-ray MT2 structure (PDB ID: 4MT2), validating the MD structures obtained.4
Similarly, in the case of apo-MT2 with seven Zn(II) equivalents only two peaks are well observed in the MALDI spectrum with one and no modification (Fig. 3h). This is also consistent with ESI-MS titration where Zn7MT2 and Zn6MT2 species are observed (Fig. S2h, ESI†). All found tryptic fragments show no modifications, except for fragment [1–22], which is singly modified (Table 1). Fig. 6 shows the comparison of two tryptic fragments [1–20] with no modification and [1–22] with one modification, which indicates that Cys21 might be modified. Moreover, the MS/MS spectrum of the singly modified [1–22] fragment strongly confirms localization of the acetamide modification on the Cys21 residue (Fig. S11, ESI†). Addition of the last Zn(II) ion caused overall structure arrangement that involved coordination of those previously non-coordinated cysteines, namely Cys21 and Cys5, and Zn(II) coordination filled it with Cys7 and Cys24 (Fig. 4g). Our molecular dynamic simulation results for Zn7MT2 species are in good agreement with the X-ray structure, thus assessing and validating the quality of the results previously reported (Fig. S12c, ESI†).
Alkylation of metal-free and Zn(II)-depleted βMT2
In order to investigate whether or not the Zn(II) binding process in the isolated β-domain of MT2 is comparable with the whole protein, we performed a MALDI-MS/MS study on non-digested and trypsinized protein. The MALDI spectrum of undigested βMT2 in the absence of Zn(II) presents a single peak corresponding to the β-domain fragment with all nine Cys modified by IAA (Fig. S13 and Table S6, ESI†). No trace of three-dimensional structure is identified, in contrast to the full apo-MT2 protein (see above Fig. 3a). An isolated β-domain is shown in Fig. S14a (ESI†) and has been obtained from molecular dynamics simulation.Addition of one molar equivalent of Zn(II) results in a maximum of seven modifications, while the most abundant signal corresponds to the species with three modified cysteines (Fig. S13 and Table S6, ESI†). The MALDI spectrum does not show the presence of a fully modified β-domain. This together suggests that two species can be formed with one (ZnS4) and two Zn(II) ions (Zn2S6–8) bound to the β-domain peptide. ESI-MS titration confirms that the system is highly dynamic due to the presence of multiple species with 0–3 bound Zn(II) ions; however, species with one and two Zn(II) predominate, which is consistent with MALDI data (Fig. S13, S15 and Table S7, ESI†). Upon addition of one Zn(II) equivalent tryptic fragments [1–20] and [1–25] are less extensively modified, with no significant changes for others; hence the first Zn(II) is probably coordinated in the [1–25] region (Fig. 7 and Table S8, ESI†). To elucidate those Cys residues involved in Zn(II) coordination, MS/MS analysis over undigested 5 M βMT2 with 1 Zn(II) eq. was performed (peak 5 M in Fig. S13b, ESI†). The produced y fragmentation series revealed that Cys19, 21, 24, 26 and 29 are modified (Table S9, ESI†), so Cys5, 7, 13 and 15 should form a coordination site for the first Zn(II) ion, which is contrary to annotations in the X-ray structure (Fig. 1b, β-cluster). The most abundant fourfold modified tryptic fragment [21–31] of βMT2 from a 1 eq. Zn(II) sample indicates modified Cys21, 24, 26 and 29 residues, and the triply modified tryptic fragment [23–31] from the same sample confirms Cys24, 26 and 29 modifications (Fig. 7). Therefore, all data consistently prove that coordination of the first Zn(II) is formed by Cys5, 7, 13 and 15, contradictory to all coordination sites for available crystal and NMR structures.4,5,7 This result is supported by the simulated structure obtained from MD calculations (Fig. S14b, ESI†).
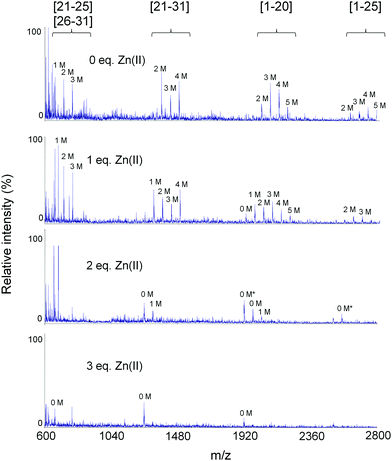 | ||
| Fig. 7 Comparison of MALDI spectra of trypsinized and differentially modified βMT2 samples incubated with 0–3 Zn(II) equivalents. All found tryptic fragments are listed in Table S7 (ESI†). Asterisk and M indicate the occurrence of two disulfide bridges and modifications in the selected fragment, respectively. | ||
Addition of two molar equivalents of Zn(II) ions causes the maximum modification of four cysteines and the singly modified form is the most abundant (Fig. S13c, ESI†). The results are consistent with ESI titration results, as for two Zn(II) molar equivalents two species exist, Zn2βMT2 and Zn3βMT2 (Fig. S15c, ESI†). Moreover, the results are consistent with observed tryptic peptides, which exist mostly in unmodified forms (Fig. 7). Specifically, the [1–25] fragment exists in unmodified form only, while [1–20] and [21–31] both demonstrate an accompanying singly modified form. The obtained structure based on MD simulations displayed Cys5 and Cys29 free of Zn(II) coordination (Fig. S14c, ESI†), supporting the MS results.
Application of the third molar equivalent of Zn(II) shows all Cys residues protected from alkylation (Fig. S13d, ESI†); hence all are involved in Zn(II) coordination. These results are in agreement with those obtained by ESI titration results in which only the Zn3βMT2 form is observed (Fig. S15d, ESI†) and with MD simulations (Fig. S14d, ESI†). Furthermore, all visible tryptic peptides are unmodified (Fig. 7).
Zn(II) transfer from MT2 to apo-SDH
Apo-SDH activity is restored relatively quickly during incubation with fully Zn(II)-loaded MT2 at room temperature and pH 7.4, in such a way that 50% of the enzyme activity is recovered after ∼17 min of incubation. Almost full recovery (more than 80%) was observed after 2 h of protein incubation (Fig. 8a). Since the SDH molecule binds one Zn(II) ion in the active centre, and both proteins (MT2 and apo-SDH) were mixed equimolarly, we assumed that one Zn(II) ion from MT2 was transferred to the metal-depleted metalloenzyme. The MALDI spectrum recorded for the sample alkylated after 120 min of incubation shows up to three possible modified cysteine residues (Fig. 8b). Tryptic peptides obtained in the subsequent experiment prove single modification in region [26–31] (Fig. S16 and Table S10, ESI†). Because the undigested protein bears three modifications, the other two should be located on Cys21 and Cys24. Similarly, the sample incubated with apo-SDH for 5 min showed no modifications in regions 1–20 and 31–61 (Table S10, ESI†), but the undigested protein bears a single modification (data not shown). Because region 21–30 was not detected, it is clear that Cys21, 24, 26 or 29 was modified. These data show that the modification pattern of Zn6MT2 species obtained by one Zn(II) ion transfer from Zn7MT2 to apo-SDH seems to be identical to Zn6MT2 obtained by addition of 6 Zn(II) equivalents to apo-MT2.CD spectra of Zn6MT2 obtained by Zn(II) association or dissociation
Independently from mass spectrometry studies we examined whether or not CD spectra of Zn6MT2 species differ depending on the preparation pathway. For this purpose, the CD spectrum of 10 μM thionein (apo-MT2) incubated with 6 equivalents of Zn(II) (60 μM) was recorded in the UV-range. Addition of the seventh Zn(II) ion almost does not change the spectrum in the applied UV range (Fig. S17a, ESI†). Because apo- and holo-SDH forms demonstrate intensive CD signals in the UV range, the use of apo-SDH enzyme as an acceptor of Zn(II) is not possible in this case due to very weak signals of zinc metallothionein species and possible effect alteration.44 To avoid this effect we used EGTA as an acceptor of Zn(II). This chelator has been shown in the past to compete with the weakly bound Zn(II) in human MT2.11,18 For that purpose fully loaded metallothionein was incubated with various concentrations of EGTA for 10 or 30 min and spectra were recorded. Fig. S17b (ESI†) shows that the CD spectrum of Zn7MT2 incubated with 20 μM EGTA does not differ from that of Zn6MT2 obtained by the addition of 6 eq. to apo-MT2. Only adding EDTA (∼3.5 order of magnitude tighter chelator than EGTA) causes virtual changes in the weak CD spectrum (Fig. S17b, ESI†).Discussion
Intramolecular interactions in metal-free MT2
Early structural studies on mammalian apo-MTs originally concluded that the lack of UV and CD signals specific for α-helices and β-sheets is proof of a disordered conformation with no secondary structural features,45,46 although obtained NMR resonance shifts suggested some residual, untypical hydrogen bonding network.47 Later on, molecular simulation studies confirmed that thionein retains a significant amount of structural features upon metal dissociation,48 which are probably held together via the hydrogen bonding network.49 Those observations were supported by the shift of charge state distribution observed in the ESI spectra of sequentially Zn(II)-depleted metallothionein,50 IM-MS results and molecular dynamics simulation studies.29The encountered problems with obtaining entirely alkylated apo-MT2 in our study confirm the existence of a residual, three-dimensional-like molecular scaffold. Furthermore, pairwise peak distribution observed in the MALDI spectra suggests the presence of intermolecular interactions involving thiol groups, lowering their solvent exposure, rotation or reactivity. Literature data suggest that an untypical hydrogen bonding network may be present, either involving the OH group from Ser residues which may contribute to steric hindrance of SH groups due to their close proximity or involving the SH group directly.29,47,51 Despite the lowered susceptibility to alkylation of thionein, literature data demonstrate that it is readily oxidized by a series of compounds, including DTNB used for the quantitative determination of thiol/thiolate group concentration.11,52 Molecular dynamics simulation studies revealed that thionein exists in multiple conformation states in solution, fluctuating between extended and globular-shaped final forms, with only a transient network of hydrogen bonds.29 Literature data as well as our results support the conclusion that apo-MT2 demonstrates residual structural features, which exist in a constant equilibrium with an extended, random coil form, although under the applied conditions the structurally ordered form seems to prevail.
Zn(II) binding to the α-domain of MT2 is sequential under cellular pH
Knowledge of the Zn(II) binding mechanism to metallothionein is a critical issue for the understanding of the zinc sites’ features and functions in Cys-rich zinc proteins. This is especially important for cellular zinc homeostasis and the zinc buffering mechanism, in which the presence of partially saturated species of metallothionein seems to function as a Zn(II) donor and acceptor buffering Zn(II) in the appropriate free concentration. In this study, with the combination of Zn(II)-to-protein titration, alkylation, enzymatic digestion and several mass spectrometry approaches, we were able to look deeper into the Zn(II) coordination mechanism of MT2 with a resolution that was not experimentally obtained before. However, due to high internal dynamics and heterogeneity of differentially Zn(II)-loaded forms, a multitude of information was not deciphered. In the following text one has to take into account several issues: (i) co-existence of several diversely saturated forms in each sample, (ii) high flexibility and internal dynamics of the system, and (iii) the observed peaks result from all present forms and provide overall information about molecular entities in each sample. Nonetheless, we attempted to elucidate the coordination site for each Zn0–7MT2 stoichiometry.The first binding event occurs by Zn(II) coordination to Cys36, 48 and 60 residues, with Cys33/Cys34/Cys37 completing the coordination sphere (Fig. 4a). Primary sequence motifs C57SCC60 and C33CSCC37, present in the α-domain, possess a well-established tendency for preferential metal ion binding and thermodynamic advantage of proximal cysteine ligands.4 Inspection of the MT2 sequence shows that Cys60 and Cys36 residues are a part of both motifs. It is worth noting that CXXC sequence motifs used for Zn(II) binding are the most widespread binding motifs in zinc proteins and form the most stable metal–protein complexes.53–55 Since experimental data revealed that Cys36 was involved in Zn(II) coordination, Cys33 could complete the Zn(II) coordination sphere ligand because it forms a CXXC binding motif with Cys36. Interestingly, this binding site does not resemble any of the sites denoted in crystal and NMR structures. It is also different from the putative nucleation centre of rabbit MT2 (49–61 region) for Cd(II)-induced folding.56 Also, molecular dynamics results indicated the prevalence of Cys37 over Cys33 and Cys34 to coordinate the first Zn(II) ion (Fig. S4a, ESI†). In this model, Cys33 and Cys34 remain far away from the coordination sphere (12.2 and 6.7 Å, respectively); moreover, the former interacts weakly, H-bonding with Lys20 (3 Å). This is not a surprising result for Zn1MT in which probably a residual network of hydrogen bonds maintains structural order of the α- and β-cluster, although flexibility and dynamic behaviour still occur. This indicates the need of Zn–S bond rearrangement during complexation of subsequent Zn(II) ions and confirms the high coordination dynamics of MT2 metal clusters.57–59
Binding of the second Zn(II) ion is associated with four more cysteine residues resistant to modification, which is clear evidence for the formation of two independent ZnS4 sites. The obtained results suggest the presence of two distinct cysteine groups that are alkylation protected to different extents, i.e. Cys48, 50 and 60 are highly protected, while Cys33, 34, 36 and 37 are less protected. Such a pattern suggests separate binding of two Zn(II) ions to CSCC and CCSCC motifs, respectively, accompanied by one Zn–S bond rearrangement upon incorporation of the second Zn(II) ion (Fig. 4b). The proposed bond re-formation is caused by steric hindrance in the C33CSCC37 motif, which could involve only three cysteine residues for coordination of the same Zn(II) ion.56,60 The second metal ion is coordinated by Cys50, Cys60 and possibly Cys36/37. Since Zn(II) shows preference towards CXXC, it is highly possible that the fourth ligand is thiolate of the Cys57 residue. Another possible model of the second Zn(II) ion coordination is its binding to Cys50, 57, 59 and 60, as has been shown by Munoz and co-workers.56 In such a scenario Cys37 remains free yet resistant to alkylation due to steric hindrance resulting from adjacent binding residues. Independent formation of two ZnS4 sites was also predicted by Rigby and co-workers in their molecular dynamics study on cadmium MT.61 Our experimental and simulation results are highly consistent with their simulation study, but differ in some Zn(II)–Cys coordination. While they proposed Cys33, 34, 44, and 48 for one out of two Zn(II) ions, slightly different results where Cys36 replaces Cys44 were obtained. As well as for the second Zn(II), in our model Cys36 acts as a bridge and Cys59 remains free. Furthermore, studies performed on cobalt MT showed that first 3–4 Co(II) ions are bound in independent sites, although their exact localization has not been studied in detail to date.62,63 However, the results obtained for Zn(II), Cd(II), Co(II) and other divalent metal ions should be treated with caution, as discussed below.
Analysis of the binding process of the third Zn(II) ion uncovers even higher sample heterogeneity, compared to the previous two ions. ESI spectra indicate the formation of Zn3–7MT2, which shows that α-cluster formation is not strictly cooperative since multiple metalated species are observed simultaneously (Fig. S2, ESI†). It is worth noting that signal intensities in the ESI spectrum should not be treated quantitatively because of the electrospray ionization operating principle and do not necessarily reflect the real distribution of species in the solution phase. This fact has been underlined recently by two recent reports proving that investigations of Zn(II)–peptide/protein equilibria by ESI in the gas phase are not quantitative due to zinc deposition or protein supermetalation during analysis.64,65 Nevertheless, such a dynamic picture shows that two possible Zn(II) binding pathways may occur “clustered” via cluster formation and “beaded” via formation of individual coordination sites prior to coalescence into a cluster, as was postulated in a recent study.14 On the other hand, our results strongly support the model in which initial metal ion binding takes place only in the α-domain. Obtained tryptic digest patterns for two and three Zn(II) equivalents do not show any major differences; hence it is reasonable to assume two independent ZnS4 sites in the Zn2MT2 and Zn3S9 cluster in Zn3MT2 form. Moreover, measured molecular masses observed in the ESI titration study confirm this hypothesis (Table S5, ESI†). Fig. 4c presents the possible location of three metal ions in the α-domain. However, it should be mentioned that other configurations of bridging cysteines may occur or several similar conformers may be present in the solution. In this conformation, Cys41 and Cys44 are not involved in coordination, although Cys41 interacts with Lys31 by weak H-bonding (3 Å) and remains far away from the coordination sphere (Fig. S4c, ESI†).
Recent semi-structural studies on Cd(II) formation intermediates of MT2 by ESI-IM-MS and MD simulations suggest the presence of several conformers of Cd3MT2 that differ in overall shape and total protein area, which is in good agreement with our experimental results.29 Molecular dynamics also allowed prediction of two independent CdS4 sites for Cd2MT2 and formation of a cluster for Cd3MT2, although in contrast to our stoichiometry, Zn3S9, they proposed a slightly different stoichiometry, Cd3S10.49
Eleven modifications in MT2 observed in the MALDI spectrum after addition of four Zn(II) equivalents may be attributed to nine alkylated cysteines in the β-domain and a fully formed α-cluster (Zn4αMT), proven by the obtained tryptic peptides (Table 1). Peaks with lower modification number (Fig. 3e), signals observed in the titration study (Fig. S2e, ESI†) and the obtained tryptic peptides (Table 1) all point to the existence of higher metalated species, i.e. Zn5–7MT2. The different modification numbers of cysteines in the [32–43] fragment (Table 1) and co-existence of several variously Zn(II)-depleted forms probably indicate high internal dynamics of Zn–S bonds, with their constant breakage and re-formation, as has been proposed in earlier studies.57–59,66 Another plausible explanation relates to the stability of particular metal binding sites. The fourth Zn(II) equivalent may be partially distributed amongst α- and β-domains, as was proposed in the literature,63 which could explain the observed protection of certain β-domain regions from alkylation (Table 1).
Overall, our results regarding α-domain folding allowed us to conclude that Zn(II) binding to the α-domain is moderate sequential rather cooperative. The first two Zn(II) ions bind to independent ZnS4 sites, the third forms the Zn3S9 cluster, and in the case of four Zn(II) ions the Zn4S11 cluster is formed with saturation of the α-domain. Association of the Zn(II) ion with the β-domain before complete filling of the α-domain shows that there is no significant difference in the affinity of the weakest site in the α-domain and the strongest in the β-domain. This is in good agreement with a pH titration study performed on Zn7MT2, where one clear absorbance increase isotherm around pH 4–5 is observed.9,30 Analogous pH-titration of Cd7MT2 shows a significant difference between α- and β-domains, which is observed as two clearly separated absorbance increase steps.9 Very early studies showed that the α-domain saturated with Cd(II) is much more stable than the β-domain,26 which was more carefully analysed in recent years. Formation and dissociation of Cd7MT2 performed by bottom-up and top-down ion-mobility mass spectrometry showed that Cd4MT2 species are highly stable and all four Cd(II) ions are located in the α-domain.67,68 A recent pH-variable study on MT1a supported the observation of Cd4MT1a high stability even at low pH.14 The same study suggested that the Zn(II) binding process is significantly less cooperative and the Zn4MT1a form is not as stable as the Cd(II) counterpart. Overall, our and the above-mentioned studies confirm the differences in Cd(II) and Zn(II) affinities towards the two respective domains in whole metallothionein.
Zn(II)-Dependent folding of the isolated β-domain differs from that of whole MT2
Metal-induced β-domain folding should be expected to resemble the α-domain folding in terms of the formation of independent and clustered sites. However, their sequence motifs differ significantly from each other. Instead of the CXCC motifs of the α-domain, there are four CXC and two CXXC motifs in the β-domain sequence. The crystal structure of hepatic rat MT2 indicates a type-I reverse turn-like conformation with an internal NH⋯Sγ hydrogen bond for all CXC motifs. According to Stout and co-workers, this type of hydrogen bonding is expected to stabilize the local conformation in apo-MT2 and thereby facilitate metal ion binding.4 Indeed, as discussed above, three cysteine residues from region 1–20 are recalcitrant to modification after addition of four Zn(II) equivalents. This fragment contains two CXC motifs and indicates quite similar affinity to the weakest binding site in the α-domain.Addition of a fifth molar equivalent to apo-MT2 sheds more light on the first Zn(II) ion bound in the β-domain due to the lack of resolution and high similarities with the previous titration step in all experimental pathways (Table 1, Fig. 3e, f and Fig. S2e, f, ESI†). Most likely this ion is bound by three thiolate ligands from the 1–20 region with one from 26–31, the exact localization of which can be assumed based on the third ion in the whole cluster (Fig. 1b and 4e). Molecular dynamics simulations performed on partially saturated cadmium metallothionein showed that the first metal ion in the β-domain occupies the site formed by Cys7, 13, 15 and 26, which agrees with our experimental and MD findings (Fig. 4e and 5a) and coordination mode in the 3D structure (Fig. 1).61 Comparison of the α-domain between the X-ray MT2 structure and MD results shows high structural similarity, which is a sign of a stable α-domain. The defined Zn(II) binding site in the β-domain observed after addition of five Zn(II) equivalents causes a decrease in coordination dynamics in 32–43 and 44–51 regions in the α-domain. It may be partially explained by an equilibrium shift of a metal ion from Zn3αZn1βMT2 to Zn4αZn0βMT2 species, which again indicates similar Zn(II) affinities of both sites and high coordination dynamics of the weakest Zn(II) ion in the β-domain. It should be noted that pronounced β-domain tryptic fragments also demonstrate a fully protected form (Table 1), and the signal intensity of the unmodified full protein in the MALDI spectrum is much higher in comparison with the previous titration step, which is a sign of co-existence of Zn6MT2 and Zn7MT2 species (Fig. 3e, f and Fig. S2f, ESI†). The high internal dynamics of Zn–S bonds and/or the presence of several partially Zn(II)-depleted MT species is maintained.
Although clustering is one possibility in handling the second Zn(II) ion in the β-domain, our results obtained for MT2 with six Zn(II) equivalents show that the formation of another separate binding ZnS4 site is much more preferred (Fig. 5b). Full protection from modification of all identified tryptic fragments (Table 1) indicates that at least seven cysteines from the β-domain are involved in Zn(II) binding. Therefore, the last cysteine that fills the coordination sphere should be either Cys21 or Cys24. Single modification of Cys21 in tryptic fragment [1–22] detected in the case of seven Zn(II) equivalents (Fig. 6) is strong evidence that the second metal ion in the β-domain is bound to the Cys24 residue. Moreover, molecular dynamics simulations strongly support this experimental result, as the Zn(II) is coordinated by Cys24 while Cys21 remains free (Fig. 5b). It should be emphasized that the employed molecular dynamics methodology makes use of a semi-bonded approach, which in contrast to the bonded approach, is able to simulate coordination changes and the ligand exchange phenomenon. Moreover, H-bonding stabilization of the non-coordinated Cys21 residue by Ser6 is observed. Localization of Zn(II) ions in the β-domain presented in Fig. 4f is based on the structure and coordination bond networks in the full cluster. Our results therefore show that the Cys21 residue more likely is the only one free cysteine in Zn6MT species. Utilization of the maximum possible number of cysteine residues in two separate binding sites in the β-domain is in agreement with previous studies indicating that there are virtually no free cysteines in Zn6MT2 species.11 This indicates that clustering does not occur until the last step of domain formation. Coordination of the last Zn(II) ion takes place by metal ion binding to Cys21 and subsequently to Cys5, 7 and 24. This binding mode causes rearrangement in the existing Zn–S bond network. Sulfur donors of Cys7, 15 and 24 become new bridging ligands. A similar conclusion came from differential modification of cadmium MT by radioactive IAA in a very early study.26,27 The authors postulated that the seventh metal ion is bound to the 20–30 region, but they did not obtain single Cys resolution. Our results demonstrate one more important feature of the last binding event in the β-domain, namely its thermodynamic difference from the other two binding sites. Full protection from alkylation should be observed for Zn7MT2 if binding sites were comparable, which is not the case (Fig. 3g and h). The singly modified peak observed for Zn7MT2 (Fig. 3h and 6) indicates partial dissociation of the last Zn(II) ion under alkylating conditions. Interestingly, Bernhard's report states that protection from modification of the N-terminal region of the β-domain causes distinctly lower affinity of the central region 20–30 for the metal ion.26,27 By identification of particular metal sites and lastly modified cysteine we may extend that conclusion forward. Binding of the first two Zn(II) ions to independent sites located on the opposite tails of the β-domain lowers the protein affinity towards the third metal ion, probably due to the significant energy-requiring conformational changes needed for Zn–S rearrangement and Zn3S9 cluster formation. Interestingly, results on a Zn(II) transfer process from Zn7MT2 to apo-SDH showed that dissociation of the first Zn(II) ion from the fully Zn(II)-loaded protein is structurally reversible to association of the last (seventh) Zn(II) ion to Zn6MT2 species. Such a mechanism is not obvious because various energetic minima can be reached during association or dissociation of a metal ion. The same modification scheme observed here for the reaction forward and back indicates that the same Zn6MT2 species is preferentially formed, which indicates its thermodynamic stability. In this case, the energetic cost related to the seventh Zn(II) ion binding is comparable with the energetic cost of its dissociation from Zn7MT2. Studies performed on Zn(II) transfer from Zn7MT2 to PTP1B or fluorescent probes FluoZin-3 and Rhod-Zin-3, which bind Zn(II) with Kd ∼ 10−8 M show that −log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kd1 of MT2 is the same as log
Kd1 of MT2 is the same as log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb7 strongly suggesting that this process is reversible.11,18 Moreover, the CD spectra recorded for Zn6MT2 species obtained by association of 6 Zn(II) ions to apo-MT2 or dissociation of the seventh Zn(II) from Zn7MT2 are identical and are not very different from the spectrum recorded for fully Zn(II)-loaded protein, which indicates that dissociation and association of weakly bound Zn(II) does not influence significantly the secondary structure of metallothionein, while being highly disordered.
Kb7 strongly suggesting that this process is reversible.11,18 Moreover, the CD spectra recorded for Zn6MT2 species obtained by association of 6 Zn(II) ions to apo-MT2 or dissociation of the seventh Zn(II) from Zn7MT2 are identical and are not very different from the spectrum recorded for fully Zn(II)-loaded protein, which indicates that dissociation and association of weakly bound Zn(II) does not influence significantly the secondary structure of metallothionein, while being highly disordered.
The isolated β-domain demonstrates a more distinct Zn(II) coordination pathway than this domain in the whole MT2 protein. The first Zn(II) ion is bound rather weakly and probably delocalized throughout the whole β-domain peptide, as the MALDI spectrum shows 6 and 7 modifications (Fig. S13b, ESI†). Based on all the obtained results, it is reasonable to assume two, not mutually exclusive, possibilities: (i) all four Zn0–3βMT2 species are present in dynamic equilibrium, with prevalence of Zn1βMT2, and (ii) distribution of the first Zn(II) throughout all binding sites, with higher appearance frequency in the N-terminal part of the peptide. The obtained MS/MS results confirmed the presence and localization of the Zn1βMT2 form and localized its binding to Cys5, 7, 13 and 15. The MD results were in agreement with the experimental results, and showed that Cys5 was H-bonding stabilized by Lys22, in contrast to Zn5MT2, where Zn(II) bound in the β-domain resulted in Cys5 free coordination (Fig. S14b, ESI†). Therefore, the data obtained for the isolated domain are contradictory to those obtained for the whole protein and do not correspond to any site in the 3D structure (Fig. 1b).
Addition of the second Zn(II) equivalent to the isolated β-domain caused a significant equilibrium shift towards Zn2βMT2 and Zn3βMT2 species, but the results did not allow the Zn(II) position to be sharply elucidated in the Zn2βMT2 form. However, there is indirect evidence for the formation of a Zn2S7–8 form, with one modified Cys from the 1–20 region and possibly another one from the 25–31 region (Table S8, ESI†). Unique behaviour of the Cys29 residue was postulated by Babu and co-workers in a study on Zn(II) release from the isolated β-domain of rat MT2.69 They proposed that Cys19 and Cys29 release Zn(II) faster than any other cysteine residues with concurrent formation of a Cys19–Cys29 disulfide bridge. Interestingly, those residues are exposed to the solvent in the isolated β-domain, but are buried in the whole protein MT2.67 Our molecular dynamics indicated that Cys5 and Cys29 are free of coordination (Fig. S14c, ESI†) in which the first one interacts with Gln23 via H-bonding (2.6 Å).
Lack of any modifications after addition of the third Zn(II) equivalent to apo-βMT2 demonstrates a more stable cluster in the isolated β-domain than in the whole protein and points to the conclusion that the isolated domain is not a good model of the whole protein. The single peak in the MALDI spectrum corresponding to the unmodified β-domain (Fig. S13d, ESI†) points to the absence of weakly coordinating cysteine and supports our statement. Lower stability of the seventh Zn(II) site in metallothioneins and the difference between isolated domains and the whole protein molecule have been shown several times in the past.11,12,18,70,71 For example, the higher affinity of Zn(II) ions in the isolated β-domain was confirmed in its competition with PAR (4-(2-pyridylazo)resorcinol).70 The most likely reason for such behaviour is higher main chain flexibility due to lack of stabilizing interdomain contacts, and lack of possible conformational restraints due to the presence of the α-domain.
The mechanism of Zn(II) and Cd(II) binding to metallothionein is entirely different
To date, our knowledge regarding the structure, chemical and biophysical properties of metallothioneins has been mostly based on studies performed with cadmium metallothionein. Historically, it is associated with the fact that metallothionein was discovered in equine kidney cortex as a cadmium protein.1 Another reason why the vast majority of metallothionein studies use this ion is that Cd(II) produces well-defined spectroscopic signals, in contrast to spectroscopically silent Zn(II) ions. Usage of Cd(II) ions allows one to study association, thermodynamic and kinetic properties, as well as the complex structure of metallothionein. However, it is now well known that metallothioneins are Zn(II) and Cu(I) metalloproteins in healthy living organisms.72–76 Comparison of NMR data obtained for zinc and cadmium metallothioneins in the early 1990s shows that both molecules are structurally similar in terms of cluster geometry, metal–sulfur bonds and overall shape, although some differences in intraprotein connectivities and protein volume were detected.77,78 Differences between these metal ions include (i) their ionic radii (109 pm for Cd(II) and 88 pm for Zn(II)), (ii) hard and soft acid–base properties – Cd(II) is a soft Lewis acid while Zn(II) is considered borderline,79 and (iii) formed M–S coordination bond lengths (averages between terminal and bridged metal centres: Cd–S ∼ 2.5 Å, Zn–S ∼ 2.3 Å).80,81To date, there are a limited number of reports focusing on the biophysical properties of zinc metallothioneins, although it is the most abundant MT form in mammals.76 Various physicochemical studies including CD, MCD, UV-Vis, and NMR spectroscopy on mammalian metallothioneins show that Cd(II) binds more tightly than Zn(II) ions, which is well understood when considering the enthalpy of M–S bond formation and highly similar structure.9,12 A number of spectroscopic studies have demonstrated that the Cd(II) binding affinity in the α-domain (Kd ∼ 10−15 M) is almost three orders of magnitude higher than in the β-domain (Kd ∼ 10−12–10−13 M).9,82–84 Contrastingly, many reports state that all seven Zn(II) ions have identical or almost identical protein affinity and both clusters are basically indistinguishable in terms of their stability, with an approximate dissociation constant of ∼10−12 M (for MT1 and MT2).9,10,24,83,85 Several recent studies on MT2 and MT3, however, demonstrated otherwise, namely that one Zn(II) ion is bound relatively weakly with Kd ∼ 10−8–10−9 M, two other ions are bound with medium affinity with Kd ∼ 10−10–10−11 M, and the remaining four ions are bound with indistinguishable high affinity of Kd ∼ 10−12 M.11,12 These discrepancies in the measured affinities of Zn(II) ions to metallothioneins are a current subject of debate.12,76,86
The results presented in this article support a model with varied Zn(II) affinities towards MT2. Binding of Zn(II) to the α-domain is sequential, whilst Cd(II) seems to be cooperative, as it is known from the literature. Observed 113Cd or 111Cd NMR signals corresponding to α-cluster formation in NMR titration studies indicate identical affinities.25,87,88 On the other hand, corresponding ESI-MS titration studies showed that intermediate forms (Cd1–3MT2) are also formed; however, Cd4MT2 is highly favourable.29,67 It is worth noting that exact identification of successive Cd(II) binding events and sites in the α-domain has not been performed to date and such studies could possibly reveal differences or the lack of differences in Cd(II) affinities toward the α-domain. Data collected to date show that both metal ions bind to the α-domain specifically with high affinity, but Zn(II) binding is more sequential.
While the binding of Zn(II) and Cd(II) to the α-domain is only slightly different, binding of these two ions to the β-domain is much more divergent. The most visible difference besides the fact that Cd(II) ions are bound more tightly than Zn(II) ions is the clear differentiation of Zn(II) affinities in the β-domain compared to Cd(II).89 This phenomenon is observed in pH-dependent binding of both metal ions to metallothioneins regardless of protein isoforms.9,25,82–84113Cd-NMR studies performed on whole protein show that Cd(II) binding to the β-domain is cooperative, although the coordination dynamics of the cluster is much more pronounced compared to the α-cluster.25 However, pH titrations clearly show that the slope of Cd(II) binding to the β-domain is significantly lower compared to the α-domain. This is due to a lower number of protons released during coordination of three ions. It also reflects more pronounced differentiation of Cd(II) affinities towards the β-domain than the α-domain. Such differentiation was observed for MT484 and is less pronounced for MT381 and least visible for MT1 and MT2 isoforms.9,82,83 This shows that the relative affinities of metal ions in the β-domain vary depending on the metallothionein isoform. It should be noted that several reports in the past also reported the presence of weaker bound Cd(II) ions in MTs. These include differential modification of cadmium MT using IAA,26,27 NMR studies on Cd(II) release from MT by EDTA85,87 and ICP-MS studies on the displacement of zinc MT with Cd(II) ions.90
Our results on the mechanism of Zn(II) association with MT2 also shed new light on Zn(II)/Cd(II) exchange in this protein. The addition of Cd(II) or Cd7MT to Zn7MT causes preferential displacement of Zn(II) ions in the α-domain and 113Cd-NMR studies show that this process is highly cooperative.91,92 If the affinity of the remaining three Zn(II) ions in the β-domain were close to each other, then Zn(II)/Cd(II) exchange should indicate cooperative character. Despite this, mixed Zn/CdMT complexes are preferentially formed in vivo and in vitro, although their compositions differ. Such formation is possible when Zn(II) ion affinities towards their sites in the β-domain vary significantly, which is the conclusion from our study. The crystal structure (Fig. 1) shows localization of Cd(II) ions in the β-domain in position IV, which indicates that the exchange of the fifth Zn(II) ion occurs exactly in this position, where the metal is bound by bridging Cys7 and Cys24 residues and terminal Cys21 and Cys5. It is reasonable that the fifth equivalent of Cd(II) displaces the weakest bound Zn(II) ion in MT. The pathway of Zn(II) metalation (Fig. 4) identified in the course of this study shows that Cys21 is the residue that more likely terminates cluster formation and is bound to the weakest Zn(II) in the β-domain. A similar conclusion comes from the competition between Zn7MT2 and apo-SDH, for which Cys21 and Cys24 residues were identified to be the weakest bound to the metal ion (Fig. 8). It should be mentioned that the composition of metal clusters in MT is believed to be a consequence of metal ion selectivity in the protein, according to which the major factor governing the cluster type is the protein structure perturbation due to the cluster volume variations. Vašák and co-workers state that metal–thiolate affinity is important in the folding process, while size-match selectivity is the dominant factor in the metal-loaded protein.93 In fact, the cluster composition strictly depends on the mechanism of the formation and conditions under which it is formed. The pathway of its formation is associated with the substrates’ stoichiometry, reaction kinetics and thermodynamics. Overall, our results give a new, higher resolution picture of Zn(II)/Cd(II) exchange and metal binding processes.
Buffering properties of metallothionein are related to sequential binding of Zn(II) and the presence of Zn(II)-depleted species
Data presented here show that although huge efforts have been made to understand the Zn(II) binding pathway in MT2, it is not fully defined, leaving the possibility for the existence of several species with the same number of metal ions per molecule. Moreover, the metal binding process may depend on several factors such as protein concentration or slightly different pH.14,94 However, our results are the first ones obtained with such a resolution and focused on biologically relevant zinc metallothionein-2. Sequential binding of Zn(II) in the β-domain supports the idea of Zn(II) affinity differentiation, pointing out the different biological function of the β-domain compared to the α-domain, which has been indicated based on MT evolution.95 According to this, the α-domain seems to function as a storage domain, and the metal binding process is highly efficient and occurs with similar affinity of Zn(II) ions. However, the data obtained so far show that Cd(II) binding to this domain is even more efficient, which is reasonable when we consider the major role of this domain in cellular detoxification. The β-domain plays another, regulatory function according to which it distributes Zn(II) among other Zn(II)-dependent or regulated proteins. Variously lowered affinities of Zn(II) ions bound in the β-domain place it either as a Zn(II) donor for other metal-depleted proteins or as an acceptor which allows binding of excess Zn(II) when it exists in partially saturated species. Cellular free Zn(II) changes and dissociation constants of Zn(II) ions bound in the β-domain (from 10−8 to 10−11 M) match each other very well, which allows metallothionein to be considered as a major component of the cellular zinc buffer.11 It is even more understandable when one takes into account the fact that this protein is easily inducible under several stimuli such as metal ions, glucocorticoids, and oxidative stress.96–98 Therefore, induction of metallothionein guarantees the binding of excess metal (biogenic and toxic) ions in such a way that the β-domain is still able to function as a Zn(II) regulatory module.The Zn(II)-binding pathway in metallothionein-2 proposed here is a relation between the structural properties of the Zn(II)-depleted species and the buffering properties of this protein. So far, our knowledge regarding partially depleted forms of MT is very limited, besides some information that the β-domain binds Zn(II) less efficiently than the α-domain. On the other hand, some reports stated that although Zn(II) binding is sequential, the affinities of particular Zn(II) ions almost do not differ.24 It is, however, in contrast with other experiments, where MT functions as a Zn(II) donor towards metal-depleted enzymes, and inhibitory and structural sites in proteins.11,12,18,20,52,70,99 By the combination of the high-resolution Zn(II)-binding process determined here together with molecular dynamics simulations and thermodynamic studies performed so far, we are able to describe structures of partially saturated species of zinc metallothionein for the first time. One needs to note that normal concentrations of freely available Zn(II) support the existence of Zn5MT2 and Zn6MT2 species.11 Therefore the understanding of this process is not only important for exploration of the structural nature of this enigmatic protein, but also critical for understanding the principles of other metallothionein functions, which we are still trying to understand 60 years after its discovery.76
Conclusions
In this study, by the combination of mass spectrometry approaches to partially saturated and alkylated human metallothionein-2 together with molecular dynamics study, we analysed the Zn(II) binding process with high resolution, which allowed us to propose the structure of Zn1–6MT species with respect to metal ions’ localization and their coordination spheres. The results show that all metallothionein species are highly dynamic even if they are clustered and the proposed structures could be a representation of one possibility, however the most prevalent under the used conditions. Our data show that the four Zn(II) ions bind preferentially to the α-domain when they are added to the apo-MT2 form and then fill the β-domain. However, this process is much less distinct than in the case of cadmium MT, which shows some thermodynamic similarities in coordination with the fourth and fifth Zn(II) ion. The binding of all Zn(II) ions is sequential, with high preference for the formation of independent ZnS4 sites in early stages of domain formation. However, the Zn(II) binding process in both domains differs in later stages of cluster formation, indicating more distinct Zn–S network rearrangement in the β-domain upon binding of the seventh Zn(II) ion. This process is interpreted as an energetic reason for significantly lower affinity of the last Zn(II) ion in the whole domain, which is in contrast to the Zn(II) binding pathway in the isolated β-domain. Our results have shown that Cys21 is more likely the last coordinating cysteine residue, which demonstrates that weakly bound Zn(II) in the β-domain corresponds to position IV (Fig. 1b). The same conclusion is reached when MT2 serves as a Zn(II) donor and supports a catalytic cofactor to Zn(II)-depleted sorbitol dehydrogenase. Then the same Cys21 residue is modified first, which shows that the Zn(II) binding and dissociation process of the weakest Zn(II) ion is fully reversible, which has also been demonstrated by fluorescent zinc probes in a previous investigation.11 The presented study sheds more light on the mechanism of zinc metallothionein-2 cluster formation with the highest resolution to date and should be useful for the investigation of metal binding and dissociation processes in other isoforms and metallothioneins from other organisms. By the comparison of various MS approaches with MD calculations, and Zn(II) release and binding equilibria in water media with other structural and thermodynamic data we provided a new opportunity to look into the function of various metallothionein proteins with high indication of the role and importance of partially or differently metal-loaded species, in which interest is constantly growing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Science Centre of Poland (NCN) under the Sonata Bis grant no. 2012/07/E/NZ1/01894. The authors thank the Wroclaw Centre of Biotechnology, Leading National Research Centre (KNOW) program for the years 2014–2018 and Wrocław Centre for Networking and Supercomputing from Wrocław University of Technology for support of open access publication and access to computational resources, respectively. The authors thank also Dr Adam Pomorski for help in the CD experiment.References
- M. Margoshes and B. L. Vallee, A cadmium protein from equine kidney cortex, J. Am. Chem. Soc., 1957, 79, 4813–4814 CrossRef CAS.
- Y. Uchida, K. Takio, K. Titani, Y. Ihara and M. Tomonaga, The growth inhibitory factor that is deficient in Alzheimer's disease brain is 68 amino acid metallothionein-like protein, Neuron, 1991, 7, 337–347 CrossRef CAS PubMed.
- C. J. Quaife, S. D. Findley, J. C. Erickson, G. J. Froelick, E. J. Kelly, B. P. Zambrowicz and R. D. Palmiter, Induction of new metallothionein isoform (MT-IV) occurs during differentiation of stratified squamous epithelia, Biochemistry, 1994, 33, 7250–7259 CrossRef CAS PubMed.
- A. H. Robbins, D. E. McRee, M. Williamson, S. A. Collett, N. H. Xuong, W. F. Furey, B. C. Wang and C. D. Stout, Refined crystal structure of Cd, Zn metallothionein at 2.0 Å resolution, J. Mol. Biol., 1991, 221, 1269–1293 CAS.
- B. A. Messerle, A. Schäffer, M. Vašák, J. H. R. Kägi and K. Wüthrich, Three-dimensional structure of human [113Cd7] metallothionein-2 in solution determined by nuclear magnetic resonance spectroscopy, J. Mol. Biol., 1990, 214, 765–779 CrossRef CAS PubMed.
- P. Schultze, E. Wörgötter, W. Braun, G. Wagner, M. Vašák, J. H. Kägi and K. Wüthrich, Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy, J. Mol. Biol., 1988, 203, 251–268 CrossRef CAS PubMed.
- A. Arseniev, P. Schultze, E. Wörgötter, W. Braun, G. Wagner, M. Vašák, J. H. Kägi and K. Wüthrich, Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance, J. Mol. Biol., 1988, 201, 637–657 CrossRef CAS PubMed.
- K. Zangger, G. Oz, J. D. Otvos and I. M. Armitage, Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy, Protein Sci., 1999, 8, 2630–2638 CrossRef CAS PubMed.
- M. Vašák and J. H. R. Kägi, in Metal Ions in Biological Systems, ed. H. Sigel, Marcel Dekker, New York, 1983, vol. 15, pp. 213–273 Search PubMed.
- J. D. Otvos, D. H. Petering and C. F. Shaw, Structure-reactivity relationships of metallothionein, a unique metal-binding protein, Comments Inorg. Chem., 1989, 9, 1–35 CrossRef CAS.
- A. Krężel and W. Maret, Dual nanomolar and picomolar Zn(II) binding properties of metallothionein, J. Am. Chem. Soc., 2007, 129, 10911–10921 CrossRef PubMed.
- M. C. Carpenter, A. S. Shah, S. DeSilva, A. Gleaton, A. Su., B. Goundie, M. L. Croteau, M. J. Stevenson, D. E. Wilcox and R. N. Austin, Thermodynamics of Pb(II) and Zn(II) binding to MT-3, a neurologically important metallothionein, Metallomics, 2016, 8, 605–617 RSC.
- P. M. Gehrig, C. You, R. Dallinger, C. Gruber, M. Brouwer, J. H. Kägi and P. E. Hunziker, Electrospray ionization mass spectrometry of zinc, cadmium, and copper metallothioneins: evidence for metal-binding cooperativity, Protein Sci., 2000, 9, 395–402 CrossRef CAS PubMed.
- G. W. Irvine, T. B. J. Pinter and M. J. Stillman, Defining the metal binding pathways of human metallothionein 1a: balancing zinc availability and cadmium seclusion, Metallomics, 2016, 8, 71–81 RSC.
- A. Krężel and W. Maret, Zinc-buffering capacity of a eukaryotic cell at physiological pZn, J. Biol. Inorg. Chem., 2006, 11, 1049–1062 CrossRef PubMed.
- A. Krężel, Q. Hao and W. Maret, The zinc/thiolate redox biochemistry of metallothionein and the control of zinc ion fluctuations in cell signaling, Arch. Biochem. Biophys., 2007, 463, 188–200 CrossRef PubMed.
- Y. Yang, W. Maret and B. L. Vallee, Differential fluorescence labeling of cysteinyl clusters uncovers high tissue levels of thionein, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 5556–5559 CrossRef CAS PubMed.
- A. Krężel and W. Maret, Thionein/metallothionein control Zn(II) availability and the activity of enzymes, J. Biol. Inorg. Chem., 2008, 13, 401–409 CrossRef PubMed.
- K. Kluska, J. Adamczyk and A. Krężel, Metal binding properties of zinc fingers with a naturally altered metal binding site, Metallomics, 2018, 10, 248–263 RSC.
- T. Kochańczyk, P. Jakimowicz and A. Krężel, Femtomolar Zn(II) affinity of minimal zinc hook peptides – a promising small tag for protein engineering, Chem. Commun., 2013, 49, 1312–1314 RSC.
- G. Roesijadi, R. Bogumil, M. Vašák and J. H. R. Kägi, Modulation of DNA binding of a tramtrack zinc finger peptide by the metallothionein-thionein conjugate pair, J. Biol. Chem., 1988, 273, 17425–17432 CrossRef.
- B. J. Murphy, T. Kimura, B. G. Sato, Y. Shi and G. K. Andrews, Metallothionein induction by hypoxia involves cooperative interactions between metal-responsive transcription factor-1 and hypoxia-inducible transcription factor-1alpha, Mol. Cancer Res., 2008, 6, 483–490 CrossRef CAS PubMed.
- J. H. Laity and G. K. Andrews, Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1), Arch. Biochem. Biophys., 2007, 15, 201–210 CrossRef PubMed.
- T. B. Pinter and M. J. Stillman, The zinc balance: competitive zinc metalation of carbonic anhydrase and metallathionein 1A, Biochemistry, 2014, 53, 6276–6285 CrossRef CAS PubMed.
- M. Good, R. Hollenstein, P. J. Sadler and M. Vašák, 113Cd NMR studies on metal-thiolate cluster formation in rabbit Cd(II)-metallothionein: evidence for pH dependence, Biochemistry, 1988, 27, 7163–7166 CrossRef CAS PubMed.
- W. R. Bernhard, M. Vašák and J. H. R. Kägi, Cadmium binding and metal cluster formation in metallothionein: a differential modification study, Biochemistry, 1986, 25, 1975–1980 CrossRef CAS PubMed.
- W. R. Bernhard, Differential modification of metallothionein with iodoacetamide, Methods Enzymol., 1991, 205, 426–433 CAS.
- J. Zaia, D. Fabris, D. Wei, R. L. Karpel and C. Fenselau, Monitoring metal ion flux in reactions of metallothionein and drug-modified metallothionein by electrospray mass spectrometry, Protein Sci., 1998, 7, 2398–2404 CrossRef CAS PubMed.
- S.-H. Chen, L. Chen and D. H. Russell, Metal-induced conformational changes of human metallothionein-2A: a combined theoretical and experimental study of metal-free and partially metalated intermediates, J. Am. Chem. Soc., 2014, 136, 9499–9508 CrossRef CAS PubMed.
- S. Hong, M. Toyama, W. Maret and Y. Murooka, High yield expression and single step purification of human thionein/metallothionein, Protein Expression Purif., 2001, 21, 243–250 CrossRef CAS PubMed.
- N. E. Wezynfeld, E. Stefaniak, K. Stachucy, A. Drozd, D. Płonka, S. C. Drew, A. Krężel and W. Bal, Resistance of Cu(Aβ4–16) to copper capture by metallothionein-3 supports a function for the Aβ4–42 peptide as synaptic CuII scavenger, Angew. Chem., Int. Ed., 2016, 55, 8235–8238 CrossRef CAS PubMed.
- G. B. Fields, Solid-Phase Peptide Synthesis, Methods Enzymol., 1997, 289, 3–780 Search PubMed.
- A. Miłoch and A. Krężel, Metal binding properties of the zinc finger metallome – insights into variations in stability, Metallomics, 2014, 6, 2015–2024 RSC.
- A. Krężel, R. Latajka, G. D. Bujacz and W. Bal, Coordination properties of tris(2-carboxyethyl)phosphine, a newly introduced thiol reductant, and its oxide, Inorg. Chem., 2003, 42, 1994–2003 CrossRef PubMed.
- P. Eyer, F. Worek, D. Kiderlen, G. Sinko, A. Stuglin, V. Simeon-Rudolf and E. Reiner, Molar absorption coefficients for the reduced Ellman reagent: reassessment, Anal. Biochem., 2003, 312, 224–227 CrossRef CAS PubMed.
- A. Kocyła, A. Pomorski and A. Krężel, Molar absorption coefficients and stability constants of metal complexes of 4-(2-pyridylazo)resorcinol (PAR): revisiting common chelating probe for the study of metalloproteins, J. Inorg. Biochem., 2015, 152, 82–92 CrossRef PubMed.
- W. Maret and D. S. Auld, Purification and characterization of human liver sorbitol dehydrogenase, Biochemistry, 1988, 27, 1622–1628 CrossRef CAS PubMed.
- D. A. Case, D. S. Cerutti, T. E. Cheatham, III, T. A. Darden, R. E. Duke, T. J. Giese, H. Gohlke, A. W. Goetz, D. Greene, N. Homeyer, S. Izadi, A. Kovalenko, T. S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, D. Mermelstein, K. M. Merz, G. Monard, H. Nguyen, I. Omelyan, A. Onufriev, F. Pan, R. Qi, D. R. Roe, A. Roitberg, C. Sagui, C. L. Simmerling, W. M. Botello-Smith, J. Swails, R. C. Walker, J. Wang, R. M. Wolf, X. Wu, L. Xiao, D. M. York and P. A. Kollman, AMBER 2017, University of California, San Francisco, 2017 Search PubMed.
- Y.-P. Pang, Successful molecular dynamics simulation of two zinc complexes bridged by a hydroxide in phosphotriesterase using the cationic dummy atom method, Proteins, 2001, 45, 183–189 CrossRef CAS PubMed.
- E. Lindahl, B. Hess and D. van der Spoel, GROMACS 3.0: a package for molecular simulation and trajectory analysis, J. Mol. Model., 2001, 7, 306–317 CrossRef CAS.
- D. C. Bas, D. M. Rogers and J. H. Jensen, Very fast prediction and rationalization of pKa values for protein-ligand complexes, Proteins, 2008, 73, 765–783 CrossRef CAS PubMed.
- E. Awoonor-Williams and C. N. Rowley, Evaluation of methods for the calculation of the pKa of cysteine residues in proteins, J. Chem. Theory Comput., 2016, 12, 4662–4673 CrossRef CAS PubMed.
- Y. M. Lee and C. Lim, Factors controlling the reactivity of zinc finger cores, J. Am. Chem. Soc., 2011, 133, 8691–8703 CrossRef CAS PubMed.
- E. Artells, O. Palacios, M. Capdevila and S. Atrian, Mammalian MT1 and MT2 metallothioneins differ in their metal biding abilities, Metallomics, 2013, 5, 1397–1410 RSC.
- M. J. Stillman, Metallothioneins, Coord. Chem. Rev., 1995, 144, 461–511 CrossRef CAS.
- Metallothioneins: synthesis, structure and properties of metallothioneins, phytochelatins and metal-thiolate complexes, ed. M. J. Stillman, C. F. Shaw III and K. I. Suzuki, VCH Publishers Inc., New York, 1992 Search PubMed.
- M. Vašák, A. Galdes, H. A. O. Hill, J. H. R. Kägi, I. Bremner and B. W. Young, Investigation of the structure of metallothioneins by proton nuclear magnetic resonance spectroscopy, Biochemistry, 1980, 19, 416–425 CrossRef.
- K. E. Rigby and M. J. Stillman, Structural studies of metal-free metallothionein, Biochem. Biophys. Res. Commun., 2004, 325, 1271–1278 CrossRef CAS PubMed.
- K. E. Rigby Duncan and M. J. Stillman, Metal-dependent protein folding: Metallation of metallothionein, J. Inorg. Biochem., 2006, 100, 2101–2107 CrossRef CAS PubMed.
- M. E. Merrifield, Z. Huang, P. Kille and M. J. Stillman, Copper speciation in the α and β domains of recombinant human metallothionein by electrospray ionization mass spectrometry, J. Inorg. Biochem., 2002, 88, 153–172 CrossRef CAS PubMed.
- N. Romero-Isart, B. Oliva and B. M. Vašák, Influence of NH – Sγ bonding interactions on the structure and dynamics of metallothioneins, J. Mol. Model., 2010, 16, 387–394 CrossRef CAS PubMed.
- C. Jacob, W. Maret and B. L. Vallee, Control of zinc transfer between thionein, metallothionein, and zinc proteins, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 3489–3494 CrossRef CAS.
- C. Andreini, I. Bertini and G. Cavallaro, Minimal functional sites allow a classification of zinc sites in proteins, PLoS One, 2011, 6, e26325 CAS.
- T. Kochańczyk, M. Nowakowski, D. Wojewska, A. Kocyła, A. Ejchart, W. Koźmiński and A. Krężel, Metal-coupled folding as the driving force for the extreme stability of Rad50 zinc hook dimer assembly, Sci. Rep., 2016, 6, 36346 CrossRef PubMed.
- T. Kochańczyk, A. Drozd and A. Krężel, Relationship between the architecture of zinc coordination and zinc binding affinity in proteins – insights into zinc regulation, Metallomics, 2015, 7, 244–257 RSC.
- A. Munoz, F. Laib, D. H. Petering and C. F. Shaw III, Characterization of the cadmium complex of peptide 49-61: a putative nucleation center for cadmium-induced folding in rabbit liver metallothionein IIA, J. Biol. Inorg. Chem., 1999, 4, 495–507 CrossRef CAS PubMed.
- P. Faller, D. W. Hasler, O. Zerbe, S. Klauser, D. R. Winge and M. Vašák, Evidence for a dynamic structure of human neuronal growth inhibitory factor and for major rearrangements of its metal–thiolate clusters, Biochemistry, 1999, 38, 10158–10167 CrossRef CAS PubMed.
- M. Vašák, G. E. Hawkes, J. K. Nicholson and P. J. Sadler, 113Cd NMR studies of reconstituted seven-cadmium metallothionein: evidence for structural flexibility, Biochemistry, 1985, 24, 740–747 CrossRef.
- W. Maret, K. S. Larsen and B. L. Vallee, Coordination dynamics of biological zinc “clusters” in metallothioneins and in the DNA-binding domain of the transciption factor Gal4, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 2233–2237 CrossRef CAS.
- A. Pomorski, J. Otlewski and A. Krężel, The high ZnII affinity of the tetracysteine tag affects its fluorescent labeling with biarsenicals, ChemBioChem, 2010, 11, 1214–1218 CrossRef CAS PubMed.
- K. E. Rigby, J. Chan, J. Mackie and M. J. Stillman, Molecular dynamics study on the folding and metallation of the individual domains of metallothionien, Proteins: Struct., Funct., Bioinf., 2006, 62, 159–172 CrossRef CAS PubMed.
- J. W. Ejnik, J. Robinson, J. Zhu, H. Försterling, C. F. Shaw III and D. H. Petering, Folding pathway of metallothionein induced by Zn2+, Cd2+ and Co2, J. Inorg. Biochem., 2002, 88, 144–152 CrossRef CAS PubMed.
- I. Bertini, C. Lucinat, C. Messori and M. Vašák, Proton NMR studies on the cobalt(II)-metallothionein system, J. Am. Chem. Soc., 1989, 111, 7296–7303 CrossRef CAS.
- H. Mattapalli, W. B. Monteith, C. S. Burns and A. S. Danell, Zinc deposition during ESI-MS analysis of peptide–zinc complexes, J. Am. Soc. Mass Spectrom., 2009, 20, 2199–2205 CrossRef CAS PubMed.
- Y. Kostyukevich, A. Kononikhin, I. Popov, M. Indeykina, S. A. Kozin, A. A. Makarov and E. Nikolaev, Supermetallization of peptides and proteins during electrospray ionization, J. Mass Spectrom., 2015, 50, 1079–1087 CrossRef CAS PubMed.
- M. Vašák, C. Berger and J. H. R. Kägi, Dynamic structure of metallothionein, FEBS Lett., 1984, 168, 174–178 CrossRef.
- S.-H. Chen, W. K. Russell and D. H. Russell, Combining chemical labeling, bottom-up and top-down ion-mobility mass spectrometry to identify metal-binding sites of partially metalated metallothionein, Anal. Chem., 2013, 85, 3229–3237 CrossRef CAS PubMed.
- S.-H. Chen and D. H. Russell, Reaction of human Cd7metallothionein and N-ethylmaleimide: kinetic and structural insights from electrospray ionization mass spectrometry, Biochemistry, 2015, 54, 6021–6028 CrossRef CAS PubMed.
- C. S. Babu, Y. M. Lee, T. Dudev and C. Lim, Modeling Zn2+ Release From Metallothionein, J. Phys. Chem. A, 2014, 118, 9244–9252 CrossRef CAS PubMed.
- L. J. Jiang, M. Vašák, B. L. Vallee and W. Maret, Zinc transfer potentials of the α- and β-clusters of metallothionein are affected by domain interactions in the whole molecule, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 2503–2508 CrossRef CAS.
- C. Capasso, V. Carginale, O. Crescenz, D. Di Maro, R. Spadaccini, P. A. Temussi and E. Parisi, Structural and functional studies of vertebrate metallothioneins: cross-talk between domains in the absence of physical contact, Biochem. J., 2005, 391, 95–103 CrossRef CAS PubMed.
- H. J. Hartmann and U. Weser, Copper-thionein from fetal bovine liver, Biochim. Biophys. Acta, 1977, 491, 211–422 CrossRef CAS.
- F. O. Brady and M. Webb, Metabolism of zinc and copper in the neonate. (Zinc, copper)-thionein in the developing rat kidney and testis, J. Biol. Chem., 1981, 256, 3931–3935 CAS.
- O. Palacios, S. Atrian and M. Capdevila, Zn- and Cu-thioneins: a functional classification for metallothioneins?, J. Biol. Inorg. Chem., 2011, 16, 991–1009 CrossRef CAS PubMed.
- L. Alvarez, H. Gonzalez-Iglesias, M. Garcia, S. Ghosh, A. Sanz-Medel and M. Coca-Prados, The stoichiometric transition from Zn6Cu1-metallothionein to Zn7-metallothionein underlies the up-regulation of metallothionein (MT) expression, J. Biol. Chem., 2012, 287, 28456–28469 CrossRef CAS PubMed.
- A. Krężel and W. Maret, The functions of metamorphic metallothioneins in zinc and copper metabolism, Int. J. Mol. Sci., 2017, 18, 1237 CrossRef PubMed.
- B. A. Messerle, A. Schäffer, M. Vašák, J. H. R. Kägi and K. Wüthrich, Comparison of the solution conformations of human [Zn7]-metallothionein-2 and [Cd7]-metallothionein-2 using nuclear magnetic resonance spectroscopy, J. Mol. Biol., 1992, 225, 433–443 CrossRef CAS PubMed.
- W. Braun, M. Vašák, A. H. Robbins, C. D. Stout, G. Wagner, J. H. Kägi and K. Wütrich, Comparison of the NMR solution structure and the x-ray crystal structure of rat metallothionein-2, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 10124–10128 CrossRef CAS.
- R. G. Pearson, Hard and soft acids and bases, J. Am. Chem. Soc., 1963, 85, 3533–3539 CrossRef CAS.
- S. S. Hasnain and C. D. Garner, Characterization of metal centers in biological systems by X-ray absorption spectroscopy, Prog. Biophys. Mol. Biol., 1987, 50, 47–65 CrossRef CAS PubMed.
- K. S. Hagen and R. H. Holm, The stereochemistry of decakis(benzenethiolato)-tetracadmate(2-) ion, a cage complex related to the cadmium-cysteinate aggregates in metallothioneins, Inorg. Chem., 1983, 22, 3171–3174 CrossRef CAS.
- Y. Wang, E. A. Mackay, M. Kurasaki and J. H. R. Kägi, Purification and Characterisation of Recombinant Sea Urchin Metallothionein Expressed in Escherichia coli, Eur. J. Biochem., 1994, 225, 449–457 CrossRef CAS PubMed.
- D. W. Hasler, L. T. Jensen, O. Zerbe, D. T. Winge and M. Vašák, Effect of the two conserved prolines of human growth inhibitory factor (metallothionein-3) on its biological activity and structure fluctuations: comparison with a mutant protein, Biochemistry, 2000, 39, 14567–14575 CrossRef CAS PubMed.
- G. Meloni, K. Zovo, J. Kazantseva, P. Palumaa and M. Vašák, Organization and assembly of metal-thiolate clusters in epithelium-specific metallothionein-4, J. Biol. Chem., 2006, 281, 14588–14595 CrossRef CAS PubMed.
- A. Munoz and A. R. Rodriguez, Electrochemical behavior of metallothioneines and related molecules. Part III: metallothionein, Electroanalysis, 1995, 7, 674–680 CrossRef CAS.
- M. A. Namdarghanbari, J. Meeusen, G. Bachowski, N. Giebel, J. Johnson and D. H. Petering, Reaction of the zinc sensor FluoZin-3 with Zn7-metallothionein: inquiry into the existence of a proposed weak binding site, J. Inorg. Biochem., 2010, 104, 224–231 CrossRef CAS PubMed.
- T. Gan, A. Munoz, C. F. Shaw III and D. H. Petering, Reaction of 111Cd7-metallothionein with EDTA, J. Biol. Chem., 1995, 270, 5339–5345 CrossRef CAS PubMed.
- C. A. Blindauer and O. I. Leszczyszyn, Metallothioneins: unparalleled diversity in structures and functions for metal ion homeostasis and more, Nat. Prod. Rep., 2010, 5, 720–741 RSC.
- F. Vazquez and M. Vašák, Comparative 113Cd-n.m.r. studies on rabbit 113Cd7-, (Zn1, Cd6) and partially metal-depleted 113Cd6-metallothionein-2a, Biochem. J., 1988, 253, 611–614 CrossRef CAS PubMed.
- K. Połeć-Pawlak, S. Palacios, M. Capdevila, P. Gonzalez-Durte and R. Łobiński, Monitoring of the displacement from the recombinant Mouse liver metallothionein Zn7-complex by capillary zone electrophoresis with electrospray MS detection, Talanta, 2002, 57, 1011–1017 CrossRef.
- E. Freisinger and M. Vašák, Cadmium in metallothioniens, Met. Ions Life Sci., 2013, 11, 339–371 CAS.
- M. J. Stillman, W. Cai and A. J. Zelazowski, Cadmium binding to metallothioneins. Domain specificity in reactions of alpha and beta fragments, apometallothionein, and zinc metallothionein with Cd2+, J. Biol. Chem., 1987, 262, 4538–4548 CAS.
- M. Good, R. Hollenstein and M. Vašák, Metal selectivity of clusters in rabbit liver metallothionein, Eur. J. Biochem., 1991, 197, 655–659 CrossRef CAS PubMed.
- J. W. Ejnik, A. Muñoz, E. DeRose, C. F. Shaw 3rd and D. H. Petering, Structural consequences of metallothionein dimerization: solution structure of the isolated Cd4-alpha-domain and comparison with the holoprotein dimer, Biochemistry, 2003, 42, 8403–8410 CrossRef CAS PubMed.
- M. Capdevila and S. Atrian, Metallothionein protein evolution: a miniassay, J. Biol. Inorg. Chem., 2011, 16, 977–989 CrossRef CAS PubMed.
- G. K. Andrews, Regulation of metallothionein gene expression by oxidative stress and metal ions, Biochem. Pharmacol., 2000, 59, 95–104 CrossRef CAS PubMed.
- J. Hernandez, J. Carrasco, E. Belloso, M. Giralt, H. Bluethmann, D. Kee Lee, G. K. Andrews and J. Hidalgo, Metallothionein induction by restraint stress: role of glucocorticoids and IL-6, Cytokine, 2000, 12, 791–796 CrossRef CAS PubMed.
- F. H. Mahoney and J. Koropatnick, Signaling events for metallothionein induction, Mutat. Res., 2003, 533, 211–226 CrossRef.
- A. Kocyła, J. Adamczyk and A. Krężel, Interdependence of free zinc changes and protein complex assembly – insights into zinc signal regulation, Metallomics, 2018, 10, 120–131 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7mt00332c |
| ‡ These authors contributed equally. |
| § Current address: Captor Therapeutics Ltd, Wrocław, Poland. |
| ¶ Current address: Department of Cell Biochemistry, University of Groningen, Groningen, The Netherlands. |
| This journal is © The Royal Society of Chemistry 2018 |