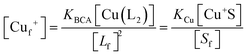A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans†
Megan L.
Straw
a,
Amanda K.
Chaplin
a,
Michael A.
Hough
 a,
Jordi
Paps
a,
Vassiliy N.
Bavro
a,
Michael T.
Wilson
a,
Erik
Vijgenboom
a,
Jordi
Paps
a,
Vassiliy N.
Bavro
a,
Michael T.
Wilson
a,
Erik
Vijgenboom
 b and
Jonathan A. R.
Worrall
b and
Jonathan A. R.
Worrall
 *a
*a
aSchool of Biological Sciences, University of Essex, Wivenhoe Park, Colchester, CO4 3SQ, UK. E-mail: jworrall@essex.ac.uk; Tel: +44(0) 1206 872095
bMicrobial Biotechnology and Health, Institute of Biology, Sylvius Laboratory, Leiden University, PO Box 9505, 2300RA Leiden, The Netherlands
First published on 19th December 2017
Abstract
Streptomyces lividans has a distinct dependence on the bioavailability of copper for its morphological development. A cytosolic copper resistance system is operative in S. lividans that serves to preclude deleterious copper levels. This system comprises of several CopZ-like copper chaperones and P1-type ATPases, predominantly under the transcriptional control of a metalloregulator from the copper sensitive operon repressor (CsoR) family. In the present study, we discover a new layer of cytosolic copper resistance in S. lividans that involves a protein belonging to the newly discovered family of copper storage proteins, which we have named Ccsp (cytosolic copper storage protein). From an evolutionary perspective, we find Ccsp homologues to be widespread in Bacteria and extend through into Archaea and Eukaryota. Under copper stress Ccsp is upregulated and consists of a homotetramer assembly capable of binding up to 80 cuprous ions (20 per protomer). X-ray crystallography reveals 18 cysteines, 3 histidines and 1 aspartate are involved in cuprous ion coordination. Loading of cuprous ions to Ccsp is a cooperative process with a Hill coefficient of 1.9 and a CopZ-like copper chaperone can transfer copper to Ccsp. A Δccsp mutant strain indicates that Ccsp is not required under initial copper stress in S. lividans, but as the CsoR/CopZ/ATPase efflux system becomes saturated, Ccsp facilitates a second level of copper tolerance.
Significance to metallomicsStreptomyces lividans is an industrially used strain with application in the heterologous production of commercially valuable biomolecules. A distinct dependence on the bioavailability of copper for S. lividans morphological development is known and knowledge of the copper uptake, storage and delivery systems has led to the creation of new strains with improved biotechnological properties. Here, we provide new insights into copper tolerance in S. lividans through the identification and characterisation of a cytosolic copper storage protein (Ccsp). Ccsp is involved in a second layer of copper resistance once the CsoR/CopZ/P1-ATPase systems become saturated. |
Introduction
Streptomyces are the largest genus of the Gram-positive phylum Actinobacteria which belong to the Terrabacteria clade.1,2 Members of this clade have evolved important adaptations to environmental hazards that enabled them to colonise on early Earth.1,2 Actinobacteria, particularly Streptomyces are of great economic importance as they are major contributors to soil ecosystems that forests depend on and they are producers of many secondary metabolites that have antibiotic, anti-tumour and anthelmintic properties.3,4 Furthermore, they hold great potential as a large-scale production host in biotechnology for the heterologous production of high value proteins and enzymes for re-use of biomass waste, therapeutic, diagnostic and agricultural purposes.5Streptomyces undergo a complex development life cycle on solid substrates. Following spore germination, a vegetative mycelium is established that in response to nutrient depletion and other signals initiates both secondary metabolite production and morphological differentiation. This leads to the formation of aerial hyphae that differentiate to produce millions of spores, which are easily dispersed into the environment.3,4Streptomyces lividans is a biotechnologically used strain that displays a distinct dependence on the bioavailability of copper (Cu) to initiate the morphological switch from vegetative to aerial growth that coincides with the production of secondary metabolites.6–10 The versatility and diverse biological roles of Cu are driven by its facile transition between reduced (Cu+) and oxidised (Cu2+) forms and its ability to form thermodynamically stable yet labile ligand-exchange coordination complexes.11 These properties whilst beneficial are also potential causes of Cu toxicity12 and so homeostatic systems have evolved to regulate the response to nutritional supply and demand.13–15 To this end our recent work has identified and characterised several mechanisms in S. lividans strain 1326 that are operable under Cu excess.16,17 In the cytosol of S. lividans, Cu toxicity is precluded through the role of a Cu-sensing regulatory transcription factor from the copper sensitive operon regulator (CsoR) family.18 Under Cu stress conditions, Cu+ ions bind to the DNA bound apo-CsoR and allosterically activate transcriptional de-repression of efflux systems involving multiple P1-type ATPases, and CopZ-like metallochaperones to rapidly remove Cu+ from the cytosol.16,17 Under Cu limiting conditions (homeostasis) an extracytoplasmic Cu pathway involving two Cu metallochaperones (Sco and ECuC) is operable.8,19 This pathway serves to metallate the dinuclear CuA site in the aa3-type cytochrome c oxidase (CcO)8 and the mononuclear Cu site in the Cu-radical oxidase GlxA.9,10 The enzymatic action of GlxA is key to the initiation of the Cu dependent morphological development between vegetative and aerial hyphae and if not correctly metallated stalls organism development.9,10
Utilisation of Cu by bacteria to ‘correctly’ metallate secreted nascent apo-enzymes or proteins is much less well understood20–22 than the Cu resistance mechanisms operative in the cytosol.23 Trafficking pathways involving metallochaperones as identified in S. lividans undoubtedly play a role, but the initial supply of Cu to periplasmic or extracytoplasmic metallochaperones is not well defined. For instance, the acquisition of extracytoplasmic Cu by ECuC in S. lividans, which can only bind Cu in the cuprous form is unknown.19 In the cyanobacterium Synechocystis, evidence has accumulated that the supply pathway for loading a periplasmic Cu metallochaperone is through routing Cu via the cytosol prior to collection from a P1-type ATPase.24 In methanotropic bacteria a designated Cu up-take system has been identified25 that acquires Cu to activate particulate (membrane-bound) methane monooxygenase (pMMO).26 Under Cu limitation siderophore-like substances known as methanobactins (Mb) are secreted that scavenge and bind with a high affinity extracellular Cu2+ and Cu+.27–29 The Cu+-loaded form is then reinternalized into the cytoplasm where it is thought to be used for activation of pMMO.30,31 Recently, the discovery of a novel family of Cu storage proteins (Csp) in the methanotroph Methylosinus trichosporium OB3b has added a new layer of intrigue to Cu handling and storage in this organism.32M. trichosporium OB3b possess three Csp members; MtCsp1 and MtCsp2 are exported after folding from the cytosol, whereas MtCsp3 lacks a twin arginine translocase (TAT) signal peptide and is therefore predicted to remain in the cytosol. MtCsp1 forms a homotetramer assembly comprised of 4 four-helix bundle units that each bind 13 Cu+ ions through coordination by 13 Cys residues that line the core of each four-helix bundle.32 Thus, MtCsp1 has the capacity to bind up to 52 Cu+ ions per homotetramer.32 Deletion of the MtCsp1 and MtCsp2 genes implicates a role for these proteins in Cu storage for methane oxidation in M. trichosporium OB3b with it also known that in vitro Mb can remove Cu+ from MtCsp1.32 More recently, MtCsp3 has been characterised revealing a homotetramer arrangement of four-helix bundles with 19 Cys residues and 19 Cu+ ions bound per protomer.33MtCsp3 homologues are reported to be widespread in bacteria, with the Csp3 from Bacillus subtilis the first Gram-positive member to be biochemically and structurally characterised, albeit in the apo state.33 Thus, this novel family of proteins with an unprecedentedly high capacity to store cytosolic Cu leads to many new questions regarding the physiological role of Cu in the bacterial cytosol.
Herein we report the identification in S. lividans of a gene encoding for a Csp3 homologue, which we have annotated ccsp (cytosolic copper storage protein). In the framework of our previous Cu tolerance and resistance studies in S. lividans, we have investigated the evolution, structural, biochemical and functional properties of Ccsp. From an evolutionary perspective, we show that the ccsp homologues are widespread in Terrabacteria and extend through into Archaea and Eukaryota. A Ccsp homotetramer is found to store up to 80 Cu+ ions and we provide evidence to indicate that Ccsp provides a second level of Cu resistance in S. lividans once the CsoR/CopZ/ATPase system becomes saturated with Cu+. Furthermore, we find that CopZ can transfer Cu+ to Ccsp and that genes surrounding Ccsp are sensitive to elevated Cu levels.
Methods
Streptomyces media and growth conditions
The agar media soya flower mannitol (SFM), R5 (complex medium) and defined medium (MM) were prepared according to Kieser et al.34 Bennett's medium contained yeast extract (Difco) 1.0 g l−1, Beef extract (LAB-LEMCO powder, Oxoid) 1.0 g l−1, N-Z Amine Type A (Sigma) 2.0 g l−1, sugars were added to a final concentration of 0.5% and agar at 1.5% for solid media. Difco nutrient agar (DNA) was prepared according to the instructions of the manufacturer and liquid R5 according to Kieser et al.34 Antibiotics were used in the following final concentrations: apramycin 50 μg μl−1, thiostrepton 5–20 μg μl−1. Agar plates and liquid cultures were incubated at 30 °C with the latter shaken at 160–250 rpm. Spore stocks were obtained from cultures grown on SFM plates and stored in 20% (v/w) glycerol at −20 °C.Generation of the Δccsp mutant of S. lividans
The parental strain used was S. lividans 1326 (John Innes Institute collection, hereafter S. lividans). The Ccsp mutant (Δccsp) was isolated essentially according to the protocol described previously by Blundell et al.8 The ccsp open reading frame (ORF) was replaced by a 62 nt scar of the lox recombination site including two XbaI sites. The mutant was analysed by PCR with genomic DNA as template to confirm the loss of the ccsp gene. For complementation of the Δccsp mutant the ccsp ORF with 150 bp upstream was cloned as a EcoRI-HindIII fragment in the moderate copy number plasmid pHJL40135 and designated pCcsp-1.Growth morphology of S. lividans
Fresh spores were isolated from SFM plates and diluted in sterile H2O to the desired concentration. Spores were spotted in 10 μl drops containing 103 spores and left to dry in a flow cabinet before incubation at 30 °C for 6 days. Spores were either spotted on standard petri dishes (diameter 9 cm) containing the indicated agar medium or in 24 well plates with 1.8 ml agar medium per well. Copper was added as Cu(II) citrate (Sigma-Aldrich) to the desired final concentration. Bennett's glucose medium and liquid R5 medium were inoculated with 2 × 106 spores in 125 ml baffled flasks and incubated with shaking (160 rpm) for 32 h. BCDA (bathocuproinedisulfonic acid; Sigma-Aldrich) was added to a final concentration of 50 μM and Cu was added as Cu(II) citrate to the desired final concentration. Samples (2 ml) were collected in duplicate in pre-weighed Eppendorf tubes after 32 h and the mycelium collected by centrifugation and the pellets dried for 48 h at 98 °C. The dry weight of the biomass was determined with an analytical balance.CcO activity
In vivo CcO activity was performed with N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD; Sigma-Aldrich) as substrate.8,36,37 Strains were spotted on DNA or Bennett's glucose agar (10 μl containing 1000 spores) and incubated for 24 h at 30 °C. A light spray of 0.3% (w/v) agarose in H2O was used to fix the mycelium spots, followed by an overlay with 10 ml of 25 mM sodium phosphate pH 7.4 solution containing 20% ethanol, 0.6% agarose, 1% sodium deoxycholate and 10 mg TMPD. The CcO activity was recorded by taking digital images every 30 s for 5–10 min. The IMAGEJ software38 was used to calculate average pixel intensities of the indophenol blue stained mycelium.Bioinformatics
Similarity searches using BLAST39 were performed as implemented in its online version in the National Center for Biotechnology Information webpage.40 The S. lividans 1326 sequence SLI_RS1725541 was used and the searches were performed against each of the major groups within Bacteria, Archaea, and Eukaryota, as listed in NCBI Taxonomy.42 The BLAST searches were performed using the e-value threshold 2 × 10−2, and the protein sequences for the top 5 results (between 1 and 5 hits) for each taxonomic group were downloaded. Multiple sequence alignments were performed using MAFFT with the “Auto” strategy.43 Positions of ambiguous alignment were removed using the online version of Gblocks44 using the “less stringent” options. Phylogenetic trees were inferred with the Maximum Likelihood approach as implemented in the program FastTree2;45 the WAG + Gamma evolutionary model of substitutions46 and a combination of parameters aimed to produce a slow and accurate tree search (-spr 4 -mlacc 2 -slownni -no2nd) were used. Local support values were calculated with the Shimodaira–Hasegawa test.47Cloning, over-expression and purification of Ccsp
The Sli3625A gene was cloned from S. lividans genomic DNA and restricted into the NdeI and HindIII sites of a pET28a vector (Novagen) to create an N-terminal His6-tagged construct. This construct, designated pET3625A, was transformed to Escherichia coli BL21(DE3) cells and single colonies were transferred to 2xYT medium (Melford) with kanamycin (50 μg per liter) (Fisher) at 37 °C. Over-expression was induced by 1 M isopropyl β-D-1-thiogalactopyranoside (IPTG; Fisher) to a final concentration of 1 mM and the temperature was decreased to 25 °C for overnight incubation. Cultures were harvested by centrifugation at 3501g for 20 min at 4 °C and the cell pellet was re-suspended in 50 mM Tris/HCl, 500 mM NaCl (Fisher) and 20 mM imidazole (Sigma) adjusted to pH 7.5 (Buffer A). The re-suspended cell suspension was lysed using an EmulsiFlex-C5 cell disrupter (Avestin) followed by centrifugation at 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 724g for 20 min at 4 °C. The clarified supernatant was loaded to a 5 ml Ni-NTA Sepharose column (GE Healthcare) equilibrated with Buffer A and eluted by a linear imidazole gradient using Buffer B (Buffer A with 500 mM imidazole). Fractions containing Ccsp were pooled and dialysed overnight at 4 °C against 50 mM Tris/HCl pH 7.0, 100 mM NaCl, (Buffer C). Following dialysis, the N-terminal His-tag was removed by incubating the protein at room temperature overnight with 125 KU of thrombin (Sigma). The protein/thrombin mixture was reapplied to the Ni-NTA Sepharose column (GE Healthcare) and the flow-through collected and concentrated using a Vivaspin centrifugal concentrator with a 30 kDa cut-off at 4 °C for application to a G75 Sephadex column (GE Healthcare) equilibrated with buffer C. Fractions eluting from the major peak of the G75 column were analysed for purity by SDS-PAGE and pooled. Far UV-CD spectroscopy using an Applied Photophysics Chirascan circular dichroism (CD) spectrophotometer (Leatherhead, UK) was used to assess whether the purified Ccsp was folded and LC-MS (denaturing and native) was used to quantify the mass and the oligomeric state.
724g for 20 min at 4 °C. The clarified supernatant was loaded to a 5 ml Ni-NTA Sepharose column (GE Healthcare) equilibrated with Buffer A and eluted by a linear imidazole gradient using Buffer B (Buffer A with 500 mM imidazole). Fractions containing Ccsp were pooled and dialysed overnight at 4 °C against 50 mM Tris/HCl pH 7.0, 100 mM NaCl, (Buffer C). Following dialysis, the N-terminal His-tag was removed by incubating the protein at room temperature overnight with 125 KU of thrombin (Sigma). The protein/thrombin mixture was reapplied to the Ni-NTA Sepharose column (GE Healthcare) and the flow-through collected and concentrated using a Vivaspin centrifugal concentrator with a 30 kDa cut-off at 4 °C for application to a G75 Sephadex column (GE Healthcare) equilibrated with buffer C. Fractions eluting from the major peak of the G75 column were analysed for purity by SDS-PAGE and pooled. Far UV-CD spectroscopy using an Applied Photophysics Chirascan circular dichroism (CD) spectrophotometer (Leatherhead, UK) was used to assess whether the purified Ccsp was folded and LC-MS (denaturing and native) was used to quantify the mass and the oligomeric state.
Over-expression and purification of CopZ-3079
CopZ-3079 (here after CopZ) was over-expressed in E. coli BL21(DE3) and purified as previously reported.17 Free thiol content was determined by the reduction of 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) monitored at 412 nm (ε = 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 M−1 cm−1).48
500 M−1 cm−1).48
Preparation of Cu(I) and Ag(I) solutions for titrations
Apo-Ccsp and apo-CopZ for Cu+-binding assays, Cu+-loading and transfer experiments were prepared together with CuCl solutions in an anaerobic chamber (DW Scientific [O2] < 2 ppm). Proteins were first incubating for 2–3 h with 2 mM DTT followed by desalting using a PD-10 column (GE Healthcare) equilibrated with 10 mM MOPS pH 7.5, 150 mM NaCl. Solid CuCl (Sigma) was dissolved in 10 mM HCl and 500 mM NaCl and diluted with 10 mM MOPS pH 7.5, 150 mM NaCl. The [Cu+] was determined spectrophotometrically using a Cary 60 UV-visible spectrophotometer (Varian) at 20 °C through step-wise addition of the stock CuCl solution into a known concentration of the Cu+ specific bidentate chelator bicinchoninic acid (BCA; Sigma). Formation of the [Cu(BCA)2]3− complex was monitored at 562 nm and the concentration determined using an ε = 7900 M−1 cm−1.49 Apo-Ccsp (4–8 μM) samples were sealed in an anaerobic quartz cuvette (Hellma) for titration with CuCl and absorbance changes in the 350 to 200 nm range monitored. A solution of AgNO3 was prepared and diluted to a 1 mM stock and titrated to apo-Ccsp with absorbance changes in the 350 to 200 nm range monitored aerobically.Determination of apparent Cu+ binding constants
Samples of apo-Ccsp (5–10 μM) in 10 mM MOPS pH 7.5, 150 mM NaCl were incubated under anaerobic conditions with various concentrations of BCA (50–1000 μM) and increasing amounts of Cu+ added. The concentration of the [Cu(BCA)2]3− complex at each titration point was determined as described above. At BCA concentrations of ≥250 μM an estimate of the apparent Cu+ binding affinity may be determined in two ways. Under the experimental conditions employed the following equilibria are present| 2Lf + Cuf+ ⇌ CuL2 = KBCA |
| Sf + Cuf+ ⇌ Cu+S = KCu |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β2 = 17.7)50 and Ccsp, respectively. Based on the above equilibria the [Cuf+] is given by
β2 = 17.7)50 and Ccsp, respectively. Based on the above equilibria the [Cuf+] is given bywhich can be rearranged to solve for KCu
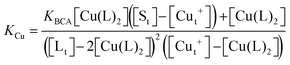 | (1) |
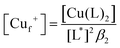 | (2) |
 | (3) |
Analytical gel filtration and monitoring of Cu+ transfer experiments
A G75 Superdex column (10/300 GL; GE-Healthcare) was equilibrated in degassed 10 mM MOPS pH 7.5, 150 mM NaCl. All protein samples were prepared anaerobically and loaded to the column with a gas tight syringe (Hamilton). Apo-CopZ samples were incubated with between 0.5 to 3 Cu+ equivalents prior to loading to the column and the elution profile monitored at 280 nm. For transfer experiments, stoichiometric equivalents of Cu+ were loaded to either CopZ or Ccsp before mixing samples in a molar ratio followed by an incubation period of up to 3 h and then loading to the column.X-ray crystallography and structure determination
Crystals of Ccsp were grown using the hanging drop vapor diffusion method at 18 °C following initial crystal hits found in commercial screens using an ARI Gryphon crystallization robot. For apo-Ccsp 1 μl of protein solution at a concentration of 15 mg ml−1 was mixed with an equal volume of reservoir solution containing 1.4 M ammonium sulfate, 0.1 M HEPES pH 7.0. Cu+-loaded Ccsp samples for crystallization were prepared through the addition of 25 equivalents of Cu+ ions to apo-Ccsp in an anaerobic chamber followed by removal of unbound Cu+ by passing the sample through a PD-10 column. Samples were concentrated to ∼15 mg ml−1 and an equal volume of protein (1 μl) and reservoir solution mixed. Crystals of Cu+–Ccsp were obtained from 1.4 M ammonium sulfate, 0.1 M MES pH 6.0. Crystals were transferred to a cryoprotectant solution consisting of 40% w/v sucrose, and flash cooled by plunging into liquid nitrogen. Apo-Ccsp crystals were measured at the Diamond Light Source on beamline I02 using an X-ray wavelength of 0.979 and a Pilatus 6M-F detector. Cu+-loaded crystals were measured at the ESRF on beamline ID29 using a Pilatus 6M detector and an X-ray wavelength of 0.976 Å. All data were indexed using XDS51 and scaled and merged using Aimless52 in the CCP4 suite with the CCP4i2 interface. The apo-Ccsp structure was solved by molecular replacement in PHASER53 using the PDB-ID 3lmf as the search model. Automated model building was carried out using the Buccaneer pipeline54 followed by cycles of model building in Coot55 and refinement in Refmac5.56 Riding hydrogen atoms were added when refinement of the protein atoms had converged. The final model of apo-Ccsp was used as the search model for Cu+–Ccsp molecular replacement. The Cu+–Ccsp data were twinned and twin refinement against intensities was performed in Refmac5 together with TLS refinement. An anomalous map for validation of Cu atom positions was generated using PHASER53 in the CCP4i2 interface from a separate dataset measured at a wavelength of 1.368 Å. Structures were validated using the Molprobity server57 the JCSG Quality Control Server and tools within Coot.55 Structural superpositions were carried out using GESAMT in CCP4i2.58 Coordinates and structure factors were deposited in the RCSB Protein Data Bank. A summary of data, refinement statistics and the quality indicators for the structures are given in Table 1.| Structure | Apo-Ccsp | Cu+–Ccsp |
|---|---|---|
| Space group | P6122 | I222 |
| Unit cell (Å) | 93.6, 93.6, 213.4 | 62.1, 64.1, 66.0 |
| Resolution (Å) | 80.3–1.34 | 45.2–1.50 |
| Unique reflections | 123![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 798 798 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 701 701 |
| Mn (I/SD) | 18.0 (0.8) | 9.7 (4.5) |
| CC1/2 | 0.999 (0.764) | 0.992 (0.958) |
| Completeness (%) | 100 (99.1) | 96.4 (96.6) |
| Redundancy | 18.2 (12.6) | 3.6 (3.5) |
| R cryst | 0.157 | 0.206 |
| R free | 0.192 | 0.231 |
| RMS dev. bond lengths (Å) | 0.015 | 0.020 |
| RMS dev. bond angles (°) | 1.63 | 2.05 |
| Ramachandran favoured (%) | 99.8 | 99.2 |
| PDB accession code | 6EI0 | 6EK9 |
Results
Identification of a cytosolic Cu storage protein in S. lividans
We identified a gene in between SLI_3625 and SLI_3626 in the original S. lividans genome sequence that transcribed on the opposite strand and thus not part of the SLI_3625/SLI_3626 operon.41 The gene was not originally annotated but was later given the locus-tag SLI_RS17255 in the Genbank annotation of S. lividans (CM001889).59 The new locus tags of the upstream and downstream genes are SLI_RS17250 (old tag SLI_3625) and SLI_RS17260 (old tag SLI_3626), respectively. Translation of SLI_RS17255 gives a protein sequence consisting of 136 amino acids with no recognisable export signal sequence and possesses 18 Cys residues of which 17 are in either CXXXC or CXXC motifs. The positional alignment of the Cys residues with Csp3 members and the absence of a signal peptide led us to naming this S. lividans protein Ccsp (cytosolic copper storage protein). Of note from a previous RNA-seq study was the observation that genes in the ccsp environment, SLI_RS17245 and SLI_RS17250 are both upregulated 6-fold following a 30 min Cu(II) pulse (400 μM) relative to Cu homeostatic growth conditions.16 Re-analysis of the RNA-seq data in light of the discovery of a ccsp gene in S. lividans reveals that ccsp is expressed at a low-level under homeostasis conditions but under Cu stress is upregulated 5-fold.16 Thus, the ccsp gene and its immediate genomic environment (i.e. SLI_RS17245 and SLI_RS17250) is sensitive to elevated Cu levels.Taxonomic distribution and evolution of Ccsp
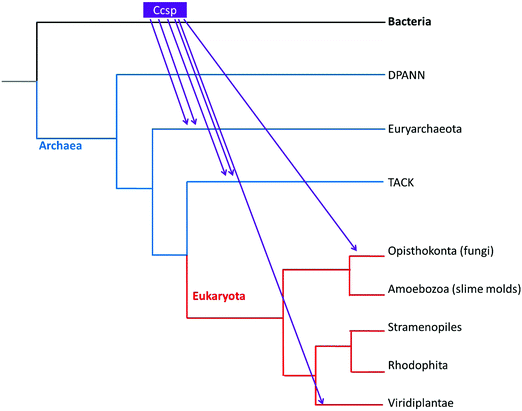
The evolutionary lineage of Ccsp was investigated. The taxonomic distribution is reported in Table S1 (ESI†) and reveals that 7 Bacterial groups contain a Ccsp. All Terrabacteria, of which streptomycetes belong, possess a Ccsp homologue, except for Tenericutes (Table S1, ESI†). Regarding the three major groups of Archaea, Ccsp is not found in the DPANN group, but is present (with a low number of hits) in the TACK group and the halophilic Euryarchaeota (Table S1, ESI†). Finally, among eukaryotes only the fungi (Opisthokonta) and land plants possess Ccsp (Table S1, ESI†). Construction of a phylogenetic tree for Ccsp reveals mainly bacterial branches with a low number of archaea and eukaryote twigs interspersed (Fig. S1, ESI†). The broad taxonomic distribution across the major Bacteria groups supports a hypothesis that Ccsp gene originated in the bacterial last common ancestor (LCA), followed by multiple gene losses in independent bacterial lineages. In contrast, the scarce presence of Ccsp in different members of Archaea and Eukaryota, together with the lack of evolutionary relationships between their sequences, points to the absence of these genes in the LCA of each of those domains and a most likely origin is multiple lateral gene transfer (LGT). Using the taxonomic occupancy together with the phylogenetic tree a reconstruction of the origin and evolutionary history of Ccsp can be created as depicted in Fig. 1. This illustrates that the bacterial Ccsp gene jumped twice to Euryarchaeota, twice to the TACK, and twice to eukaryotes (fungi and plants).Solution properties and X-ray crystal structure of S. lividans Ccsp
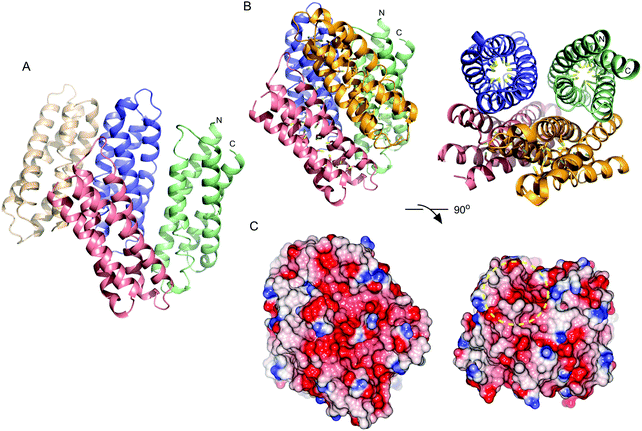
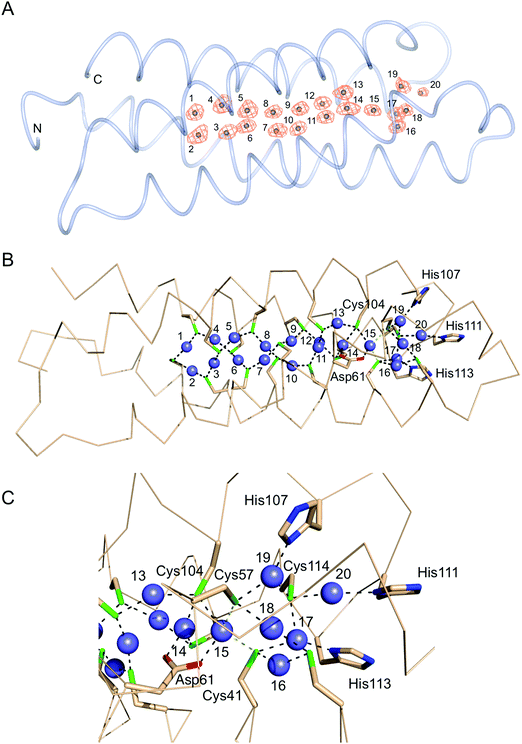
Purified recombinant S. lividans Ccsp (residues 1–136) gave a mass under denaturing conditions of 14604.6 Da (as expected following His6-tag cleavage) and a far UV-CD spectrum consistent for a folded protein with α-helical secondary structure content (Fig. S2, ESI†). Ccsp eluted from an analytical gel filtration column with a retention volume consistent with a higher order species, which was confirmed from native mass spectrometry where a mass of 58418.16 Da was obtained. This together with the gel filtration is consistent with a homotetramer assembly in solution for Ccsp as reported for other members of this protein family.33,60 The X-ray structure of Ccsp was determined to 1.34 Å resolution (Table 1). Four Ccsp protomers (Chains A to D) were identified in the crystallographic asymmetric unit (Fig. 2A), with unbroken electron density observed for residues 17–136 in chain A, 20–135 in chain B, 19–135 in chain C and 16–136 in chain D. Thus, in all four protomers electron density corresponding to residues 1–15 was not observed. Each protomer is made-up of four α-helices arranged to form a four helix bundle motif (Fig. 2A). Chains A, C and D together with a symmetry related molecule create the functional quaternary structure (Fig. 2B). The core of each four helix-bundle protomer creates a solvent shielded pore or channel that is lined with the 18 Cys residues (Fig. 2B), none of which participate in disulfide bonds or exhibit any modifications. Generation of the electrostatic surface potential of the quaternary homotetramer reveals large stretches of negative charge spanning essentially the length of the protomer interaction site (Fig. 2C). Notably, from the surface representation it is apparent that there is an asymmetry of negative charge between the two ends of the pore opening (Fig. 2C) and may have consequences for Cu+ loading.Ccsp has a high capacity to bind Cu+ and Ag+ ions
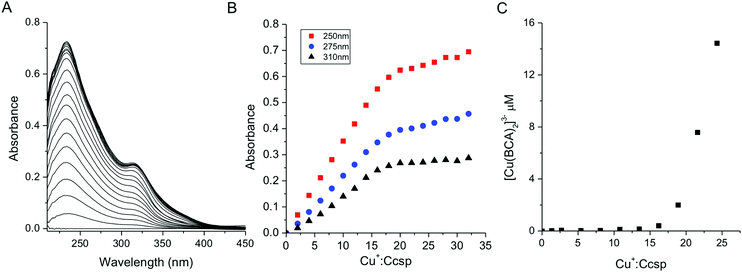
Binding of the Group 11 monovalent Cu+ and Ag+ ions to Ccsp was characterised. Addition of Cu+ ions to Ccsp under anaerobic conditions led to the appearance of absorbance bands in the UV-spectrum (Fig. 3A). These are attributed to (Cys)Sγ → Cu+ ligand to metal charge transfer (LMCT) bands, which increased concomitantly with the Cu+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ccsp ratio until a saturation point was reached coinciding with a stoichiometry of between ∼18–20 Cu+ ions per Ccsp protomer (Fig. 3B). This indicates that 72 to 80 Cu+ ions may be stored per Ccsp tetramer. Stoichiometric loading of Ccsp with Cu+ ions to create the holo-Ccsp resulted in a far UV-CD spectrum (Fig. S2, ESI†) and gel filtration profile that were not significantly different from apo-Ccsp, demonstrating that bound Cu+ ions do not grossly alter the secondary or quaternary assembly. Addition of Ag+ ions to Ccsp also led to changes in the UV-spectrum which may be attributed to (Cys)Sγ → Ag+ LMCT bands (Fig. S3, ESI†). A less well defined break point is reached than in the case of Cu+ but nevertheless is consistent with a stoichiometry of >15 Ag+ ions bound per Ccsp protomer (Fig. S3, ESI†).
Ccsp ratio until a saturation point was reached coinciding with a stoichiometry of between ∼18–20 Cu+ ions per Ccsp protomer (Fig. 3B). This indicates that 72 to 80 Cu+ ions may be stored per Ccsp tetramer. Stoichiometric loading of Ccsp with Cu+ ions to create the holo-Ccsp resulted in a far UV-CD spectrum (Fig. S2, ESI†) and gel filtration profile that were not significantly different from apo-Ccsp, demonstrating that bound Cu+ ions do not grossly alter the secondary or quaternary assembly. Addition of Ag+ ions to Ccsp also led to changes in the UV-spectrum which may be attributed to (Cys)Sγ → Ag+ LMCT bands (Fig. S3, ESI†). A less well defined break point is reached than in the case of Cu+ but nevertheless is consistent with a stoichiometry of >15 Ag+ ions bound per Ccsp protomer (Fig. S3, ESI†).
Ccsp binds Cu+ ions in a cooperative manner
An estimation of Cu+ binding affinities for cuproproteins is possible through competition experiments using high affinity chromogenic Cu+ bidentate ligands such as BCA.50,61 At low BCA concentrations (50–100 μM) Ccsp binds all Cu+ ions until >15 Cu+ equivalents have been added and the formation of the [Cu(BCA2)]3− complex occurs (Fig. 3C). Upon increasing the BCA concentration (250–1000 μM) competition for the titrated Cu+ ions between Ccsp and BCA is now observed (Fig. 4A), with a maximum Cu+ occupancy in a Ccsp protomer estimated to be 15 Cu+ equivalents. Using eqn (1) apparent Cu+ dissociation constants (KCu) for each titration of Cu+ into Ccsp at BCA concentrations of 250, 500 and 1000 μM can be determined, with an average KCu = 3.3 ± 1.3 × 10−17 M. Alternatively, the data in Fig. 4A at BCA concentrations ≥250 μM can be used to calculate the [Cufree+] using eqn (2) (see Methods). Plots of fractional occupancy of Cu+ sites versus the [Cufree+] at two set BCA concentrations are illustrated in Fig. 4B. The data clearly show a sigmoidal dependence and given that the system is at equilibrium (see Methods) then this implies cooperativity of Cu+ binding. Therefore, these data have been fitted accordingly using a nonlinear form of the Hill equation (eqn (3)) (Fig. 4B). From triplicate experiments with set BCA concentrations ranging between 250–1000 μM an average KCu = 2.9 ± 0.2 × 10−17 M and a Hill coefficient, n = 1.9 ± 0.2 are determined. Thus, Cu+ binding to S. lividans Ccsp appears to be a cooperative process with a binding affinity in line with a role in sequestering and storing cytosolic Cu+ ions.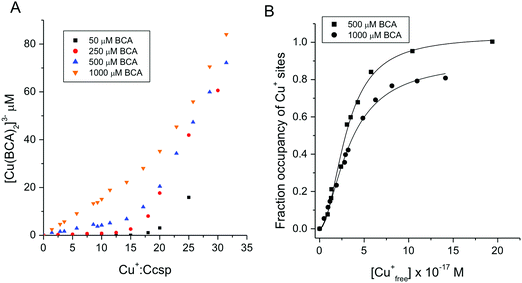 | ||
Fig. 4 Affinity of Ccsp for Cu+ ions (A) plots of [Cu(BCA)2]3− concentration versus the Cu+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ccsp concentration ratio upon titrating Cu+ ions into Ccsp (5–10 μM) in the presence of different concentrations of BCA as indicated on the plot. As the BCA concentration increases a competition with Ccsp for Cu+ ions occur. (B) Plots of fractional occupancy of Cu+ binding sites in Ccsp at varying Cu+free concentrations determined from the data in (A) at 500 and 1000 μM BCA concentrations. The solid lines through the data points are fits to a non-linear form of the Hill equation (eqn (3)) to give a KCu = 3.0 ± 0.1 × 10−17 M and a Hill coefficient, n = 2.0 ± 0.2 at 500 μM BCA and a KCu = 3.4 ± 0.2 × 10−17 M and n = 1.7 ± 0.1 at 1000 μM BCA. All experiments were performed in 10 mM MOPS pH 7.5, 150 mM NaCl. Ccsp concentration ratio upon titrating Cu+ ions into Ccsp (5–10 μM) in the presence of different concentrations of BCA as indicated on the plot. As the BCA concentration increases a competition with Ccsp for Cu+ ions occur. (B) Plots of fractional occupancy of Cu+ binding sites in Ccsp at varying Cu+free concentrations determined from the data in (A) at 500 and 1000 μM BCA concentrations. The solid lines through the data points are fits to a non-linear form of the Hill equation (eqn (3)) to give a KCu = 3.0 ± 0.1 × 10−17 M and a Hill coefficient, n = 2.0 ± 0.2 at 500 μM BCA and a KCu = 3.4 ± 0.2 × 10−17 M and n = 1.7 ± 0.1 at 1000 μM BCA. All experiments were performed in 10 mM MOPS pH 7.5, 150 mM NaCl. | ||
The X-ray crystal structure of Cu+-loaded Ccsp reveals 20 Cu+ ions bind per protomer
The structure of the Cu+-loaded Ccsp was determined to 1.5 Å resolution with a single Ccsp protomer found in the asymmetric unit (Table 1). Unbroken electron density was observed for residues 16–136. Strong anomalous scattering that is attributed to the presence of bound Cu+ ions is observed (Fig. 5A). The anomalous electron density map, plotted from a dataset measured at a wavelength of 1.368 Å, reveals 20 Cu+ ions coordinated in the core of the Ccsp four-helix bundle (Fig. 5A), thus corroborating the estimate from the titration data (Fig. 3B). Therefore, the Ccsp homotetramer has the capacity to bind a total of 80 Cu+ ions. Inspection of the Cu+–Ccsp structure reveals that Cu+ ions 1 to 13 all have bis-cysteinate coordination with (Cys)–Sγ–Cu+ bond distances of between 2.0 and 2.3 Å and each Cys residue bridging two different Cu+ ions (Fig. 5B). Out of these 13 Cu+ ions seven (Cu2, Cu4, Cu5, Cu7, Cu10, Cu12 and Cu13) are coordinated by CXXXC motifs, with the remainder coordinated by Cys residues that are on different helices of the bundle. No Cu+ ions are coordinated by CXXC motifs. The bis-cysteinate coordination pattern is broken at Cu+ ion 14 which has a third coordinate bond from the Oδ1 atom (2.2 Å) of Asp61 (Fig. 5B and C). Similarly, the Cu+ ion 15 has a coordinate bond with the Oδ2 atom of Asp61 (2.1 Å) as well as thiolate ligation from Cys104, which also participates in ligation with the Cu+ ions 13 and 14. (Fig. 5B and C). It is possible that the Cu+ ion 15 is further coordinated by Cys41 and Cys57 (the latter being the only Cys residue not in either a CXXC or CXXXC motif), to create a distorted tetrahedral coordination geometry, however we note that the (Cys)–Sγ–Cu+ bond distances of 2.5 and 2.7 Å (Fig. 5C), respectively, are longer than for other thiolate Cu+ interactions. The remaining 5 Cu+ ions, 16 to 20, cluster beyond Cu+ ion 15 towards the entrance of the pore (Fig. 5B and C) and if a coordinate bond from Cys41 and Cys57 to Cu+ ion 15 is absent it may be considered as a separate cluster. None of these remaining Cu+ ions are coordinated in CXXXC motifs. Cu+ ions 16 and 18 have bis-cysteinate coordination (Fig. 5C), with the Cu+ ions 17 and 19 having bis-cysteinate coordination as well as ligation from the Nδ of His113 (1.9 Å) and His107 (2.1 Å), respectively. Finally, Cu+ ion 20 is coordinated by Cys114 and the Nδ of His111 (2.2 Å). Cys114 is the only other Cys residue to participate in coordination with three different Cu+ ions (Fig. 5C). Finally, a total of nine Cu+–Cu+ interactions are identified with distances between 2.5 and 2.8 Å.S. lividans Ccsp is required for growth under extreme Cu conditions in vivo
In the knowledge that S. lividans Ccsp can tightly bind significant numbers of Cu+ ions, its role in the morphological development and growth of S. lividans was next investigated. The wild type parent S. lividans strain has a very high Cu tolerance, which varies depending on the type of media used.7,8 This is illustrated in Fig. 6 using defined medium with either glucose or mannitol as the sole carbon sources. In the presence of glucose (a reducing sugar) Cu tolerance exceeds 5 mM but the development of aerial mycelium and spores is reduced or absent at Cu concentrations above 1 mM (Fig. 6A). By contrast growth on mannitol (a sugar alcohol) is less Cu tolerant by ∼5-fold (Fig. 6B). Growth on complex media such as R5 or Bennetts-glucose also shows a high Cu tolerance and is again media dependent (Fig. S4, ESI†). On all media tested, the Δccsp mutant consistently revealed a reduced tolerance for Cu compared to the parent strain (Fig. 6 and Fig. S4, ESI†). For example, on glucose, growth becomes significantly inhibited at 500 μM Cu whereas on mannitol inhibition is observed at 200 μM Cu (Fig. 6A and B). This demonstrates that Ccsp is required for growth and development at high Cu concentrations. In contrast, at low Cu levels the Δccsp mutant growth and development is like wild type, suggesting that Ccsp is not needed under these conditions (Fig. 6 and Fig. S4, ESI†). Introduction of ccsp on a plasmid (pCcsp-1) under transcriptional control of its own promoter, restores Cu tolerance in the Δccsp mutant to wild type levels in all media tested (Fig. 6 and Fig. S4, ESI†). In liquid media, a similar pattern to that observed on solid media is found (Fig. S4, ESI†) with a reduced tolerance for Cu in terms of the biomass produced in the Δccsp mutant as illustrated for Bennetts-glucose medium (Fig. 6C).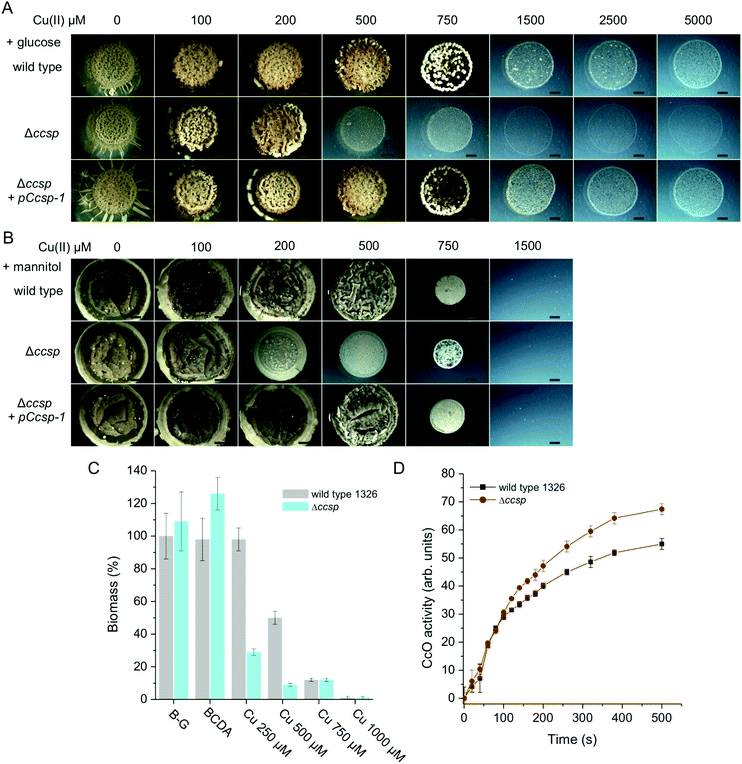 | ||
| Fig. 6 The effect of Ccsp on the growth and development of S. lividans at 30 °C. Cu tolerance after 6 days growth of the wild type parent strain, the Δccsp mutant strain and the Δccsp mutant strain complemented with the pCcsp-1 plasmid on defined agar media with (A) glucose and (B) mannitol as the sole carbon source. Cu(II) concentrations as indicated. All images are the same magnification with a scale bar of 2 mm. (C) Biomass production after 32 h in liquid Bennetts-glucose (B-G) cultures for the wild type and the Δccsp mutant strain in the presence of the Cu chelator BCDA and various concentrations of Cu(II) citrate. The dry weight biomass of the wild type strain in the B-G culture was set at 100%. (D) CcO oxidase activity at 24 h growth on B-G agar detected by the TMPD assay. Average pixel intensity of the indophenol blue stained mycelium were calculated using ImageJ software38 and expressed in arbitrary units. Experiments were carried out in triplicate. | ||
Under Cu homeostasis, enzymes requiring Cu for their activity such as CcO or GlxA in S. lividans obtain their Cu from a Cu trafficking pathway that involves at least two Cu metallochaperones, ECuC and Sco.8–10,19 To investigate whether Ccsp influences this extracytoplasmic Cu trafficking pathway the activity of CcO under low exogenous Cu concentrations was determined in the wild type strain and the Δccsp mutant on various media. As illustrated on Bennetts-glucose agar (Fig. 6D) the CcO activity of the Δccsp mutant is identical to the wild type, therefore demonstrating that Ccsp is not participating in the Cu trafficking pathway for maturation of CcO.
A cytosolic Cu metallochaperone can load Cu+ ions to Ccsp
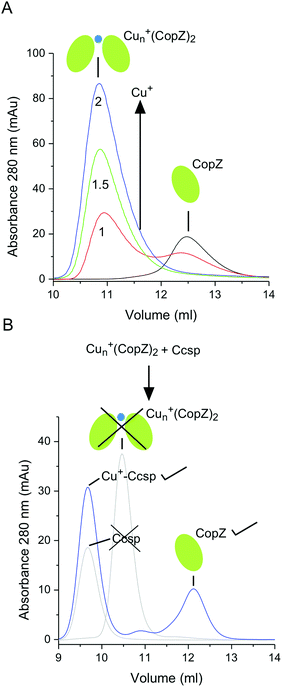
The movement and trafficking of Cu in the cytosol of S. lividans under homeostasis and stress has been shown to involve CopZ-like Cu metallochaperones.16,17 These Atx-1 homologues possess a βαββαβ-fold and a MXCXXC metal binding motif that utilizes the two Cys residues for bis-cysteinate Cu+ ion coordination.62 Therefore, we investigated in vitro whether a CopZ plays a role in Cu+ trafficking to and from Ccsp. In the absence of Cu+, CopZ-3079 (hereafter CopZ) elutes from a size exclusion column at a volume of 12.5 ml (Fig. 7A), which based on the column calibration is consistent with a monomer species (Mr ∼ 8 kDa). At Cu+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CopZ ratios higher than 1
CopZ ratios higher than 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 a shift in the elution profile (∼11 ml) is observed, which based on the column calibration is consistent with the presence of a dimer species. Binding of Cu+ to Bacillus subtilis CopZ has been shown to be a very complex process with initial binding of Cu+ resulting in dimerization to form a Cu+–(CopZ)2 species which has the capacity to bind three further Cu+ ions at the monomer interface to create a Cu4+–(CopZ)2 species as the addition of stoichiometric Cu+ ions increase.63–65 In the present work with S. lividans CopZ we have not determined how many Cu+ ions, n, are at the monomer interface (Cun+–(CopZ)2). However, based on the size exclusion chromatography profile a dimer species is predominately formed at >1 Cu+/CopZ.
1 a shift in the elution profile (∼11 ml) is observed, which based on the column calibration is consistent with the presence of a dimer species. Binding of Cu+ to Bacillus subtilis CopZ has been shown to be a very complex process with initial binding of Cu+ resulting in dimerization to form a Cu+–(CopZ)2 species which has the capacity to bind three further Cu+ ions at the monomer interface to create a Cu4+–(CopZ)2 species as the addition of stoichiometric Cu+ ions increase.63–65 In the present work with S. lividans CopZ we have not determined how many Cu+ ions, n, are at the monomer interface (Cun+–(CopZ)2). However, based on the size exclusion chromatography profile a dimer species is predominately formed at >1 Cu+/CopZ.
Metal trafficking from a donor to an acceptor has been shown in vitro to occur through transient interactions that initiate a ligand-exchange mechanism to facilitate the transfer of the metal (i.e. the metal is never dissociated into solution). Therefore, by simply mixing a donor and acceptor in vitro and having a robust readout, it can be determined whether metal transfer between partners occurs. Ccsp and Cu+–Ccsp elute from the size exclusion column at a volume of ∼9 ml (Fig. 7B) and thus do not overlap with the elution profiles of CopZ or the Cun+–(CopZ)2. Upon mixing Cun+–(CopZ)2 with Ccsp the elution profile shown in Fig. 7B is observed. This indicates that the Cun+–(CopZ)2 donor no longer has Cu+ bound and instead Cu+ is transferred to Ccsp. The reverse experiment, whereby Cu+–Ccsp is mixed with CopZ was conducted in a similar manner, with incubation periods of >1 h, but with no indication that Cu transfer occurs (i.e. no elution peak at ∼11 ml for a Cun+–(CopZ)2 species). Taken together these results support the notion that in vitro CopZ can transfer Cu+ ions to Ccsp, but the release of Cu+ from Ccsp to CopZ does not occur under the conditions employed.
Discussion
Many organisms possess Cu resistance systems that have been identified and characterised to varying extents. The bacterial cytosol has no known metabolic requirement for Cu and therefore Cu storage proteins had been thought not to exist. The unprecedented discovery of the Csp family and the extensive distribution of cytosolic Csp3 members in the Tree of Life (Fig. 1) offers the tantalising possibility that new layers of Cu resistance or a possible cytosolic requirement for Cu exists amongst many bacteria that are yet to be fully understood. S. lividans is highly dependent on the bioavailability of Cu for its development.8 In this context, we have previously reported the transcriptional response of the CsoR/CopZ/P1-type ATPase trafficking and efflux system during Cu overload and undertaken an extensive structural, thermodynamic and kinetic characterisation of this system.16,17,66,67 A notable finding from the transcriptional studies (RNA-seq) was that many more genes were up- and down-regulated in response to Cu overload other than just the cytosolic CsoR/CopZ/ATPase efflux system.16 At that time the Csp family had not been discovered (neither its gene in S. lividans annotated), but we revealed a 6-fold upregulation of SLI_RS17250 (DUF4396) under Cu stress, which based on transcriptional data with the ΔcsoR mutant, was determined not to be under transcriptional control of CsoR.16 Analysis of the SLI_RS17250 protein sequence predicts at least four transmembrane helices with a CXXXC motif at the start of the first transmembrane helix as well as a His-rich N-terminal sequence (13 His in total). BLAST search against the PDB did not reveal any close homologues of the SLI_RS17250 sequence with known structure, and it likely presents a distinct family. Intriguingly, however, some remote homology can be established with the substrate binding S-subunits of the energy coupling factor (ECF) family of micronutrient transporters,68 and there is a tentative link to the SLC11/NRAMP family, which are involved in transition metal ion transport.69,70 Whether this uncharacterised membrane protein has a functional link with Ccsp warrants further investigation.The overall structure of the Ccsp protomer shows high similarity with other Csp3 members that have recently had their structures determined.33 No evidence for disulfide bond formation is observed and a maximum of 20 Cu+ ions are found to be coordinated in a Ccsp protomer. Therefore Ccsp has the highest Cu+ binding capacity of any Csp3 member so far characterised.33 Furthermore, Ccsp is the first Csp3 member to reveal a unique coordination role of a highly-conserved Asp residue (Asn in MtCsp333). This residue is positioned at the end of the Cys cage that harnesses Cu+ ions 1 to 13, which are all coordinated by two Cys ligands (Fig. 5A and B). Asp61 is situated such that it breaks this coordination trend by providing coordination to Cu+ ions 14 and 15 through its Oδ1 and Oδ2 carboxylate atoms, respectively. The remaining 5 Cu+ ions form a separate cluster utilizing coordination by three conserved His residues and four Cys residues. The three His residues clearly form an entrance to the more solvent accessible end of the four helix bundle core and in addition to their coordination role may also play a part in partner recognition to assist Cu+-loading into Ccsp.
Apparent KCu values for Cu metallochaperones utilising bis-cysteinate coordination are in the range of 10−17 to 10−18 M.61 It is perhaps therefore no surprise that Csp members with multiple bis-cysteinate coordination also have KCu values in this region.32,33 Ccsp is no different, although we note that our KCu values using BCA as the affinity probe are at the lower end to those determined for other Csp3 members (3.1 × 10−17 M vs. 6 × 10−18 M, average for MtCsp3 and BsCsp333). Interestingly, cooperativity of Cu+ ion binding is observed for Ccsp with an average Hill coefficient (n) value of 1.9. Cooperative binding has been reported for Csp1 but perhaps surprisingly is not observed for MtCsp3 or BsCsp3.33 A discussion of the apparent cooperativity of Cu+ binding to Ccsp maybe prefixed with a note of caution given that the [Cu+free], which are exceedingly low (Fig. 4B), are calculated indirectly from binding to BCA. However, putting such caveats aside and concluding cooperativity is present for Ccsp, then we may suggest the mechanistic basis for this phenomenological result may lie in the observation by Dennison and co-workers that Cu+ clusters of the type [Cu4(S-Cys)4] are thermodynamically favoured.60
The ability of the Ccsp tetramer to bind 80 Cu+ ions leads to the question of how Ccsp acquires Cu+ in the stringently Cu controlled surroundings of the cytosol. Under normal growth conditions three CopZ/ATPase couples are present in S. lividans at a basal level that act to buffer the Cu+ ion concentration.16,17 Two of these couples are under direct transcriptional control by a CsoR, whereas control of the third couple is not presently known, but is upregulated under Cu stress.16 A fourth couple is encoded on the genome and is also under control of the CsoR but under homeostasis is not constitutively present.17In vitro two of the CopZ proteins have been determined to transfer Cu+ to the CsoR in a unidirectional manner.17 Therefore in vivo this would induce transcription of the three CopZ/ATPases couples under CsoR control, which together with the fourth non-CsoR controlled couple creates a high capacity for Cu resistance under Cu stress. The ability of CopZ to traffic Cu+ to CsoR and P1-type ATPases indicates an inherent promiscuity in these small chaperone proteins for off-loading their metal cargo. This indiscriminate nature is also apparent for CopZ with Ccsp, where Cu+ is transferred from the Cun+–(CopZ)2 species to Ccsp as evidenced by the reformation of the apo-CopZ monomer (Fig. 7B). In contrast, under stoichiometric conditions apo-CopZ is unable to remove Cu+ from Cu+–Ccsp in a physiologically meaningful time frame as has also reported for BsCsp3 with its cognate BsCopZ.33 A KCu value of 3.9 × 10−18 M has been determined for this particular S. lividans CopZ used in this work under the same buffer conditions as employed in the transfer experiments with Ccsp.17 This value is lower than the KCu value determined for Ccsp and thus from a thermodynamic perspective the transfer can be viewed as being unfavourable. However, other factors such as the tuning of the reactivity of the ligands involved in the transfer of Cu+ from donor to acceptor are important considerations and have been shown to influence the directionality of transfer. Such an example has been reported for the transfer of Cu+ from CopZ to the CsoR, where the Cys residues in the CXXXC motif of CopZ have been optimised to favour Cu release to the acceptor (CsoR).17
Building on the above discussion it is now pertinent to address the contribution of Ccsp to Cu resistance in S. lividans. The in vivo data show that Ccsp is not required for Cu homeostasis at Cu concentrations up to several hundred μM depending on the medium used (Fig. 6A and B). This observation is further demonstrated by the CcO activity data, which precludes a downstream role for Ccsp in supplying Cu+ to the extracytoplasmic environment to be utilised by the Cu-chaperones Sco and ECuC for metalation of CcO and GlxA.8,10,19 The CopZ/ATPase couples appear sufficient to maintain the control of cytoplasmic Cu levels. However, as exogenous Cu concentrations rise above 200 μM, a clear phenotype for Ccsp is observed albeit with a range limit that is strongly medium dependent (Fig. 6). Re-analysed RNA-seq data16 reveals transcription of ccsp is up-regulated 5-fold in liquid defined medium supplemented with 400 μM Cu and thus fits with the phenotype in Fig. 6 showing that Ccsp becomes essential for growth and development in the 200–500 μM Cu range. Importantly, in contrast to three out of the four CopZ/ATPase couples, the Ccsp expression is not under the control of CsoR as demonstrated by the absence of a consensus CsoR binding site in the ccsp promoter region and expression induction in the csoR mutant.16 This all suggests that a second layer of Cu responsive transcription is operating on top of the CsoR regulon in S. lividans and becomes operative at more extreme Cu concentrations to express ccsp.
Whilst the up-regulation of ccsp may well be an act of last resort to survival under high Cu stress in S. lividans, there remains several lines of further enquiry. Is Ccsp simply acting in a storage capacity, taking delivery of Cu+ from CopZ when the CsoR regulon becomes saturated? If this is the case then on returning to homeostasis there will be a large store of Cu in the cytosol for which the requirement and mechanism of Cu release from Ccsp is presently unclear. In addition to a possible interaction with SLI_RS17250 (DUF4396), it is worth considering that nonmethanotrophic bacteria may possess Cu scavenging systems, like Mb in methanotrophs. To this end a diisonitrile compound produced from a non-ribosomal peptide synthetase in S. thioluteus has recently been reported and shown to have a chalkophore function (i.e. Cu-import into the cytosol).71 This discovery could suggest that chalkophores are more widespread than originally considered and an interplay with Ccsps in nonmethanotrophic bacteria certainly requires further investigation.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
MLS acknowledges the Eastern Arc consortium for a SynBio PhD studentship. AKC was funded by a University of Essex Silberrad PhD scholarship. The authors would like to thank Diamond Light Source for beamtime (BAG proposal mx13467), and the staff of beamline I02 for assistance with data collection. We acknowledge the European Synchrotron Radiation Facility (ESRF) for provision of synchrotron radiation facilities at beamline ID29, accessed via the “Fundamental and Applied Aspects of Radiation Damage” BAG.References
- F. U. Battistuzzi, A. Feijao and S. B. Hedges, BMC Evol. Biol., 2004, 4, 44 CrossRef PubMed.
- F. U. Battistuzzi and S. B. Hedges, Mol. Biol. Evol., 2009, 26, 335–343 CrossRef CAS PubMed.
- K. Flardh and M. J. Buttner, Nat. Rev. Microbiol., 2009, 7, 36–49 CrossRef PubMed.
- D. Claessen, D. E. Rozen, O. P. Kuipers, L. Sogaard-Andersen and G. P. van Wezel, Nat. Rev. Microbiol., 2014, 12, 115–124 CrossRef CAS PubMed.
- J. Anne, B. Maldonado, J. Van Impe, L. Van Mellaert and K. Bernaerts, J. Biotechnol., 2012, 158, 159–167 CrossRef CAS PubMed.
- B. J. Keijser, G. P. van Wezel, G. W. Canters, T. Kieser and E. Vijgenboom, J. Mol. Microbiol. Biotechnol., 2000, 2, 565–574 CAS.
- M. Fujimoto, A. Yamada, J. Kurosawa, A. Kawata, T. Beppu, H. Takano and K. Ueda, Microb. Biotechnol., 2012, 5, 477–488 CrossRef PubMed.
- K. L. Blundell, M. T. Wilson, D. A. Svistunenko, E. Vijgenboom and J. A. Worrall, Open Biol., 2013, 3, 120163 CrossRef PubMed.
- A. K. Chaplin, M. L. Petrus, G. Mangiameli, M. A. Hough, D. A. Svistunenko, P. Nicholls, D. Claessen, E. Vijgenboom and J. A. Worrall, Biochem. J., 2015, 469, 433–444 CrossRef CAS PubMed.
- M. L. Petrus, E. Vijgenboom, A. K. Chaplin, J. A. Worrall, G. P. van Wezel and D. Claessen, Open Biol., 2016, 6, 150149 CrossRef PubMed.
- Y. Fu, F. M. Chang and D. P. Giedroc, Acc. Chem. Res., 2014, 47, 3605–3613 CrossRef CAS PubMed.
- L. Macomber and J. A. Imlay, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 8344–8349 CrossRef CAS PubMed.
- B. E. Kim, T. Nevitt and D. J. Thiele, Nat. Chem. Biol., 2008, 4, 176–185 CrossRef CAS PubMed.
- C. Rademacher and B. Masepohl, Microbiology, 2012, 158, 2451–2464 CrossRef CAS PubMed.
- J. M. Arguello, D. Raimunda and T. Padilla-Benavides, Front. Cell. Infect. Microbiol., 2013, 3, 73 Search PubMed.
- S. Dwarakanath, A. K. Chaplin, M. A. Hough, S. Rigali, E. Vijgenboom and J. A. Worrall, J. Biol. Chem., 2012, 287, 17833–17847 CrossRef CAS PubMed.
- A. K. Chaplin, B. G. Tan, E. Vijgenboom and J. A. Worrall, Metallomics, 2015, 7, 145–155 RSC.
- T. Liu, A. Ramesh, Z. Ma, S. K. Ward, L. Zhang, G. N. George, A. M. Talaat, J. C. Sacchettini and D. P. Giedroc, Nat. Chem. Biol., 2007, 3, 60–68 CrossRef CAS PubMed.
- K. L. Blundell, M. A. Hough, E. Vijgenboom and J. A. Worrall, Biochem. J., 2014, 459, 525–538 CrossRef CAS PubMed.
- K. J. Waldron and N. J. Robinson, Nat. Rev. Microbiol., 2009, 7, 25–35 CrossRef CAS PubMed.
- K. J. Waldron, J. C. Rutherford, D. Ford and N. J. Robinson, Nature, 2009, 460, 823–830 CrossRef CAS PubMed.
- A. W. Foster, D. Osman and N. J. Robinson, J. Biol. Chem., 2014, 289, 28095–28103 CrossRef CAS PubMed.
- Z. Ma, F. E. Jacobsen and D. P. Giedroc, Chem. Rev., 2009, 109, 4644–4681 CrossRef CAS PubMed.
- K. J. Waldron, S. J. Firbank, S. J. Dainty, M. Perez-Rama, S. Tottey and N. J. Robinson, J. Biol. Chem., 2010, 285, 32504–32511 CrossRef CAS PubMed.
- H. J. Kim, D. W. Graham, A. A. DiSpirito, M. A. Alterman, N. Galeva, C. K. Larive, D. Asunskis and P. M. Sherwood, Science, 2004, 305, 1612–1615 CrossRef CAS PubMed.
- R. Balasubramanian, S. M. Smith, S. Rawat, L. A. Yatsunyk, T. L. Stemmler and A. C. Rosenzweig, Nature, 2010, 465, 115–119 CrossRef CAS PubMed.
- A. El Ghazouani, A. Basle, S. J. Firbank, C. W. Knapp, J. Gray, D. W. Graham and C. Dennison, Inorg. Chem., 2011, 50, 1378–1391 CrossRef CAS PubMed.
- A. El Ghazouani, A. Basle, J. Gray, D. W. Graham, S. J. Firbank and C. Dennison, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 8400–8404 CrossRef CAS PubMed.
- A. A. DiSpirito, J. D. Semrau, J. C. Murrell, W. H. Gallagher, C. Dennison and S. Vuilleumier, Microbiol. Mol. Biol. Rev., 2016, 80, 387–409 CrossRef PubMed.
- R. Balasubramanian, G. E. Kenney and A. C. Rosenzweig, J. Biol. Chem., 2011, 286, 37313–37319 CrossRef CAS PubMed.
- L. M. Dassama, G. E. Kenney, S. Y. Ro, E. L. Zielazinski and A. C. Rosenzweig, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 13027–13032 CrossRef CAS PubMed.
- N. Vita, S. Platsaki, A. Basle, S. J. Allen, N. G. Paterson, A. T. Crombie, J. C. Murrell, K. J. Waldron and C. Dennison, Nature, 2015, 525, 140–143 CrossRef CAS PubMed.
- N. Vita, G. Landolfi, A. Basle, S. Platsaki, J. Lee, K. J. Waldron and C. Dennison, Sci. Rep., 2016, 6, 39065 CrossRef CAS PubMed.
- T. Kieser, M. J. Bibb, M. J. Buttner, K. F. Chater and D. A. Hopwood, Pratcical Streptomyces Genetics, John Innes Foundation, Norwich, UK, 2000 Search PubMed.
- J. L. Larson and C. L. Hershberger, Plasmid, 1986, 15, 199–209 CrossRef CAS PubMed.
- G. N. Green and R. B. Gennis, J. Bacteriol., 1983, 154, 1269–1275 CAS.
- J. P. Mueller and H. W. Taber, J. Bacteriol., 1989, 171, 4967–4978 CrossRef CAS PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS PubMed.
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers and D. J. Lipman, J. Mol. Biol., 1990, 215, 403–410 CrossRef CAS PubMed.
- M. Johnson, I. Zaretskaya, Y. Raytselis, Y. Merezhuk, S. McGinnis and T. L. Madden, Nucleic Acids Res., 2008, 36, W5–W9 CrossRef CAS PubMed.
- P. Cruz-Morales, E. Vijgenboom, F. Iruegas-Bocardo, G. Girard, L. A. Yanez-Guerra, H. E. Ramos-Aboites, J. L. Pernodet, J. Anne, G. P. van Wezel and F. Barona-Gomez, Genome Biol. Evol., 2013, 5, 1165–1175 CrossRef PubMed.
- E. W. Sayers, T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, M. Feolo, L. Y. Geer, W. Helmberg, Y. Kapustin, D. Landsman, D. J. Lipman, T. L. Madden, D. R. Maglott, V. Miller, I. Mizrachi, J. Ostell, K. D. Pruitt, G. D. Schuler, E. Sequeira, S. T. Sherry, M. Shumway, K. Sirotkin, A. Souvorov, G. Starchenko, T. A. Tatusova, L. Wagner, E. Yaschenko and J. Ye, Nucleic Acids Res., 2009, 37, D5–D15 CrossRef CAS PubMed.
- K. Katoh, K. Misawa, K. Kuma and T. Miyata, Nucleic Acids Res., 2002, 30, 3059–3066 CrossRef CAS PubMed.
- J. Castresana, Mol. Biol. Evol., 2000, 17, 540–552 CrossRef CAS PubMed.
- M. N. Price, P. S. Dehal and A. P. Arkin, PLoS One, 2010, 5, e9490 Search PubMed.
- S. Whelan and N. Goldman, Mol. Biol. Evol., 2001, 18, 691–699 CrossRef CAS PubMed.
- H. Shimodaira and M. Hasegawa, Mol. Biol. Evol., 1999, 16, 1114–1116 CrossRef CAS.
- G. L. Ellman, Arch. Biochem. Biophys., 1959, 82, 70–77 CrossRef CAS PubMed.
- Z. Xiao, P. S. Donnelly, M. Zimmermann and A. G. Wedd, Inorg. Chem., 2008, 47, 4338–4347 CrossRef CAS PubMed.
- P. Bagchi, M. T. Morgan, J. Bacsa and C. J. Fahrni, J. Am. Chem. Soc., 2013, 135, 18549–18559 CrossRef CAS PubMed.
- W. Kabsch, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 125–132 CrossRef CAS PubMed.
- P. R. Evans and G. N. Murshudov, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2013, 69, 1204–1214 CrossRef CAS PubMed.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef CAS PubMed.
- K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2006, 62, 1002–1011 CrossRef PubMed.
- P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- G. N. Murshudov, A. A. Vagin and E. J. Dodson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1997, 53, 240–255 CrossRef CAS PubMed.
- I. W. Davis, A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall, 3rd, J. Snoeyink, J. S. Richardson and D. C. Richardson, Nucleic Acids Res., 2007, 35, W375–W383 CrossRef PubMed.
- E. Krissinel, J. Mol. Biochem., 2012, 1, 76–85 Search PubMed.
- D. A. Benson, M. Cavanaugh, K. Clark, I. Karsch-Mizrachi, D. J. Lipman, J. Ostell and E. W. Sayers, Nucleic Acids Res., 2013, 41, D36–D42 CrossRef CAS PubMed.
- A. Basle, S. Platsaki and C. Dennison, Angew. Chem., Int. Ed. Engl., 2017, 56, 8697–8700 CrossRef CAS PubMed.
- Z. Xiao, J. Brose, S. Schimo, S. M. Ackland, S. La Fontaine and A. G. Wedd, J. Biol. Chem., 2011, 286, 11047–11055 CrossRef CAS PubMed.
- A. C. Rosenzweig, D. L. Huffman, M. Y. Hou, A. K. Wernimont, R. A. Pufahl and T. V. O'Halloran, Structure, 1999, 7, 605–617 CrossRef CAS PubMed.
- M. A. Kihlken, A. P. Leech and N. E. Le Brun, Biochem. J., 2002, 368, 729–739 CrossRef CAS PubMed.
- S. Hearnshaw, C. West, C. Singleton, L. Zhou, M. A. Kihlken, R. W. Strange, N. E. Le Brun and A. M. Hemmings, Biochemistry, 2009, 48, 9324–9326 CrossRef CAS PubMed.
- K. L. Kay, C. J. Hamilton and N. E. Le Brun, Metallomics, 2016, 8, 709–719 RSC.
- B. G. Tan, E. Vijgenboom and J. A. Worrall, Nucleic Acids Res., 2014, 42, 1326–1340 CrossRef CAS PubMed.
- T. V. Porto, M. A. Hough and J. A. Worrall, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2015, 71, 1872–1878 CAS.
- D. J. Slotboom, Nat. Rev. Microbiol., 2014, 12, 79–87 CrossRef CAS PubMed.
- I. A. Ehrnstorfer, E. R. Geertsma, E. Pardon, J. Steyaert, R. Dutzler and D. J. Slotboom, Nat. Struct. Mol. Biol., 2014, 21, 990–996 CAS.
- I. A. Ehrnstorfer, C. Manatschal, F. M. Arnold, J. Laederach and R. Dutzler, Nat. Commun., 2017, 8, 14033 CrossRef CAS PubMed.
- L. Wang, M. Zhu, Q. Zhang, X. Zhang, P. Yang, Z. Liu, Y. Deng, Y. Zhu, X. Huang, L. Han, S. Li and J. He, ACS Chem. Biol., 2017, 12, 3067–3075 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7mt00299h |
| This journal is © The Royal Society of Chemistry 2018 |