Tracking iron-associated proteomes in pathogens by a fluorescence approach†‡
Nan
Jiang§
a,
Tianfan
Cheng§
b,
Minji
Wang§
 a,
Godfrey Chi-Fung
Chan
c,
Lijian
Jin
a,
Godfrey Chi-Fung
Chan
c,
Lijian
Jin
 *b,
Hongyan
Li
a and
Hongzhe
Sun
*b,
Hongyan
Li
a and
Hongzhe
Sun
 *a
*a
aDepartment of Chemistry, The University of Hong Kong, Pokfulam Road, Hong Kong SAR, P. R. China. E-mail: hsun@hku.hk; Tel: +852 2859 8974
bDiscipline of Periodontology, Faculty of Dentistry, The University of Hong Kong, Hong Kong SAR, P. R. China. E-mail: ljjin@hku.hk
cDepartment of Paediatrics and Adolescent Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, 21 Sassoon Road, Pokfulam, Hong Kong
First published on 14th December 2017
Abstract
Porphyromonas gingivalis is a key oral anaerobic bacterium involved in human periodontitis, which may affect up to 15% adults worldwide. Using the membrane permeable fluorescent probe Fe-TRACER, we identified 17 iron-associated proteins in Porphyromonas gingivalis. We demonstrated the specific binding of the probe towards iron-associated proteins using transferrin as an example and provided an X-ray structure of the fluorescent probe-bound transferrin. Our study provides a basis for the understanding of iron homeostasis in pathogens, and our approach based on the integration of fluorescence imaging with proteomics and bioinformatics can be readily extended to mine other metalloproteomes in microbials.
Significance to metallomicsPorphyromonas gingivalis is a keystone oral pathogen in periodontitis that highly relies on the acquisition of iron. To obtain a more complete spectrum for the iron–proteome, a fluorescent probe has been utilized to image living cells and visualize iron–proteomes on a SDS-Page for subsequent protein identification. The probe–protein binding mechanism is validated by transferrin with a crystal structure, whereas the probe can be further anchored on the targeted protein for stronger fluorescence. The finding extended our existing fluorescent probe-based approach to tracking iron-associated proteomes in living pathogens. |
Introduction
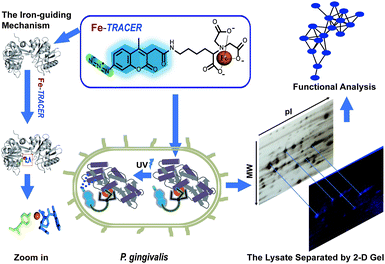
Periodontitis is a chronic inflammatory disease that affects approximately 10–15% adults worldwide.1Porphyromonas gingivalis (P. gingivalis), a pathogenic Gram-negative bacterium, is considered as a keystone microbial species that causes periodontitis and related systematic diseases.2 It strictly requires heme as a source of iron as well as protoporphyrin IX (PPIX), obtained from hemoproteins, hemoglobin, heme-albumin, and hemopexin,3 for its survival and pathogenesis in the progress of periodontitis.4P. gingivalis is also equipped with a full arsenal of heme-binding proteins.3,5 Although multiple dimension capillary HPLC and tandem mass spectrometry6,7 allows the detection of proteomes in P. gingivalis, the proteome-wide identification of iron-associated proteins has not been achieved to date. Several fluorescent iron probes, including PG-SK8 and calcein,9 are commercially available; however, these probes can only detect chelatable, or free, iron and are unable to track iron-associated proteins.Recently, a novel fluorescent probe with a genetically fused His6-tag (Ni2+-NTA-AC, renamed as Ni2+-TRACER) has been successfully utilized to image and track metal-binding proteins in live cells.10–12 Herein, we developed an iron fluorescent probe, i.e., Fe-TRACER, to track iron-associated proteins in P. gingivalis (ATCC 33277). We show that the probe can specifically recognize iron-associated proteins, as evidenced by the X-ray structure of probe-bound transferrin, leading to significant fluorescence enhancement. We demonstrate that Fe-TRACER can readily enter live P. gingivalis cells to target iron-associated proteins. Upon the photoactivation of the arylazide group of the probe, the labelled proteins can be anchored to the probe, thereby allowing subsequent protein identification and analysis (Fig. 1).
Results and discussions
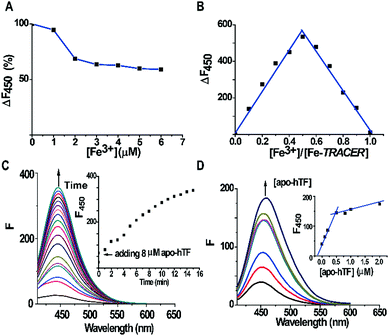
The fluorescent ligand TRACER was synthesized by conjugating a nitrilotriacetate (NTA) moiety with a coumarin fluorophore and an arylazide group as previously reported.11 The Fe-TRACER probe was then generated by reacting TRACER with equimolar amounts of Fe3+ (as FeCl3) in an aqueous solution (pH 7.4). It exhibits emission at 450 nm upon excitation at 350 nm (Fig. S2, ESI‡). The stepwise addition of Fe3+ to 5 μM TRACER solution caused a gradual decrease in fluorescence; at equimolar amounts of Fe3+, the fluorescence was quenched by ca. 40% (Fig. 2A). By fitting the titration curve with the Ryan–Weber equation,13 the dissociation constant (Kd) was determined to be 66 ± 16 nM, whereas the dissociation kinetics (koff) was determined to be 0.020 ± 0.001 s−1 by fitting the time-dependent change of fluorescence at 450 nm with the addition of NTA to 5 μM Fe-TRACER solution (Fig. S3, ESI‡). Both values indicate strong binding between TRACER and Fe3+. In addition, the probe is kinetically stable, which is a prerequisite for tracking iron-associated proteomes. The formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex between TRACER and Fe3+ was evidenced by Job's plot (Fig. 2B) based on the fluorescence response of TRACER to Fe3+ at 450 nm. The binding stoichiometry was further confirmed by the observation of a peak at m/z 591.1 in electrospray ionization mass spectrometry (ESI-MS), in accordance with the calculated m/z value of 592.3 (Fig. S1, ESI‡).
1 complex between TRACER and Fe3+ was evidenced by Job's plot (Fig. 2B) based on the fluorescence response of TRACER to Fe3+ at 450 nm. The binding stoichiometry was further confirmed by the observation of a peak at m/z 591.1 in electrospray ionization mass spectrometry (ESI-MS), in accordance with the calculated m/z value of 592.3 (Fig. S1, ESI‡).
To examine the ability of Fe-TRACER to label iron-associated proteins in vitro, an iron-binding protein, human transferrin (hTF),14 was selected as a showcase study. A gradual increase in fluorescence intensity at 450 nm was noted upon incubation of 8 molar equivalents (8 μM) of apo-hTF with the probe for 15 min, resulting in a 13-fold increase in fluorescence (Fig. 2C). Stepwise titration of apo-hTF to 1 μM Fe-TRACER solution resulted in an increase in fluorescence at 450 nm, until a plateau is reached at a molar ratio of apo-hTF to Fe-TRACER of 0.5 (Fig. 2D), suggesting that the binding stoichiometry of apo-hTF to Fe-TRACER is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
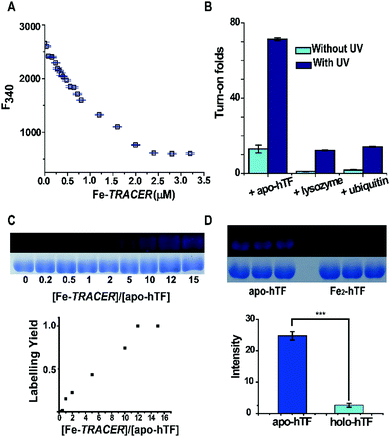
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, in agreement with the known binding of two Fe3+ ions to one hTF protein. Subsequently, the binding affinity (Kd) of apo-hTF to Fe-TRACER was determined to be 0.68 ± 0.11 μM by fitting the curve of Fe-TRACER induced fluorescence decrease of tryptophan of apo-hTF at 340 nm (Fig. 3A). In contrast, negligible changes in fluorescence were observed when Fe-TRACER was incubated with Fe2-hTF (i.e., the two iron-binding sites being fully occupied) under identical conditions (Fig. S4, ESI‡). These data strongly suggest that the probe specifically targets proteins through Fe3+, leading to significant fluorescence increase.
2, in agreement with the known binding of two Fe3+ ions to one hTF protein. Subsequently, the binding affinity (Kd) of apo-hTF to Fe-TRACER was determined to be 0.68 ± 0.11 μM by fitting the curve of Fe-TRACER induced fluorescence decrease of tryptophan of apo-hTF at 340 nm (Fig. 3A). In contrast, negligible changes in fluorescence were observed when Fe-TRACER was incubated with Fe2-hTF (i.e., the two iron-binding sites being fully occupied) under identical conditions (Fig. S4, ESI‡). These data strongly suggest that the probe specifically targets proteins through Fe3+, leading to significant fluorescence increase.
To further validate the selectivity of Fe-TRACER towards iron-binding proteins, the probe was incubated with lysozyme and ubiquitin under identical conditions. As shown in Fig. 3B, the fluorescence was slightly enhanced for both lysozyme (1 fold) and ubiquitin (2 fold). Interestingly, upon UV irradiation of the samples at 365 nm for 20 min, about 71-fold increase in fluorescence was found for apo-hTF upon labelling by Fe-TRACER due to full activation of the arylazide group of TRACER.11 We have previously demonstrated15 that the activation of the arylazide group is prerequisite for fluorescence activation upon protein labelling by the probe. The relative low turn-on effects without additional UV irradiation are due to inefficient activation of the arylazide group during fluorescence spectral recording as the excitation wavelength falls in the UV range. The labelling efficiency was evaluated by monitoring the fluorescence intensities of 10 μM apo-hTF relative to those of Coomassie Blue staining after incubation with different amounts of the probe, followed by UV irradiation for 20 min prior to SDS-PAGE and fluorescence imaging. It is noted that about 50% of the apo-hTF is labelled in the presence of 5 molar equivalents (50 μM) of Fe-TRACER (Fig. 3C). In contrast, the labelling efficiency for Fe2-hTF by the probe is very low under identical conditions as judged by more than 9-fold of apo-hTF being labelled by Fe-TRACER as compared to Fe2-hTF based on the protein intensities exhibited in the fluorescence images (Fig. 3D). Considering the reducing environment inside cells,16 we examined the effects of reduced glutathione (GSH) on the labelling capability of Fe-TRACER towards apo-hTF. Even in the presence of 100 molar equivalents of GSH (5 mM), about 30% of apo-hTF still remained to be labelled by the probe (Fig. S5, ESI‡).
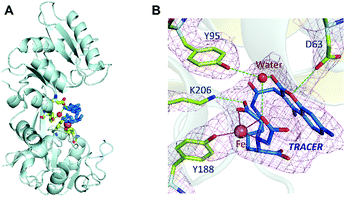
Binding of Fe-TRACER to hTF was further investigated by X-ray crystallography. We first crystalized mono-ferric (C-lobe) hTF, i.e., FeC-hTF (PDB: 4× 1B)17 as described previously, and Fe-TRACER was added to a cryo-protectant for FeC-hTF crystals. The structure of the Fe-TRACER-bound hTF (Fe-TRACER-hTF) was then resolved at 2.85 Å. The TRACER sites were identified based on the positive peaks present on the mFo–DFc map after molecular replacement. Similar to FeC-hTF, the C-lobe of Fe-TRACER-hTF remains closed, with Fe3+ situated at the metal-binding pocket. A distinguished positive peak (12σ) on mFo–DFc map was found near Tyr188 after molecular replacement and was confirmed as Fe3+via examination of the mFo–DFc and anomalous maps after removing the metal ions from the final model (Table S2, ESI‡). Moreover, two very large areas of positive electron density were observed near Tyr188 in the N-lobe and on the surface of the C-lobe near Arg602 and were identified as TRACER (Fig. S7, ESI‡). No intact azide group was observed in crystal structure and its remnant nitrogen atom was modelled as NH2.
Surprisingly, the NTA moiety of the probe coordinates to Fe3+ by only two carboxylates and one nitrogen, with the bond lengths of 2.0–2.2 Å. At the iron-binding site, Tyrosine (Tyr188) coordinates to Fe3+ with an Fe⋯O bond length of 2.1 Å. The large size of the probe may restrict other amino acids of hTF from participating in coordination. The binding of the probe to hTF was further stabilized via hydrogen bonds with Lys206, Asp63, and Tyr95 (through a water molecule, Fig. 4B). Stabilization of the bond by Asp63 was also observed with our previously reported BiNFeC-hTF probe.18 However, in the current structure, the site for a synergistic bicarbonate ion is sacrificed by the spatial needs of the bulky ligand of TRACER. Notably, the Fe-TRACER molecule folds like a sandwich such that the NTA group flips back onto the coumarin moiety via the flexible (CH2)4-linker. To our surprise, even the TRACER molecule located on the surface of hTF exhibits the sandwich-like structure (Fig. S7, ESI‡) although this phenomenon cannot be ascribed to the spatial availability of lobe gap. Based on the X-ray crystallography study, we demonstrate clearly that Fe-TRACER targets iron-associated proteins through iron-guiding.
We then applied the Fe-TRACER probe to track iron-associated proteins in pathogens that might directly or potentially bind with the probe. Due to the importance of iron for the survival and pathogenies of P. gingivalis,4,19 we selected this pathogen as a showcase study. Previously, we have demonstrated that only the metal-TRACER complex but not the TRACER itself can enter live cells to label intracellular proteins.15 This advantage enabled the non-specific binding of TRACER inside cells to be eliminated. P. gingivalis cells were incubated with 50 μM Fe-TRACER for 30 min, washed, and then subjected to confocal imaging, and the LIVE/DEAD BacLight Bacterial Viability Kit was also applied to detect the cell viability. Almost all bacterial cells, which typically exhibit green fluorescence, have been found to emit blue fluorescence, indicating that Fe-TRACER enters the live bacterial cells to light up the intracellular proteins (Fig. 5A). Owing to the small size of P. gingivalis (0.5 μm × 1–2 μm rods or coccobacilli)20 and its prokaryotic structure, differentiation of subcellular localization of Fe-TRACER is not possible. After UV irradiation at 365 nm for 15 min, the cells were then subjected to lysis and separation by 2-D gel electrophoresis. Multiple fluorescent spots were observed on the 2-D gel, and most of them were located at molecular weights (MW) ranging from 35 to 63 kDa. The protein spots appearing in both the fluorescence image and the silver stained gel were excised and subjected to protein identification by MALDI-TOF-MS (Table S4, ESI‡). The MW and isoelectric point (pI) values of some proteins with multimeric domains were shifted as compared to their theoretical values; this might be attributed to the 2-D gel assay, the labeling by the probe, and the post-translational process.21
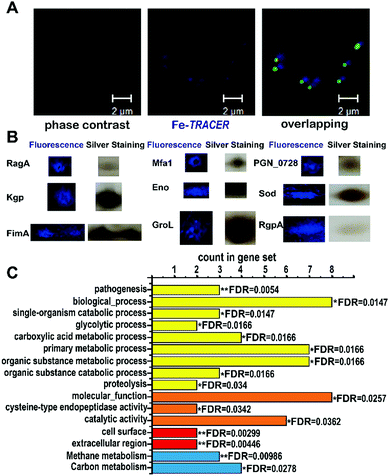 | ||
| Fig. 5 (A) Confocal imaging of P. gingivalis bacterial cells upon incubation of 50 μM of Fe-TRACER. Scale bar: 2 μm. (B) MALDI-TOF-MS confirmed spots of iron-associated proteins labelled by Fe-TRACER selected from 2D-gel; the whole 2D-gel is shown in Fig. S9, ESI.‡ (C) The enrichment false discovery rate (FDR) was calculated with STRING (*0.01 < FDR < 0.05; **0.001 < FDR < 0.01) with the categories of functional enrichments shown in different colors (yellow bars for biological process, orange bars for molecular function, red bars for cellar component and blue bars for KEGG pathways). | ||
Using this strategy, we have identified 17 iron-associated proteins, among which eight proteins have been reported to be associated either with iron (including [Mn/Fe] (SOD)22) or heme, the iron source of P. gingivalis, such as Lys-gingipain (Kgp), Gingipain R1 (RgpA), and Receptor antigen A (RagA), which have been shown to be involved in heme uptake, transfer, and utilization.23 Furthermore, fimbrilin (Mfa1), the OmpA protein family (PGN_0728), enolase (Eno), and fimbrilin (FimA) have been reported to be related to the formation of biofilms, which highly correlates with the uptake of heme in P. gingivalis.24,25 The other nine iron-associated proteins, however, are observed in P. gingivalis for the first time; they have been reported to be both iron-binding, e.g. GroEL (GroL) secreted by Helicobacter pylori,26 and iron-associated, e.g. aminomethyltransferase (GcvT), and glutamate dehydrogenase (Gdh), whose expressing levels change in Sinorhizobium meliloti27 and in rat brains,28 respectively, under iron-limited conditions. Moreover, two putative iron-binding sites consisting of Glu44, Glu45, and Asp48 and Arg79, Glu129, and Tyr133 were found on the surface of the class I fructose-bisphosphate aldolase (Fda) according to its crystal structure (PDB id: 2iqt). It was notable that proteins of Gdh, GcvT, GroL, Eno, SOD, and Fda were mapped into the Markov Cluster Algorithm (MCL) clustering network, indicative of the closed functional linkers among them (Fig. S10, ESI‡).
The 17 identified proteins were further subjected to GO enrichment analysis by STRING. In total, 9 biological processes, 3 molecular functions, 2 cellular components and 2 KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways were associated with the identified proteins (Fig. 5C). Most of the proteins are found to be involved in primary metabolic process (FDR = 0.0166) and organic substance metabolic process (FDR = 0.0166), and are associated with one molecular function, i.e., catalytic activity (FDR = 0.0362). The high significance of cellular component terms with FDR less than 0.01 was noted for cell surface and extracellular region, which might result from the close relationship between heme uptake and bio-film formation.24 Detailed examination of the identified iron-associated proteins reveals that RgpA and Kgp, which play vital roles in the degradation of hemoglobin,3 are involved in 9 categories of STRING analysis. Importantly, Kgp and RgpA secreted by P. gingivalis play the vital role of protecting P. gingivalis from being killed by neutrophils.29 Moreover, Eno is associated with 16 categories from STRING, and P. gingivalis depends on FimA fimbriae for adhesion and colonization.30 Our results clearly demonstrate the vital role of iron and iron-associated proteomes for the survival and pathogenesis of P. gingivalis.
Methods
Formation of Fe-TRACER and materials
The synthesis of the fluorescent ligand TRACER was carried out based on our former report.15Fe-TRACER was generated by reacting Fe3+ with equimolar amounts of TRACER in aqueous solution. The other chemicals and solvents (Sigma-Aldrich or otherwise stated) are either in analytical grades or carry the BioUltra label and were used directly.The Porphyromonas gingivalis strain ATCC 33277 was used, and its maintaining details are described in ESI.‡
Fluorescence spectroscopic measurements and 1-D SDS-PAGE
All fluorescence measurements were performed using a Hitachi F-7000 fluorescence spectrophotometer with a 1000 W xenon lamp source using a 0.5 cm × 1 cm × 3 cm quartz cuvette. Protein samples were diluted in a buffer containing 10 mM HEPES, 100 mM NaCl, and 5 mM NaHCO3 (pH 7.4). All samples were measured at 350 nm excitation and 450 nm emission with slits of 5 nm and PMT at 600 Volts.All 1-D SDS-PAGE analyses were performed using 4% stacking gel with 12% separation gel and imaged by MY ECL Imager (Thermo Scientific). The intensities of the protein bands on the SDS-PAGE gel or in the fluorescence images were quantified and analyzed with the ImageJ software, and the staining of Coomassie blue was used for comparison.
Crystallization of Fe-TRACER-hTF
In brief, the crystals of mono-ferric FeC-hTF protein were prepared by the sitting-drop method and was soaked in a cryo-protectant containing Fe-TRACER. Selected crystals were subjected to X-ray diffraction at beam line BL17U1, Shanghai Synchrotron Radiation Facility, using 0.93001 Å radiation.Confocal imaging of P. gingivalis
The fluorescence imaging of P. gingivalis cells stained by Fe-TRACER and the LIVE/DEAD BacLight Bacterial Viability Kit (Thermo Scientific) were carried out using a Carl Zeiss LSM700 Inverted Confocal Microscope with a Plan-Apochromat 63 × 1.40 NA oil-immersion objective. The emission was measured at 405, 488, and 555 nm for Fe-TRACER, SYTO 9 and propidium iodide (PI), respectively.Identification of iron-associated proteome in P. gingivalis
Briefly, the total cell lysate of P. gingivalis labelled by Fe-TRACER was cleaned up with the ReadyPrep 2-D Cleanup Kit (BioRad) and then subjected to 2-D SDS-PAGE. The gels were imaged using the ChemiDoc™ XRS+ System (BioRad) with Filter for Hoechst/coumarin; silver staining was then performed to visualize protein spots. Only the intense blue fluorescent spots that align with the silver-stained gel were excised and subjected to trypsin digestion then identified by MALDI-TOF-MS.Bioinformatics
The STRING database (version 10.5) was used to obtain the categories of functional enrichments, related FDR and for the MCL clustering analysis.Conclusion
In summary, we have developed an approach to track and analyze iron-associated proteomes in live bacteria cells using a fluorescent probe in combination with proteomics and bioinformatics. We validate that the Fe-TRACER specifically binds to human transferrin at the Fe3+ binding site by X-ray crystallography. The probe can readily enter cells and label the iron proteome of P. gingivalis. Upon photo-activation, 17 iron-associated proteins are subsequently identified, including 8 proteins that have been previously reported to be related to iron or heme; the remaining 9 iron-associated proteins were identified for the first time in this pathogen. This approach can be extended to mine iron-associated proteomes in other microbials and pathogens, thereby offering a basis for understanding iron homeostasis in pathogens and for the development of new antimicrobials.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Research Grants Council of Hong Kong (17304614P) and the University of Hong Kong (for an e-SRT on Integrative Biology and for a scholarship to NJ). The crystal diffraction data were collected at beam line BL17U1, Shanghai Synchrotron Radiation Facility (SSRF) at Shanghai Institute of Applied Physics, the Chinese Academy of Sciences. We thank the staff at BL17U1 beamline of SSRF for their generous help.References
- P. E. Petersen and H. Ogawa, The global burden of periodontal disease: towards integration with chronic disease prevention and control, Periodontology 2000, 2012, 60, 15 CrossRef PubMed.
- G. Hajishengallis, R. P. Darveau and M. A. Curtis, The keystone-pathogen hypothesis, Nat. Rev. Microbiol., 2012, 10, 717 CrossRef CAS PubMed.
- J. Ciuraszkiewicz, M. Smiga, P. Mackiewicz, A. Gmiterek, M. Bielecki, M. Olczak and T. Olczak, Fur homolog regulates Porphyromonas gingivalis virulence under low-iron/heme conditions through a complex regulatory network, Mol. Oral Microbiol., 2014, 29, 333 CrossRef CAS PubMed.
- J. W. Smalley and T. Olczak, Heme acquisition mechanisms of Porphyromonas gingivalis – strategies used in a polymicrobial community in a heme-limited host environment, Mol. Oral Microbiol., 2017, 32, 1 CrossRef CAS PubMed.
- G. Hajishengallis, Periodontitis: from microbial immune subversion to systemic inflammation, Nat. Rev. Immunol., 2015, 15, 30 CrossRef CAS PubMed.
- Q. Xia, T. Wang, Y. Park, R. J. Lamont and M. Hackett, Differential quantitative proteomics of Porphyromonas gingivalis by linear ion trap mass spectrometry: non-label methods comparison, q-values and LOWESS curve fitting, Int. J. Mass Spectrom., 2007, 259, 105 CrossRef CAS PubMed.
- Q. Xia, T. Wang, F. Taub, Y. Park, C. A. Capestany, R. J. Lamont and M. Hackett, Quantitative proteomics of intracellular Porphyromonas gingivalis, Proteomics, 2007, 7, 4323 CrossRef CAS PubMed.
- F. Petrat, H. de Groot and U. Rauen, Subcellular distribution of chelatable iron: a laser scanning microscopic study in isolated hepatocytes and liver endothelial cells, Biochem. J., 2001, 356, 61 CrossRef CAS PubMed.
- M. Shvartsman, E. Fibach and Z. I. Cabantchik, Transferrin-iron routing to the cytosol and mitochondria as studied by live and real-time fluorescence, Biochem. J., 2010, 429, 185 CrossRef CAS PubMed.
- K. P. Carter, A. M. Young and A. E. Palmer, Fluorescent sensors for measuring metal ions in living systems, Chem. Rev., 2014, 114, 4564 CrossRef CAS PubMed.
- Y.-T. Lai, Y. Yang, L. Hu, T. Cheng, Y.-Y. Chang, M. Koohi-Moghadam, Y. Wang, J. Xia, J. Wang, H. Li and H. Sun, Integration of fluorescence imaging with proteomics enables visualization and identification of metallo-proteomes in living cells, Metallomics, 2017, 9, 38 RSC.
- Y. Wang, L. Hu, F. Xu, Q. Quan, Y.-T. Lai, W. Xia, Y. Yang, Y.-Y. Chang, X. Yang, Z. Chai, I.-K. Chu, H. Li and H. Sun, Integrative approach for the analysis of the proteome-wide response to bismuth drugs in Helicobacter pylori, Chem. Sci., 2017, 8, 4626 RSC.
- D. K. Ryan and J. H. Weber, Fluorescence quenching titration for determination of complexing capacities and stability constants of fulvic acid, Anal. Chem., 1982, 54, 986 CrossRef CAS.
- H. Sun, H. Li and P. J. Sadler, Transferrin as a metal ion mediator, Chem. Rev., 1999, 99, 2817 CrossRef CAS PubMed.
- Y.-T. Lai, Y.-Y. Chang, L. Hu, Y. Yang, A. Chao, Z.-Y. Du, J. A. Tanner, M.-L. Chye, C. Qian, K.-M. Ng, H. Li and H. Sun, Rapid labeling of intracellular His-tagged proteins in living cells, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2948 CrossRef CAS PubMed.
- R. C. Cumming, N. L. Andon, P. A. Haynes, M. Park, W. H. Fischer and D. Schubert, Protein disulfide bond formation in the cytoplasm during oxidative stress, J. Biol. Chem., 2004, 279, 21749 CrossRef CAS PubMed.
- M. Wang, T. P. Lai, L. Wang, H. Zhang, N. Yang, P. J. Sadler and H. Sun, “Anion clamp” allows flexible protein to impose coordination geometry on metal ions, Chem. Commun., 2015, 51, 7867 RSC.
- N. Yang, H. Zhang, M. Wang, Q. Hao and H. Sun, Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe, Sci. Rep., 2012, 2, 999 CrossRef PubMed.
- J. P. Lewis, Metal uptake in host–pathogen interactions: role of iron in Porphyromonas gingivalis interactions with host organisms, Periodontology 2000, 2010, 52, 94 CrossRef PubMed.
- X. Zhou and Y. Li, Atlas of Oral Microbiology: From Healthy Microflora to Disease, Academic Press, 2015 Search PubMed.
- J. Potempa and K. A. Nguyen, Purification and characterization of gingipains, Curr. Protoc. Protein Sci., 2007, 49, 21.20:1 Search PubMed.
- A.-F. Miller, Superoxide dismutases: ancient enzymes and new insights, FEBS Lett., 2012, 586, 585 CrossRef CAS PubMed.
- T. Olczak, W. Simpson, X. Liu and C. A. Genco, Iron and heme utilization in Porphyromonas gingivalis, FEMS Microbiol. Rev., 2005, 29, 119 CrossRef CAS PubMed.
- K. Maeda, H. Nagata, M. Ojima and A. Amano, Proteomic and transcriptional analysis of interaction between oral microbiota Porphyromonas gingivalis and Streptococcus oralis, J. Proteome Res., 2014, 14, 82 CrossRef PubMed.
- K. L. Naylor, M. Widziolek, S. Hunt, M. Conolly, M. Hicks, P. Stafford, J. Potempa, C. Murdoch, C. Douglas and G. P. Stafford, Role of OmpA2 surface regions of Porphyromonas gingivalis in host–pathogen interactions with oral epithelial cells, MicrobiologyOpen, 2017, 6, e00401 CrossRef PubMed.
- M. A. González-López, N. Velázquez-Guadarrama, M. E. Romero-Espejel and J. de Jesús Olivares-Trejo, Helicobacter pylori secretes the chaperonin GroEL (HSP60), which binds iron, FEBS Lett., 2013, 587, 1823 CrossRef PubMed.
- T.-C. Chao, J. Buhrmester, N. Hansmeier, A. Pühler and S. Weidner, Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation, Appl. Environ. Microbiol., 2005, 71, 5969 CrossRef CAS PubMed.
- V. Taneja, K. Mishra and K. Agarwal, Effect of maternal iron deficiency on GABA shunt pathway of developing rat brain, Indian J. Exp. Biol., 1990, 28, 466 CAS.
- T. Maekawa, Jennifer L. Krauss, T. Abe, R. Jotwani, M. Triantafilou, K. Triantafilou, A. Hashim, S. Hoch, Michael A. Curtis, G. Nussbaum, John D. Lambris and G. Hajishengallis, Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis, Cell Host Microbe, 2014, 15, 768 CAS.
- K. Nagano, Y. Abiko, Y. Yoshida and F. Yoshimura, Porphyromonas gingivalis FimA fimbriae: Roles of the fim gene cluster in the fimbrial assembly and antigenic heterogeneity among fimA genotypes, J. Oral Biosci., 2012, 54, 160 CrossRef CAS.
Footnotes |
| † The coordinates and structure factors for Fe-TRACER have been deposit at Protein Data Bank (wwpdb) with accession code: 5Y6K. |
| ‡ Electronic supplementary information (ESI) available. See DOI: 10.1039/c7mt00275k |
| § These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |