Highly efficient antibody purification with controlled orientation of protein A on magnetic nanoparticles†
Sunghyun
Kim
,
Daekyung
Sung
and
Jeong Ho
Chang
 *
*
Center for Convergence Bioceramic Materials, Korea Institute of Ceramic Engineering and Technology, Chungbuk 28160, Republic of Korea. E-mail: jhchang@kicet.re.kr; Tel: +82 43 913 1501
First published on 2nd November 2017
Abstract
In this study, we prepared protein A grafted magnetic nanoparticles for the industrial large-scale purification of antibodies with enhancement of binding capacity and immobilization by controlled orientation with chlorophenylsilane (CPTMS) on the surface. For site-specific immobilization of protein A, genetically modified protein A with a cysteine residue was expressed in E. coli and purified by affinity chromatography. To improve the surface area to volume ratio and increase the immobilization amount of protein A, chlorophenylsilane functionalized magnetic nanoparticles (CPTMS@MNPs) were prepared, which are smaller nanoparticles with an average diameter of 20 nm compared to commercial magnetic microparticles (Dynabeads) with an average size of 2.8 μm. The CPTMS@MNPs showed the enhancement of protein A immobilization and binding capacity to antibodies, being 11.5-fold and 7-fold higher than those of commercial Dynabeads, respectively. In addition, the CPTMS@MNPs retained about 80% of the initial protein binding capacity until the third stage of recycling. Therefore, protein A grafted CPTMS@MNPs may be useful for the industrial large-scale purification of antibodies.
Introduction
Antibodies are the most important molecules in biopharmaceuticals. Nowadays, there are approximately 70 therapeutic monoclonal antibodies for use in clinical practice and 200 antibodies in clinical development.1 Protein A-based affinity chromatography is the gold standard to purify monoclonal antibodies due to its high selectivity and robustness.2 Protein A has the ability to recognize specifically the Fc region of an antibody without interrupting its antigen-binding ability.3 Commercial protein A-based affinity chromatography provides a product purity of up to 95% in a single step. However, this chromatographic operation has several bottlenecks such as low binding capacity, a slow purification process and expensive protein A-beads.4–6 Therefore, a large volume of cell culture supernatants requires sequential purification cycles, which increase the process time and purification cost. Consequently, in order to overcome these limitations, different purification strategies have received great interest.7–9Iron oxide magnetic particles are attractive particles in many applications such as biosensors, MRI agents, drug delivery carriers, as well as materials for purification processes.10–14 In particular, magnetic separations offer fast processing and higher binding capacity compared to protein A affinity column resins due to small particles of generally 2 μm.15–20 Now, protein A functionalized magnetic particles with micrometer size are already commercially available and showed good performance for the separation of biological molecules such as enzymes and antibodies.21 However, these magnetic separations are not yet sufficient in industrial applications. The main cause of this problem is the antibody binding capacity. The most popular product, Dynabead® protein A, consists of uniform, 2.8 μm superparamagnetic beads which have 8 μg per mg beads of antibody binding capacity. This low binding capacity results in a high price, limiting the large-scale industrial application. Furthermore, it is desirable to develop efficient protein A magnetic particles to achieve good performance with high binding capacity which is cost-effective for antibody purification.
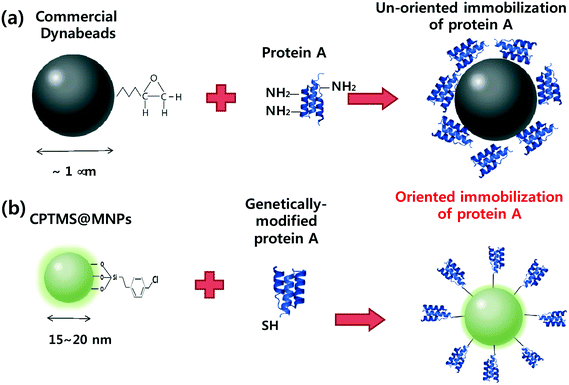
Here, we report well-oriented protein A on magnetic nanoparticles with excellent binding capacity to antibodies and cost-effectiveness. Fig. 1a represents the conventional immobilization procedures of commercial protein A magnetic microparticles (Dynabeads). The total number of amine groups on lysine residues and thiol groups on cysteine residues in protein A is 27 and 0, respectively. Therefore, several lysine residues react with the epoxy group of magnetic particles and produce non-oriented immobilization which reduces the functional activity of protein A. In addition, the large size of magnetic particles on the micrometer scale results in a low surface area to volume ratio and produces low binding capacity. To address these problems, we developed oriented protein A magnetic nanoparticles (CPTMS@MNPs@protein A) using genetically modified protein A with a cysteine residue and chloromethylphenylsilane functionalization (Fig. 1b). We expected that the CPTMS@MNPs can provide a highly efficient purification method of antibodies compared to commercial Dynabeads because they retain the intact functional activity of protein A by site-specific immobilization and offer higher antibody binding capacity by means of an increased surface area to volume ratio in magnetic nanoparticles.
Experimental procedures
The CPTMS@MNPs were prepared by a previously reported method.22 The gene of genetically modified protein A with a cysteine residue was synthesized and cloned into the pET28b prokaryotic expression vector. Protein A-cys was overexpressed using isopropyl β-D-thiogalactopyranoside (IPTG) induction and was purified by Ni-NTA affinity chromatography. The immobilization conditions of protein A-cys on CPTMS@MNPs were applied; different amounts of protein A-cys (25–300 μg) and different pH values (pH 5–9). The protein A amounts in CPTMS@MNPs@protein A and Dynabead@protein A were determined by the bicinchoninic acid (BCA) assay, respectively. The binding capacity of CPTMS@MNPs@protein A and Dynabead@protein A was measured in 100, 200, 400, 600, and 800 μg of monoclonal antibodies, respectively. After magnetic separations, the antibodies were released into the glycine solution (pH 2.7) for 10 min and neutralized in 1 M Tris buffer (pH 9.0). The amount of eluted antibodies was confirmed using the BCA assay and SDS-PAGE electrophoresis.Results and discussion
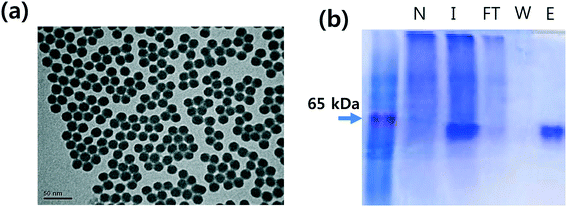
The Fe3O4 magnetites were synthesized by co-precipitation of Fe2+ and Fe3+ salts in a basic solution. The silica-coated magnetic nanoparticles were produced by the sol–gel reaction of tetraethyl orthosilicate (TEOS). The CPTMS@MNPs were prepared by a previously reported method.22 To increase the surface area to volume ratio of the particles, we generated smaller magnetic nanoparticles than commercial Dynabeads. Transmission electron microscopy (TEM) indicates that the CPTMS@MNPs have a spherical shape with an average size of 20 nm and are well-dispersed (Fig. 2a).To achieve the oriented protein A immobilization on magnetic nanoparticles, protein A containing a cysteine residue at the c-terminus (protein A-cys) was prepared by genetic engineering. The gene of protein A-cys was synthesized using a DNA synthesizer and cloned into the pET28b prokaryotic expression vector. The recombinant protein A-cys was successfully overexpressed in E. coli using IPTG induction and purified using Ni-NTA agarose resin which binds to the recombinant protein with His-tag. As shown in the figure, the purity of recombinant protein A-cys was above 95% (Fig. 2b).
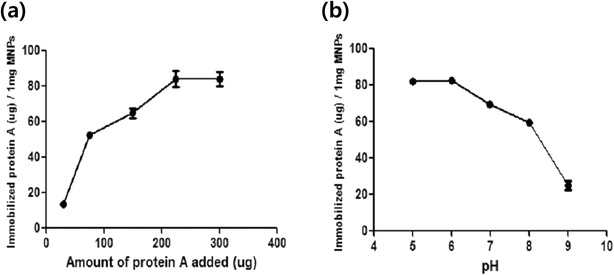
The optimal immobilization conditions of protein A-cys on CPTMS@MNPs were investigated. To quantify the immobilization of protein A-cys, the BCA assay was used. The BCA assay is capable of determining the protein amounts via a change in color from green to purple depending on the given protein concentration. To maximize the immobilization of protein A-cys on magnetic nanoparticles, two parameters were optimized, i.e. the reaction concentration and buffer conditions. First of all, reaction concentrations from 25 to 300 μg of protein A in 20 mM PB buffer (pH 7) were tested. In Fig. 3a, the immobilized protein amount was increased depending on the increased reaction concentration. Protein A-cys on CPTMS@MNPs begins to saturate at a protein A amount of 200 μg. The maximum amount of immobilized protein A-cys is 70 μg per 1 mg of magnetic nanoparticles. To examine the optimal pH, we demonstrated several buffer conditions between pH 5 and pH 9 using 200 μg of protein A-cys (Fig. 3b). At pH 6, the immobilization of protein A-cys on CPTMS@MNPs is maximum and decreased depending on the increase in pH. Protein A-cys is a single chain polypeptide with 307 amino acids and its isoelectric point is at around 5.0. Above pH 7, protein A-cys has a negative net charge, producing a repulsion force with CPTMS@MNPs because the silica group of MNPs also has a negative surface charge. However, at pH 5 and pH 6, the surface charge of protein A-cys is neutralized allowing the accessible contact for conjugation between protein A-cys and CPTMS@MNPs, producing highly efficient immobilization of protein A-cys on CPTMS@MNPs.
Cysteine-mediated immobilization of protein A-cys on CPTMS@MNPs was investigated. For site-specific conjugation, we immobilized protein A-cys on CPTMS@MNPs at pH 6. At pH 6, the amines in lysine and arginine remain protonated and are not nucleophilic to react with benzyl chloride. Therefore, the thiol group of cysteine in protein A can react with benzyl chloride specifically. To demonstrate this reaction, we performed the immobilization of wild-type protein A (WT protein A) and genetically modified protein A (protein A-cys) on CPTMS@MNPs at pH 6 (Fig. S1†). The immobilization amount of WT protein A and protein A-cys on CPTMS@MNPs was 24 μg and 82 μg, respectively. Therefore, protein A-cys can react with benzyl chloride efficiently and specifically using the thiol group of cysteine at pH 6. Consequently, these values, i.e. the concentration of protein A above 200 μg mL−1 and pH 6, were used as the optimized conditions for all experiments. Using these conditions, CPTMS@MNPs achieved a maximum immobilization amount of 82 μg protein A and oriented protein–cys functionalized magnetic nanoparticles (CPTMS@MNPs@protein A) were prepared.
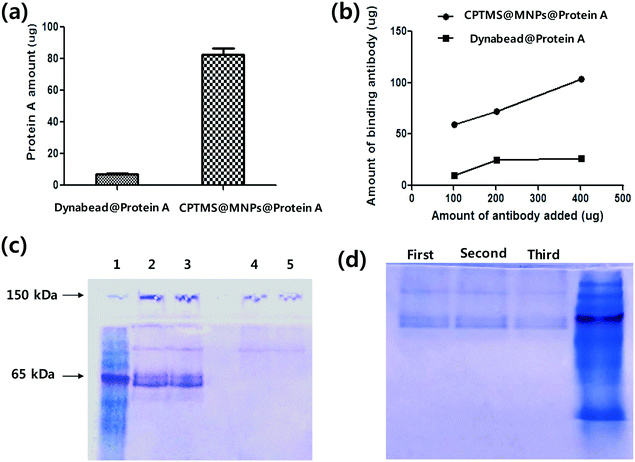
The magnetic properties of CPTMS@MNPs@protein A were examined using a vibrating sample magnetometer (VSM). The saturated magnetizations of CPTMS@MNPs were measured at 40 emu g−1 at 300 K and quickly separated by using a magnet within 15 s (Fig. S2†). We also characterized the protein A amount in Dynabead@protein A as a positive control. In the case of Dynabeads, only 7.1 μg of protein A were immobilized on 1 mg of magnetic microparticles (Fig. 4a). Therefore, CPTMS@MNPs had an 11.5-fold higher immobilization amount than Dynabead. This high immobilized amount of protein A on CPTMS@MNPs is due to smaller particles with a higher surface area to volume ratio. The antibody binding capacity of CPTMS@MNPs@protein A and Dynabead@protein A was measured, respectively. After the binding between the antibodies and protein A functionalized magnetic particles for 30 min, the antibodies bound to magnetic particles were separated using the magnet. The antibodies were released into the glycine solution (pH 2.7) and the amounts of antibodies were confirmed using the BCA assay and SDS-PAGE electrophoresis. As shown in Fig. 4b, Dynabead@protein A has a maximum quantity of 20 mg g−1 of particles. However, the maximum quantities of CPTMS@MNPs@protein A were shown to be 60 (mg g−1 of particles) for low and 103 (mg g−1 of particles) for higher concentrations.
Antibodies are 150 kDa proteins consisting of four polypeptide chains, with two identical heavy chains and two identical light chains linked together by disulfide bonds. The SDS-PAGE showed an intact band of IgG at 150 kDa and a reduced form at 55 kDa of a heavy chain (Fig. 4c). CPTMS@MNPs@protein A showed thicker bands compared to Dynabeads. To analyze the band thickness, we used Image J software.23 The bands of our nanoparticles were 7-fold thicker than Dynabead@protein A. Finally, we examined the reusability of CPTMS@MNPs@protein A by repetitively performing the regeneration and reuse of the particles. As shown in Fig. 4d, this magnetic nanoparticle retained about 80% of the initial protein binding capacity until the third stage of recycling.
Conclusion
We have developed oriented protein A immobilized magnetic nanoparticles which have an 11.5-fold improved immobilization amount of protein A and offer 7-fold higher binding capacity to antibodies compared to commercial magnetic particles. In addition, the functional activity of our magnetic nanoparticles retained about 80% of the initial protein binding capacity after the third re-use. Therefore, protein A grafted CPTMS@MNPs may be useful for industrial large-scale purification of antibodies.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This research was supported by the Industrial Core-Technology Development projects funded by the Ministry of Trade, Industry and Energy (MOTIE) and the Basic Science Research Program through the National Research of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2016R1C1B11008096).Notes and references
- D. M. Ecker, S. D. Jones and H. L. Levine, mAbs, 2015, 7, 9 CrossRef CAS PubMed
.
- U. Gottshalk, K. Brorson and A. Shukla, Nat. Methods, 2012, 30, 489 Search PubMed
.
- A. Jungbauer and R. Hahn, Curr. Opin. Drug Discovery Dev., 2004, 7, 248 CAS
.
- T. Ishihara, N. Nakajima and T. Kadoya, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2010, 878, 2141 CrossRef CAS PubMed
.
- R. Hahn, R. Schlegel and A. Jungbauer, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2003, 790, 35 CrossRef CAS
.
- R. Mallik and D. Hage, J. Sep. Sci., 2006, 29, 1686 CrossRef CAS PubMed
.
- A. M. Azevedo, P. A. J. Rosa, I. F. Ferreira, A. M. M. O. Pisco, J. de Vries, R. Korporaal, T. J. Visser and M. R. Aires-Barros, Sep. Purif. Technol., 2009, 65, 31 CrossRef CAS
.
- M. Kuczewski, E. Schirmer, B. Lain and Z. Gregory, J. Biotechnol., 2011, 6, 56 CrossRef CAS PubMed
.
- I. Safarik and M. Safarikova, Biomagn. Res. Technol., 2004, 2, 7 CrossRef PubMed
.
- Y. Zhang and D. Zhou, Expert Rev. Mol. Diagn., 2012, 12, 565 CrossRef CAS PubMed
.
- J. Park, S. Kim, P. E. Saw, I. H. Lee, M. K. Yu, M. Kim, K. Lee, Y. C. Kim, Y. Y. Jeong and S. Jon, J. Controlled Release, 2012, 163, 111 CrossRef CAS PubMed
.
- M. Arruebo, R. Fernández-Pacheco, M. R. Ibarra and J. Santamaría, Nano Today, 2007, 2, 22 CrossRef
.
- S. Y. Lee, C. Y. Ahn, J. Lee and J. H. Chang, Anal. Biochem., 2010, 405, 135 CrossRef PubMed
.
- M. I. J. Denison, S. Raman, N. Duraisamy, R. M. Thangavelu, S. U. M. Riyaz, D. Gunasekaran and K. Krishnan, RSC Adv., 2015, 5, 99820 RSC
.
- H. Xuemei, Z. Changjie, T. Yanlong, D. Shuliang, Z. Xiang and Z. Jiupeng, Chem. Res. Chin. Univ., 2016, 32, 889 CrossRef
.
- V. L. Dhadge, A. Hussain, A. M. Azevedo, R. Aires-Barros and A. C. A. Roque, J. R. Soc., Interface, 2014, 11, 20130875 CrossRef PubMed
.
- M. Lattuada and A. T. Hatton, Langmuir, 2007, 23, 2158 CrossRef CAS PubMed
.
- S. Laurent, D. Forge, M. Port, A. Roch, C. Robic, E. L. Vander and R. N. Muller, Chem. Rev., 2008, 108, 2064 CrossRef CAS PubMed
.
- A. M. G. C. Dias, A. Hussain, S. Marcos and C. A. Roque, Biotechnol. Adv., 2011, 29, 142 CrossRef CAS PubMed
.
- K. Holschuh and A. Schwämmle, J. Magn. Magn. Mater., 2005, 293, 345 CrossRef CAS
.
- I. M. Cristea and B. T. Chait, Cold Spring Harb. Protoc., 2011, 5, 534 Search PubMed
.
- J. Lee and J. H. Chang, Nanoscale Res. Lett., 2014, 9, 664 CrossRef PubMed
.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7md00468k |
| This journal is © The Royal Society of Chemistry 2018 |