Generation of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides during the enzymatic hydrolysis of tropical banded cricket (Gryllodes sigillatus) proteins†
Alice B.
Nongonierma
 ab,
Candice
Lamoureux
a and
Richard J.
FitzGerald
ab,
Candice
Lamoureux
a and
Richard J.
FitzGerald
 *ab
*ab
aDepartment of Biological Sciences, University of Limerick, Limerick, Ireland. E-mail: dick.fitzgerald@ul.ie; Fax: +353-61331490; Tel: +353-61202598
bFood for Health Ireland (FHI), University of Limerick, Limerick, Ireland
First published on 27th November 2017
Abstract
Tropical banded crickets (Gryllodes sigillatus) were studied for their ability to yield hydrolysates with dipeptidyl peptidase IV (DPP-IV) inhibitory properties. A cricket protein isolate (CPI) was prepared following extraction of the water soluble proteins from G. sigillatus powder (CP). The extraction yield and purity were 20.90 ± 0.35% and 57.0 ± 2.23%, respectively. Endogenous proteinase activities were detected in the CP, which were linked to the significant protein breakdown seen in this sample. Fifteen CPI hydrolysates (H1–H15) were generated with Protamex™ using a design of experiments (DOE) approach combining three parameters, temperature (40, 50 and 60 °C), enzyme to substrate ratio (E![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S, 0.50, 1.25 and 2.00% (w/w)) and hydrolysis time (60, 150 and 240 min). The DPP-IV half maximal inhibitory concentrations (IC50) of the CPI hydrolysates ranged from 0.40 ± 0.03/0.40 ± 0.02 (H2/H3) to 1.01 ± 0.07 mg mL−1 (H7). Following simulated gastrointestinal digestion (SGID), the DPP-IV IC50 of CPI decreased (>3.57 vs. 0.78 ± 0.04 mg mL−1) while that of H5 increased (0.47 ± 0.03 vs. 0.71 ± 0.06 mg mL−1). This study has demonstrated for the first time that G. sigillatus protein hydrolysates are able to inhibit DPP-IV. The study of these hydrolysates in vivo is needed to evaluate their potential role in glycaemic management.
S, 0.50, 1.25 and 2.00% (w/w)) and hydrolysis time (60, 150 and 240 min). The DPP-IV half maximal inhibitory concentrations (IC50) of the CPI hydrolysates ranged from 0.40 ± 0.03/0.40 ± 0.02 (H2/H3) to 1.01 ± 0.07 mg mL−1 (H7). Following simulated gastrointestinal digestion (SGID), the DPP-IV IC50 of CPI decreased (>3.57 vs. 0.78 ± 0.04 mg mL−1) while that of H5 increased (0.47 ± 0.03 vs. 0.71 ± 0.06 mg mL−1). This study has demonstrated for the first time that G. sigillatus protein hydrolysates are able to inhibit DPP-IV. The study of these hydrolysates in vivo is needed to evaluate their potential role in glycaemic management.
Introduction
Type 2 diabetes (T2D) incidences have reached epidemic status worldwide. T2D diabetes represents around 91% of diabetes cases. Estimates of diabetes cases worldwide in 2015 were in the region of 415 million adults (20–79 years). Projections indicate that 642 million diabetics will exist in 2050.1 Dietary modifications are often used as the first approach in the management of T2D. Interestingly, the ingestion of food proteins has been shown to modify insulin secretion in humans.2 Hydrolysed food proteins and peptides have also been shown to be able to modulate the activity of a range of enzymes (i.e., α-amylase and dipeptidyl peptidase IV (DPP-IV) inhibition) associated with the regulation of insulin secretion and glycaemia.3–6Incretins such as a glucagon-like peptide-1 (GLP-1), glucose inhibitory polypeptide (GIP) and peptide YY (PYY) induce insulin secretion during the post-prandial phase by a mechanism known as the incretin effect. DPP-IV is responsible for the degradation and inactivation of incretins, which is detrimental to their insulinotropic activity. However, the incretin effect can be restored following inhibition of DPP-IV.7,8 The positive role of food protein-derived bioactive peptides in the inhibition of DPP-IV has been shown in several studies.9,10 DPP-IV inhibitory peptides are found in a wide range of food proteins. In silico approaches have shown that several staple proteins such as dairy, meat, plant and marine proteins are a good source of DPP-IV inhibitory peptides.11,12 A recent publication has demonstrated that a water soluble extract from housefly larvae (Musca domestica) was able to inhibit DPP-IV in vitro.13 However, edible insect protein hydrolysates do not appear to have been employed for the generation of DPP-IV inhibitory peptides, to date.
Edible insects are a rich protein source and have potential uses for animal feed and human nutrition.14,15 Edible insects are part of the normal human diet in several countries mainly located in Asia, Africa and Latin America. An increasing number of Western countries have also recognised the potential role of edible insects as a source of high quality nutrients for animal feed and human food. Therefore, legislation is evolving around the globe to provide a framework for the safe production and utilisation of edible insects.16–18 For example, the European Union (EU) plans to include edible insects as novel foods in its Regulation in January 2018, (EU, 2015/2283). Proteins from a range of edible insects have been hydrolysed for the generation of bioactive peptides. To date, most of the studies describing the generation of bioactive peptides have been carried out with silkworm (Bombyx mori).16 In particular the generation of angiotensin converting enzyme (ACE) inhibitory hydrolysates has been described in a number of studies.19–21
Tropical banded crickets (Gryllodes sigillatus) belong to the Orthoptera order and the Gryllidae family. G. sigillatus has been reported for its relatively high protein content, which can reach up to 70% (w/w dry matter).22 Recently, the microbial quality of fresh G. sigillatus obtained from different rearing companies in Belgium has been evaluated.23,24 The microbial contamination found in the samples emphasised the necessity to apply heat treatments for the manufacture of safe products. Enzymatic hydrolysis of tropical banded crickets (G. sigillatus) has not been extensively studied. The first study reporting on G. sigillatus hydrolysates has recently been published by Hall, Jones, O'Haire, and Liceaga.25 In this study, hydrolysis of G. sigillatus proteins with Alcalase™ resulted in an improvement of their technofunctional properties (i.e., solubility, emulsification and foamability). The first report on the biofunctional properties (antioxidant and anti-inflammatory) of hydrolysates of G. sigillatus obtained following in vitro gastrointestinal digestion was recently published.26
The DPP-IV inhibitory properties of G. sigillatus hydrolysates have not been evaluated, to date. Therefore, the aim of this study was to investigate the potential of G. sigillatus to yield DPP-IV inhibitory peptides. This was achieved by preparing a cricket protein isolate (CPI). The CPI was hydrolysed with Protamex™ using a design of experiments (DOE) to optimise the generation of DPP-IV inhibitory hydrolysates. The hydrolysates generated were assessed for their DPP-IV inhibitory properties. Their physicochemical characteristics (peptide profile and molecular mass distribution) were also determined. Subsequently, the effect of simulated gastrointestinal digestion (SGID) on the DPP-IV inhibitory properties of the CPI and CPI hydrolysates was studied.
Materials and methods
Reagents
Hydrochloric acid (HCl), sodium hydroxide (NaOH), high performance liquid chromatography (HPLC) grade water and acetonitrile (ACN) were from VWR (Dublin, Ireland). 2,4,6-Trinitrobenzenesulfonic acid (TNBS) and a micro bicinchoninic acid (BCA) protein assay kit from Pierce Biotechnology were obtained from the Medical Supply Company (Dublin, Ireland). Gly-Pro-AMC was purchased from Bachem (Bubendorf, Switzerland). Coomassie Brilliant Blue, methanol, acetic acid, β-mercaptoethanol, trifluoroacetic acid (TFA), trichloro acetic acid (TCA), azocasein, tris(hydroxymethyl)aminomethane (TRIS), sodium phosphate monobasic, sodium phosphate dibasic, sodium dodecyl sulphate (SDS), Leu, diprotin A (Ile-Pro-Ile), porcine DPP-IV (≥10 units per mg protein), standards for molecular mass distribution (i.e., bovine serum albumin, β-lactoglobulin, α-lactalbumin, aprotinin, bacitracin, Leu-Trp-Met-Arg and Asp-Glu), mass spectrometry (MS) grade water and ACN were from Sigma-Aldrich (Dublin, Ireland). The commercial organic cricket (G. sigillatus) powder (CP) for human consumption (60% proteins, 20% fats and 10% carbohydrates), obtained following a roasting step for 150–270 min at 230–250 °C (Entomo Farms, personal communication) was a gift from Entomo Farms (Norwood, ON, Canada). Protamex™ was provided by Novozymes (Bagsvaerd, Denmark), pepsin by Biocatalysts (Cefn, Wales, UK) and Corolase PP™ by AB enzymes (Darmstadt, Germany).Preparation of the CPI
Non-protein nitrogen and protein components from edible insects may be tightly bound to the chitin present in their exoskeleton.16 In order to avoid interferences from chitin during enzymatic hydrolysis and subsequent bioactivity determination, it was decided within this study to focus on the water soluble protein fraction of the CP. Water soluble proteins were extracted from the CP. The CP was dispersed in distilled water at a concentration of 1% (w/w). The pH of this mixture was adjusted to 10.0 with 1 M NaOH under agitation using a stirring plate. Agitation was carried out at room temperature (21 °C) for 60 min. The mixture was centrifuged at 4190g for 15 min at 4 °C (Sorvall RC-5, Fisher Scientific, Dublin, Ireland). The supernatant was retained and adjusted to pH 7.0 using 0.1 M HCl. This solution was freeze-dried (FreeZone 18L, Labconco, Kansas City, MO, USA) and stored at −20 °C until utilisation.The protein content of the CPI was determined using a micro BCA kit. This allowed calculation of the protein extraction yield and purity of the CPI.27
Determination of the general endoproteinase activity within the CP
The azocasein assay was performed as per Kilcawley, Wilkinson, and Fox28 in triplicate (n = 3). Briefly, the CP was incubated with the azocasein substrate for 30 min at 50 °C. The reaction was terminated by the addition of 2 M TCA. Samples were centrifuged at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 255g for 5 min (Hettich Universal 320R, Hettich, Tuttlingen, Germany). The absorbance of the azo dye within the supernatant was read at 440 nm using a microplate reader (Biotek Synergy HT, Winoosky, VT, USA). The azocasein activity was expressed as Abs min−1 mg−1 sample.
255g for 5 min (Hettich Universal 320R, Hettich, Tuttlingen, Germany). The absorbance of the azo dye within the supernatant was read at 440 nm using a microplate reader (Biotek Synergy HT, Winoosky, VT, USA). The azocasein activity was expressed as Abs min−1 mg−1 sample.
Hydrolysis of the CPI
The CPI was hydrolysed with Protamex™. CPI hydrolysates were produced following a DOE as previously outlined in Nongonierma, Mazzocchi, Paolella, and FitzGerald.29 The parameters of the DOE were the incubation temperature (40, 50 and 60 °C), enzyme to substrate ratio (E![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S; 0.50, 1.25 and 2.00% (w/w)) and hydrolysis time (60, 150 and 240 min) which were evaluated at three different levels (−1, 0 and +1, centred and reduced values (z-centred), respectively, Table 1). Hydrolysates (H1–H14) were generated once within the DOE. The central point hydrolysate (50 °C, 1.25% E
S; 0.50, 1.25 and 2.00% (w/w)) and hydrolysis time (60, 150 and 240 min) which were evaluated at three different levels (−1, 0 and +1, centred and reduced values (z-centred), respectively, Table 1). Hydrolysates (H1–H14) were generated once within the DOE. The central point hydrolysate (50 °C, 1.25% E![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S and 150 min) was generated as three independent replicates (n = 3, H15A, H15B and H15C) for reproducibility purposes.
S and 150 min) was generated as three independent replicates (n = 3, H15A, H15B and H15C) for reproducibility purposes.
| Sample | Variable levela | ANb (mg NH3 per g) | DPP-IV IC50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b,c (mg mL−1) b,c (mg mL−1) |
||
|---|---|---|---|---|---|
| Temperature (°C) | E![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (%) S (%) |
Time (min) | |||
| a The z-centred values for each variable of the experimental design are provided in brackets. b Mean ± SD (n = 3). Superscript lower case and capital letters were used to compare hydrolysates H5 and H15, respectively. Values with different superscript letters are significantly different (p < 0.05). nd: not determined. c IC50: concentration inducing 50% inhibition of DPP-IV, expressed in mg protein equivalents per mL (mg mL−1). The IC50 value of the positive control Ile-Pro-Ile was 3.26 ± 0.55 μM. | |||||
| CP | — | — | — | — | >3.57 |
| CPI | — | — | — | — | 0.66 ± 0.03 |
| C40 | 40 (−1) | — | 240 (+1) | — | 0.81 ± 0.08 |
| C50 | 50 (0) | — | 240 (+1) | — | 0.79 ± 0.05 |
| C60 | 60 (+1) | — | 240 (+1) | — | 0.90 ± 0.07 |
| H1 | 40 (−1) | 0.50 (−1) | 60 (−1) | 0.18 ± 0.07 | 0.44 ± 0.02 |
| H2 | 40 (−1) | 0.5 (−1) | 240 (+1) | 0.19 ± 0.11 | 0.40 ± 0.03 |
| H3 | 40 (−1) | 1.25 (0) | 150 (0) | 0.25 ± 0.04 | 0.40 ± 0.02 |
| H4 | 40 (−1) | 2.00 (+1) | 60 (−1) | 0.23 ± 0.02 | 0.42 ± 0.03 |
| H5 | 40 (−1) | 2.00 (+1) | 240 (+1) | 0.37 ± 0.04a | 0.43 ± 0.04a |
| H6 | 60 (+1) | 0.50 (−1) | 60 (−1) | 0.14 ± 0.04 | 0.50 ± 0.20 |
| H7 | 60 (+1) | 0.50 (−1) | 240 (+1) | 0.29 ± 0.05 | 1.01 ± 0.07 |
| H8 | 60 (+1) | 1.25 (0) | 150 (0) | 0.39 ± 0.01 | 0.54 ± 0.01 |
| H9 | 60 (+1) | 2.00 (+1) | 60 (−1) | 0.38 ± 0.04 | 0.55 ± 0.02 |
| H10 | 60 (+1) | 2.00 (+1) | 240 (+1) | 0.53 ± 0.06 | 0.57 ± 0.07 |
| H11 | 50 (0) | 1.25 (0) | 60 (−1) | 0.73 ± 0.01 | 0.49 ± 0.05 |
| H12 | 50 (0) | 0.50 (−1) | 150 (0) | 0.30 ± 0.06 | 0.42 ± 0.04 |
| H13 | 50 (0) | 1.25 (0) | 240 (+1) | 0.69 ± 0.14 | 0.43 ± 0.02 |
| H14 | 50 (0) | 2.00 (+1) | 150 (0) | 0.44 ± 0.06 | 0.43 ± 0.01 |
| H15A | 50 (0) | 1.25 (0) | 150 (0) | 0.32 ± 0.05A | 0.51 ± 0.04A |
| H15B | 50 (0) | 1.25 (0) | 150 (0) | 0.32 ± 0.01A | 0.54 ± 0.05A |
| H15C | 50 (0) | 1.25 (0) | 150 (0) | 0.31 ± 0.02A | 0.54 ± 0.05A |
| H16 | 40 (−1) | 1.25 (0) | 60 (−1) | 0.02 ± 0.01 | 0.48 ± 0.07 |
| H17 | 40 (−1) | 1.25 (0) | 240 (+1) | 0.15 ± 0.03 | 0.50 ± 0.08 |
| H18 | 60 (+1) | 1.25 (0) | 60 (−1) | 0.28 ± 0.03 | 0.52 ± 0.10 |
| H19 | 60 (+1) | 1.25 (0) | 240 (+1) | 0.40 ± 0.02 | 0.50 ± 0.04 |
| H5B | 40 (−1) | 2.00 (+1) | 240 (+1) | 0.41 ± 0.04a | 0.48 ± 0.05b |
| H5C | 40 (−1) | 2.00 (+1) | 240 (+1) | 0.37 ± 0.08a | 0.47 ± 0.03b |
| H5_SGID | — | — | — | nd | 0.71 ± 0.06 |
| CPI_SGID | — | — | — | nd | 0.78 ± 0.04 |
CPI (5% (w protein equivalents/w)) was resuspended in distilled water and incubated for 60 min in a water bath (Lauda E100, Lauda Brinkmann, Lauda-Königshofen, Germany) set at the hydrolysis temperature (40, 50 or 60 °C). The pH was then adjusted to 7.0 with 0.5 M NaOH. Temperature, E![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S and hydrolysis time were set as defined by the DOE (Table 1). At the end of each reaction, the enzyme was heat inactivated (90 °C, 20 min) in a water bath. A control sample (C40, C50 or C60) without enzyme addition was prepared by incubation of the CPI for 240 min at each hydrolysis temperature. Samples were freeze-dried (Labconco) and stored at −20 °C until utilisation.
S and hydrolysis time were set as defined by the DOE (Table 1). At the end of each reaction, the enzyme was heat inactivated (90 °C, 20 min) in a water bath. A control sample (C40, C50 or C60) without enzyme addition was prepared by incubation of the CPI for 240 min at each hydrolysis temperature. Samples were freeze-dried (Labconco) and stored at −20 °C until utilisation.
The free amino group content (AN) of the CPI hydrolysates was determined in triplicate (n = 3) using the TNBS method as per Le Maux, Nongonierma, Barre, and FitzGerald.30 Absorbance values (350 nm) were measured with a microplate reader (Biotek Synergy HT). The AN was determined using eqn (1).
| AN = AN2 − AN1 | (1) |
Multilinear regression (MLR) model linking the hydrolytic parameters to the DPP-IV IC50 values of the CPI hydrolysates
The DPP-IV IC50 value of the hydrolysates was linked to the parameters of the DOE using a MLR model (eqn (2)). The MLR was built with Matlab (version R2014b) as previously described.31,32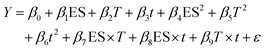 | (2) |
The response surface methodology (RSM) curves were plotted using the MLR model within the boundaries of the DOE.31,33
In vitro SGID of intact and hydrolysed CPI
The CPI and H5 sample were subjected to in vitro SGID as outlined by Walsh et al.34 Briefly, samples were resuspended in distilled water to 2% (w protein equivalents/w) for 30 min at 37 °C and the pH was adjusted to 2.0 using 1 M HCl. Pepsin (E![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S 2.5% (w/w)) was used to hydrolyse the samples under pH regulation (2.0) with HCl (pH stat Titrando 843, Tiamo 1.4 Metrohm, Dublin, Ireland) for 90 min at 37 °C. Pepsin was subsequently heat inactivated (90 °C, 20 min). The pH of the peptic hydrolysate was adjusted to 7.5 using 1 M NaOH. This sample was then hydrolysed with Corolase PP (E
S 2.5% (w/w)) was used to hydrolyse the samples under pH regulation (2.0) with HCl (pH stat Titrando 843, Tiamo 1.4 Metrohm, Dublin, Ireland) for 90 min at 37 °C. Pepsin was subsequently heat inactivated (90 °C, 20 min). The pH of the peptic hydrolysate was adjusted to 7.5 using 1 M NaOH. This sample was then hydrolysed with Corolase PP (E![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S 1% (w/w)), a porcine pancreatic enzyme preparation, for 150 min at 37 °C, pH 7.5 using a pH stat (Metrohm). The reaction was terminated by thermal treatment (90 °C, 20 min). Samples were freeze-dried and stored at −20 °C until utilisation.
S 1% (w/w)), a porcine pancreatic enzyme preparation, for 150 min at 37 °C, pH 7.5 using a pH stat (Metrohm). The reaction was terminated by thermal treatment (90 °C, 20 min). Samples were freeze-dried and stored at −20 °C until utilisation.
Dipeptidyl peptidase IV (DPP-IV) inhibition assay
The freeze-dried samples were dispersed in HPLC grade water at concentrations ranging from 3.57 × 10−2 to 3.57 mg mL−1 (final concentration expressed in mg protein equivalents per mL). Ile-Pro-Ile was used as positive control at final concentrations ranging 1.25 × 10−4 to 1.25 × 10−2 mg mL−1. The DPP-IV inhibition assay was carried out in triplicate as outlined by Lankas et al.35 and Nongonierma and FitzGerald,4 with modifications. All reagents were diluted in 100 mM Tris-HCl buffer (pH 8.0). Briefly, samples were mixed with Gly-Pro-AMC (final concentration 0.200 mM) in 96-well microplates (Thermo Fisher Scientific, Waltham, MA, USA). DPP-IV (final concentration 0.0025 U mL−1) was added to the wells. The microplate was incubated for 30 min at 37 °C in a microplate reader (Biotek Synergy HT). Fluorescence of the AMC was monitored at excitation and emission wavelengths of 360 and 460 nm, respectively. The DPP-IV IC50 values were determined by plotting the percentage inhibition as a function of the test compound concentration. Each analysis was conducted in triplicate (n = 3).Peptide profile of the CPI hydrolysates by reverse-phase ultra-performance liquid chromatography (RP-UPLC)
Peptide profiles were determined by RP-UPLC (UPLC Acquity – Waters, Milford, MA, USA) as described previously by Nongonierma and FitzGerald,36 with modifications. Briefly, samples were resuspended (0.29% w protein equivalents/v) in solvent A (0.1% (v/v) TFA in MS grade water) and filtered with 0.2 μm cellulose acetate filters (VWR). Solvent B consisted of 0.1% (v/v) TFA and 80% MS grade ACN in water. Peptide separation was carried out at 30 °C with a flow rate of 0.3 mL min−1 and an injection volume of 10 μL. An Acquity UPLC BEH C18, 130 Å column (2.1 mm × 50 mm × 1.7 μm) equipped with an Acquity BEH C18 (1.7 μm) vanguard pre-column, both from Waters, were used. Peptides and proteins were eluted using a linear gradient: 0–0.28 min: 100% A; 0.28–30 min: 100–85% A, 30–44 min: 85–20% A. Absorbance was monitored at 214 nm.Molecular mass distribution of the CPI hydrolysates by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and gel permeation high performance liquid chromatography (GP-HPLC)
Samples were analysed with SDS-PAGE using a Mini Protean II electrophoresis system (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions. Samples at 5% (w/w) in distilled water were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in SDS solution (1% (w/v)). Samples were resuspended 1
1 in SDS solution (1% (w/v)). Samples were resuspended 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) ratio with loading buffer (protein loading buffer blue 2X – National Diagnostics, Atlanta, GA, USA) in reducing conditions (β-mercaptoethanol). Precast polyacrylamide gels (TruPage®, gradient 4–20%, Sigma-Aldrich) were loaded with the sample and then placed in Mini Protean Tetra Cell (Bio-Rad). The molecular weight markers range was between 6500 and 200
1 (v/v) ratio with loading buffer (protein loading buffer blue 2X – National Diagnostics, Atlanta, GA, USA) in reducing conditions (β-mercaptoethanol). Precast polyacrylamide gels (TruPage®, gradient 4–20%, Sigma-Aldrich) were loaded with the sample and then placed in Mini Protean Tetra Cell (Bio-Rad). The molecular weight markers range was between 6500 and 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da (SigmaMarker®, Sigma-Aldrich). A constant voltage of 100 V was applied during 50 min until the dye front reached the end of the separating gel. After electrophoresis, gels were stained with a Coomassie Brilliant Blue solution (0.025% (w/v) Coomassie Brilliant Blue and 10% (v/v) acetic acid) and de-stained (40% methanol and 10% (v/v) acetic acid).
000 Da (SigmaMarker®, Sigma-Aldrich). A constant voltage of 100 V was applied during 50 min until the dye front reached the end of the separating gel. After electrophoresis, gels were stained with a Coomassie Brilliant Blue solution (0.025% (w/v) Coomassie Brilliant Blue and 10% (v/v) acetic acid) and de-stained (40% methanol and 10% (v/v) acetic acid).
Samples were analysed by GP-HPLC as described earlier37 using an HPLC system (Waters model 600 binary pump, model 2707 autosampler and model 2489 dual λ absorbance detector interfaced with Empower™). Separation of compounds was conducted in isocratic mode at 21 °C using a TSK G2000 SW separating column (600 × 7.5 mm ID – Tosoh Bioscience, Tokyo, Japan) connected to a TSKGEL SW guard column (75 × 7.5 mm ID – Tosoh Bioscience) with a mobile phase made of 0.1% (v/v) TFA and 30% HPLC grade ACN in HPLC water. The flow rate was 0.5 mL min−1 for 60 min. A volume of 20 μL sample (0.14% (w protein equivalents/v) in mobile phase filtered through 0.2 μm PTFE syringe filters (VWR)) was injected. The absorbance was monitored at 214 nm. The molecular mass distribution of compounds >10, 10–5, 5–1 and <1 kDa was determined. Bovine serum albumin, β-lactoglobulin, α-lactalbumin, aprotinin, bacitracin, Leu-Trp-Met-Arg and Asp-Glu (Sigma-Aldrich) were used as standards.
Statistical analysis
Differences between mean AN or mean DPP-IV IC50 values were analysed with a one-way analysis of variance (ANOVA) at a significance level p < 0.05. Multiple means comparison (p < 0.05) was conducted using a post-hoc Student–Newman–Keuls test with SPPS (version 22, SPSS Inc., Chicago, IL, USA). The lack of significant difference between the experimentally determined and predicted DPP-IV IC50 values of hydrolysate H5 was assessed with Matlab using a one-sample Student test (t-test) with a significance of p < 0.05.Results
Physicochemical and DPP-IV inhibitory properties of the CP, CPI and the negative controls
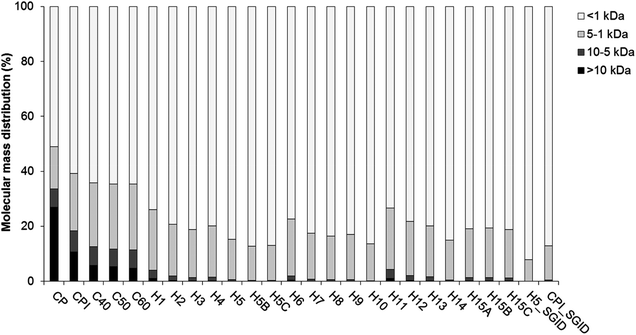
The protein yield and purity of the CPI were 20.90 ± 0.35% and 57.04 ± 2.23% (w/w), respectively.Both CP and CPI contained significant amounts of peptides (material < 10 kDa) as can be seen from the GPC profile (Fig. 1) and the SDS-PAGE profile (ESI Fig. S1†). This suggested that the cricket proteins were degraded during the processing steps leading to CP, i.e., freezing, roasting and grinding. Additional protein/peptide degradation was seen during the extraction process yielding CPI. This is illustrated by the reduction in the proportion of large molecular mass components (>10 kDa) in CPI compared to CP (Fig. 1). The SDS-PAGE profile also reveals a more intense smear below 10 kDa in the CPI compared to the CP (ESI Fig. S1†), indicating proteolytic/peptidolytic degradation of components within this sample.
The degradation of proteins within both CP and CPI are likely to arise from the presence of endogenous proteinases within the crickets. The azocasein assay was performed in order to understand if CP contained endogenous proteolytic activities. An azocasein activity of 0.0027 ± 0.0007 Abs min−1 mg−1 was obtained in the CP. This indicated the presence of endogenous proteinase(s) within the original substrate. Furthermore, when CPI was incubated at 40, 50 and 60 °C for 240 min to generate the negative controls for the different hydrolysates, additional hydrolysis was seen, as illustrated by the protein (>10 kDa) breakdown within CPI (Fig. 1).
The CP, CPI and the negative controls (C40, C50 and C60) were assessed for their DPP-IV inhibitory properties. All of these samples were able to inhibit DPP-IV in vitro. Their DPP-IV IC50 value ranged from 0.66 ± 0.03 to >3.57 mg mL−1 for CPI and CP, respectively (Table 1).
Physicochemical and DPP-IV inhibitory properties of the CPI hydrolysates
The reproducibility of the hydrolysis reaction was assessed by the generation of triplicate hydrolysates H15A, H15B and H15C. The AN of these samples as well as their DPP-IV IC50 values were not significantly different (p > 0.05, Table 1). These results indicated that hydrolysis of the CPI with Protamex™ was reproducible.The hydrolysates generated within the DOE were produced using Protamex™. The increase in AN (Table 1) was due to further protein breakdown within the CPI as can be seen on the molecular mass distribution profile (Fig. 1). In contrast with the negative controls (C40, C50 and C60), most hydrolysates did not contain material >10 kDa. In addition, hydrolysates contained a higher proportion of short peptides (<1 kDa) than the negative controls (Fig. 1).
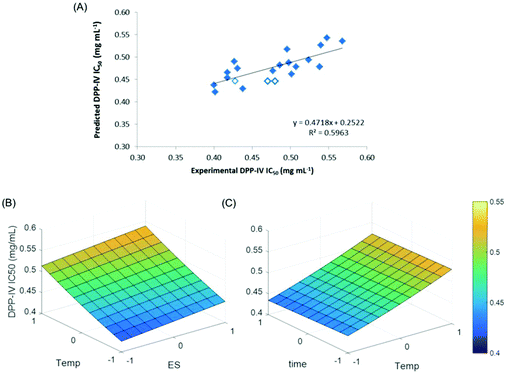
The DPP-IV IC50 values of the hydrolysates of the DOE varied between 0.40 ± 0.03/0.40 ± 0.02 and 1.01 ± 0.07 mg mL−1 for H2/H3 and H7, respectively (Table 1). The DPP IC50 values were used to generate the MLR model #1 (eqn (2)). The coefficients of this model are provided in Table 2. While a R2 of 0.758 was determined, the p-value model (0.126) was not significant. The only parameter of the model which was significant (p < 0.05) was the temperature (Table 2). The model was therefore simplified, taking into account the three main parameters and the DOE and T2. The p-value of this model was however still not significant (data not shown). The DPP-IV IC50 value obtained for H7 was an outlier, therefore it was removed from the model. Four additional hydrolysates (H16–H19, Table 2) were produced. A new model (model #2) was built, taking into account the data obtained with hydrolysates H1–H19 while excluding H7 (Table 2). Model #2 was statistically significant (p < 0.05, Table 2). In addition, the non-significance of the lack of fit (p > 0.05, Table 2) indicated that model #2 was relevant. The MLR between the experimental and predicted DPP-IV IC50 value is illustrated in Fig. 2A.
| Parametera | Coefficient | Estimate value | Standard error | t value | p | |
|---|---|---|---|---|---|---|
| a T: temperature; ES: enzyme to substrate ratio and t: time. b Parameters having a p < 0.05 are significantly different from 0. | ||||||
| Model #1 | Intercept | β 0 | 0.47 | 0.05 | 10.41 | 1.64 × 10−5 |
| ES | β 1 | −0.04 | 0.03 | −1.13 | 0.30 | |
| T | β 2 | 0.11 | 0.03 | 3.21 | 0.01 | |
| t | β 3 | 0.05 | 0.03 | 1.36 | 0.22 | |
| ES2 | β 4 | −0.01 | 0.06 | −0.15 | 0.89 | |
| T 2 | β 5 | 0.04 | 0.06 | 0.59 | 0.57 | |
| t 2 | β 6 | 0.03 | 0.06 | 0.41 | 0.69 | |
| ES × T | β 7 | −0.05 | 0.04 | −1.33 | 0.23 | |
| ES × t | β 8 | −0.06 | 0.04 | −1.50 | 0.18 | |
| T × t | β 9 | 0.07 | 0.04 | 1.87 | 0.10 | |
| Root mean squared error = 0.106; R2 = 0.758; p-value model = 0.126; p-value lack of fit = 0.019 | ||||||
| Model #2 | Intercept | β 0 | 0.478 | 0.02 | 29.20 | 1.24 × 10−14 |
| ES | β 1 | 0.012 | 0.01 | 0.85 | 0.41 | |
| T | β 2 | 0.044 | 0.01 | 3.65 | 2.39 × 10−3 | |
| t | β 0 | −0.003 | 0.01 | −0.28 | 0.78 | |
| T 2 | β 5 | 0.004 | 0.02 | 0.20 | 0.85 | |
| Root mean squared error = 0.0433; R2 = 0.502; p-value model = 0.026; p-value lack of fit = 0.134 | ||||||
The positive coefficient associated with temperature (Table 2) indicated that the DPP-IV IC50 value would be minimal at low temperature. The effect of temperature on the DPP-IV IC50 value is illustrated on the RSM curve, indicating a decrease in the IC50 value at the lowest (−1, z-centred value) temperature (Fig. 2B and C). Therefore, one of the experimental hydrolysates generated at low temperature, H5 (40 °C (−1), E![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S 2.00% (+1) and 240 min (+1)), was regenerated twice independently, yielding H5B and H5C. The AN values of the three samples (H5, H5B and H5C) were not significantly different (p > 0.05 Table 1). While significant differences were found for their DPP-IV IC50 values (p < 0.05), these were of the same order, ranging from 0.43 ± 0.04 to 0.48 ± 0.05 (for H5 and H5B, respectively, Table 1). The DPP-IV IC50 value of H5 was predicted using model #2 to be 0.45 mg mL−1. There was no significant difference (p > 0.05) between the predicted (0.45 mg mL−1) and experimental DPP-IV IC50 value of H5 (mean value 0.46 ± 0.04 mg mL−1).
S 2.00% (+1) and 240 min (+1)), was regenerated twice independently, yielding H5B and H5C. The AN values of the three samples (H5, H5B and H5C) were not significantly different (p > 0.05 Table 1). While significant differences were found for their DPP-IV IC50 values (p < 0.05), these were of the same order, ranging from 0.43 ± 0.04 to 0.48 ± 0.05 (for H5 and H5B, respectively, Table 1). The DPP-IV IC50 value of H5 was predicted using model #2 to be 0.45 mg mL−1. There was no significant difference (p > 0.05) between the predicted (0.45 mg mL−1) and experimental DPP-IV IC50 value of H5 (mean value 0.46 ± 0.04 mg mL−1).
SGID of CPI and H5
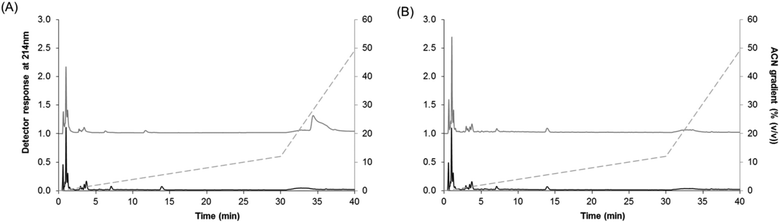
H5 was subjected to SGID as it was predicted to yield the most potent DPP-IV inhibitory hydrolysate. Following SGID, the physicochemical properties of both CPI and H5 were modified. The proportion of the larger molecular mass components (>5 kDa) was significantly reduced following SGID. As a consequence, the proportion of short peptides (<1 kDa) increased to >87% after SGID (Fig. 1). The peptide profiles of CPI and H5 before and after SGID are illustrated on Fig. 3. All samples contained intense peaks eluting before 5 min (2.5% ACN), which is indicative of the presence of hydrophilic compounds. Very few differences in terms of peptide peak profiles were seen following SGID of both samples. Differences between CPI before and after SGID were more evident (Fig. 3A). In particular, peaks eluting at 12 and 35 min in CPI were absent in the CPI_SGID sample.The DPP-IV inhibitory potency of CPI increased following SGID, resulting in a decrease in the DPP-IV IC50 value from >3.57 to 0.78 ± 0.04 mg mL−1 (Table 1). The opposite was seen with H5, where an increase in the DPP-IV IC50 value from 0.47 ± 0.03 to 0.71 ± 0.06 mg mL−1 was seen (Table 1). The SGID of CPI and of H5 both yielded samples having DPP-IV IC50 values of the same order.
Discussion
To date, the protein composition of G. sigillatus is essentially unknown. However, the protein content of G. sigillatus, reported to be as high as 70% (w/w),22 makes it interesting to investigate as a starting substrate for the generation of bioactive peptides. This is particularly the case, since assessment of the bioactive properties of G. sigillatus protein hydrolysates have not been extensively investigated. To our knowledge, one publication currently exists, which studied the antioxidant and anti-inflammatory properties of G. sigillatus proteins hydrolysed using an SGID protocol.26Extraction of proteins from edible insects has been conducted using a wide range of protocols.16 Alkaline solubilisation followed by isoelectric precipitation at acidic pH has been applied to recover water soluble proteins from G. sigillatus.26 In the present study, isoelectric precipitation resulted in a decrease in the extraction yield (data not shown). Therefore, this step was not applied during protein extraction. The extraction yield for proteins was 20.90 ± 0.35%, this is consistent with the solubility of G. sigillatus proteins as reported earlier. i.e., ∼25% at pH 10.0.25
The starting CP displayed significant levels of protein breakdown as evidenced by the molecular mass distribution profile (Fig. 1 and ESI Fig. S1†). These results are in contrast with the protein characterisation carried out earlier on G. sigillatus which revealed relatively high molecular mass proteins ranging from 6.5 to 212 kDa.25,26 These studies were however carried out with whole frozen25 or starved and then heat treated (boiled or baked)26 crickets. Heat treatments have been applied during the production of microbiologically safe edible insects for human consumption.24 However, heat treatment (boiling for 10 min at 100 °C or baking for 10 min at 150 °C) has been shown to break down large molecular mass proteins in G. sigillatus.26 The CP used herein was obtained following a roasting step (150–270 min at 230–250 °C). This may have contributed to some protein degradation. In addition, the presence of endoproteinase activities within the CP sample was demonstrated herein using the azocasein assay. The endogenous enzymes degraded the proteins within CP during the generation of CPI and also during incubation of CPI at 40, 50 and 60 °C (Table 1 and Fig. 1). The exact nature of these endogenous enzymes within G. sigillatus is not known. However, they may be similar to those identified within other species of crickets. For instance, tryspin- and chymotrypsin-like activities have been reported in the gastrointestinal tract of other cricket species.38–42 This assumption is further supported by the fact that the endogenous proteinases were active within the optimum range of trypsin and chymotrypsin (i.e., pH 7.0 and temperatures between 40 and 60 °C, Table 1).
Interestingly, the degradation of insect proteins by the endogenous proteinases resulted in an increase in the DPP-IV inhibitory potency. A relatively low DPP-IV IC50 value was obtained with CPI (0.66 ± 0.03 mg mL−1, Table 1). This DPP-IV IC50 value is lower than that recently reported for a water soluble extract of M. domestica larvae, i.e., 3.52 mg mL−1.13 In the previous study, insects were heated for 10 min until reaching boiling point. This treatment step may have deactivated endogenous proteinases from the larvae. Earlier studies have also demonstrated that endogenous proteinases may play a role in the release of DPP-IV inhibitory peptides within other biological samples. This was the case during the extraction of quinoa proteins where endogenous enzymes degraded the proteins, resulting in a sample displaying DPP-IV inhibitory activity in vitro.27
The DPP-IV inhibitory potency of most cricket protein hydrolysates was generally higher than that of CPI and the negative controls, except for H7 (Table 1). The lower DPP-IV IC50 value of the hydrolysates was linked to the release of short peptides following further hydrolysis with Protamex™. To date, most potent DPP-IV inhibitory peptides (IC50 value < 100 μM) have been shown to possess ≤10 amino acids.9,43 The cricket protein hydrolysates mostly contained peptides with a molecular mass <1 kDa (peptide length ≤ 10 amino acids). Protamex™, a Bacillus-derived enzyme preparation, contains subtilisin-like activities which cleave at the C-terminal side of hydrophobic amino acids.44 The positive role of hydrophobic amino acids within DPP-IV inhibitory peptides has been suggested.45–47 These hydrophobic amino acids are likely to interact with the S1 hydrophobic subsite of DPP-IV active site.48,49 Owing to its enzyme specificity, Protamex™ should release peptides possessing hydrophobic amino acids at their C-terminus. Several relatively potent DPP-IV inhibitors, having IC50 values <100 μM have been identified to possess hydrophobic amino acids at their C terminal side.43 Therefore, Protamex™ appeared as an interesting enzyme preparation to use during CPI hydrolysis. Protamex™ has been described elsewhere for the generation of food protein hydrolysates with relatively potent DPP-IV inhibitory properties. Various plant proteins (hemp, pea, rice and soy) hydrolysed with Protamex™ were shown to possess DPP-IV inhibitory properties, with DPP-IV IC50 values ranging from 0.73 ± 0.11 to 3.80 ± 0.13 mg mL−1 for pea and hemp proteins, respectively.50 DPP-IV IC50 values of 0.75 ± 0.15 (ref. 32) and 1.29 ± 0.21 mg mL−1 (ref. 51) were reported for Protamex™ hydrolysates of bovine caseins and whey proteins, respectively. A DPP-IV IC50 value of 2.43 ± 0.10 mg mL−1 was obtained with Barbel skin gelatine hydrolysed with Protamex™.52 The details of previously published potent DPP-IV inhibitory food protein hydrolysates, having IC50 values <1.0 mg mL−1, were recently compiled in a review article.43 The most potent food protein hydrolysate reported, to date, is a peptic bovine α-lactalbumin hydrolysate having a DPP-IV IC50 value of 0.036 mg mL−1.6 The cricket protein hydrolysates generated herein have a potency ∼10 times less than this α-lactalbumin hydrolysate. The cricket protein hydrolysates were however relatively potent DPP-IV inhibitory samples in comparison to other food protein hydrolysates reported in the literature. Interestingly, this study is the first reporting on the DPP-IV inhibitory properties of G. sigillatus protein hydrolysates. There is little to no information in the scientific literature regarding the protein composition and sequences of G. sigillatus. Therefore, it is not possible to predict the specific protein sources of the DPP-IV inhibitory peptides. However, muscle proteins such as actin and myosin have been described in other cricket species, e.g., Acheta domestica.53 Proteins including actin, mysosin and collagen have also been reported in various edible insects.54In silico studies have classified collagen as a protein source particularly rich in DPP-IV inhibitory peptides.12 Pro-containing peptides have been shown to be relatively potent DPP-IV inhibitory peptides.45,55 Collagen has been used for the generation of DPP-IV inhibitory hydrolysates with Pro-containing peptides.56,57 Given the cleavage specificity of Protamex™, Pro-rich peptides may be released from collagen.
The MLR regression model #2 generated within the DOE employed herein was statistically significant (Table 2). The R2 obtained with model #2 was relatively low compared to MLR models obtained earlier using a DOE linking hydrolysis parameters to the DPP-IV IC50 of the resultant hydrolysates.29,32,58,59 This may come from the fact that other parameters which were not taken into account in the model had an important role in peptide release. One of these parameters may be the impact of endogenous proteolytic enzymes which played a significant role in protein breakdown and therefore had an effect on the DPP-IV inhibitory activity of the resultant hydrolysates. The only hydrolysis parameter which was shown to have an effect (p < 0.05, Table 1) on the generation of DPP-IV inhibitory peptides was the temperature. The potency of cricket protein hydrolysates was highest with samples generated at 40 °C (Table 1, Fig. 2B and C). A similar effect of temperature on DPP-IV inhibition by Protamex™ hydrolysates of bovine sodium caseinate was reported earlier.32 This effect of temperature on Protamex™ was explained by its influence on the enzyme kinetics. In our study, temperature had an overall effect on both Protamex™ and the endogenous enzyme activities present within the CP.
Following SGID of H5, the DPP-IV inhibitory properties were still observed. In addition, SGID increased the DPP-IV inhibitory potency of CPI. Recently, a good agreement between peptide release during the digestion of milk proteins by SGID and in vivo digestion in humans60 and in pigs61 has been demonstrated. If the in vitro data reported herein translates in vivo this may suggest that G. sigillatus proteins have potential to generate agents with glucoregulatory properties in humans. The antioxidant and anti-inflammatory properties of the SGID of G. sigillatus have been reported earlier.26 All these results may form the basis for the development of multifunctional hydrolysates relevant to the management of various conditions of the metabolic syndrome.
The peptides present within G. sigillatus hydrolysates developed herein are not known. However, the identification of such peptides may be challenging as the G. sigillatus proteome is essentially unknown. The only detailed information currently available on G. sigillatus proteins concerns the reproductive proteins from this insect. However, future work may include peptide identification based on protein similarity with other insects belonging to the Orthoptera order. The non-water soluble proteins from G. sigillatus were not analysed in this study. These may also contain DPP-IV inhibitory peptide motifs which may be released using enzymatic hydrolysis. Future work may therefore study non-water soluble proteins from G. sigillatus. In addition, other enzyme preparations may be employed to verify their efficiency in the generation of more potent DPP-IV inhibitory samples.
Conclusions
Hydrolysis of G. sigillatus with Protamex™ has been described for the first time for the generation of DPP-IV inhibitory samples. The CP contains endogenous enzymes which were able per se to hydrolyse G. sigillatus proteins and yield samples with DPP-IV inhibitory properties. However, the addition of Protamex™ generally increased the DPP-IV inhibitory potency of the resultant hydrolysates. Relatively potent DPP-IV inhibitory hydrolysates were produced within this study. Interestingly, SGID of both the CPI and H5 generated samples having equivalent DPP-IV inhibitory potencies. The future evaluation of these hydrolysates in vivo is of interest as this may lead to the identification of novel food-grade samples with glycaemic regulatory properties.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
The work described herein was supported by Enterprise Ireland under Grant Number TC2013-0001. Candice Lamoureux was funded by the ERASMUS Program. The authors wish to thank Dr Goldin from Entomo Farms for supplying the CP and Aurélien Le Gouic for his assistance with the SDS-PAGE.Notes and references
- IDF, IDF diabetes atlas, 7th edn, 2015 Search PubMed.
- A. B. Nongonierma and R. J. FitzGerald, Peptides, 2015, 73, 20–34 CrossRef CAS PubMed.
- S. T. Silveira, D. Martínez-Maqueda, I. Recio and B. Hernández-Ledesma, Food Chem., 2013, 141, 1072–1077 CrossRef CAS PubMed.
- A. B. Nongonierma and R. J. FitzGerald, Peptides, 2013, 39, 157–163 CrossRef CAS PubMed.
- G. Tulipano, V. Sibilia, A. M. Caroli and D. Cocchi, Peptides, 2011, 32, 835–838 CrossRef CAS PubMed.
- I. M. Lacroix and E. C. Y. Li-Chan, J. Agric. Food Chem., 2013, 61, 7500–7506 CrossRef CAS PubMed.
- E. S. Andersen, C. F. Deacon and J. J. Holst, Diabetes, Obes. Metab., 2017 DOI:10.1111/dom.13018 , in press.
- L. Juillerat-Jeanneret, J. Med. Chem., 2014, 57, 2197–2212 CrossRef CAS PubMed.
- I. M. E. Lacroix and E. C. Y. Li-Chan, Trends Food Sci. Technol., 2016, 54, 1–16 CrossRef CAS.
- A. B. Nongonierma and R. J. FitzGerald, Curr. Opin. Food Sci., 2016, 8, 19–24 CrossRef.
- A. B. Nongonierma and R. J. FitzGerald, Food Chem., 2014, 165, 489–498 CrossRef CAS PubMed.
- I. M. E. Lacroix and E. C. Y. Li-Chan, J. Funct. Foods, 2012, 4, 403–422 CrossRef CAS.
- H. Li, A. Inoue, S. Taniguchi, T. Yukutake, K. Suyama, T. Nose and I. Maeda, PharmaNutrition, 2017, 5, 119–126 CrossRef.
- A. van Huis, Annu. Rev. Entomol., 2013, 58, 563–583 CrossRef CAS PubMed.
- A. van Huis, Proc. Nutr. Soc., 2016, 75, 294–305 CrossRef PubMed.
- A. B. Nongonierma and R. J. FitzGerald, Innovative Food Sci. Emerging Technol., 2017, 43, 239–252 CrossRef CAS.
- A. Halloran, P. Vantomme, Y. Hanboonsong and S. Ekesi, Food Secur., 2015, 7, 739–746 CrossRef.
- v. d. M. Spiegel, M. Noordam and H. Fels-Klerx, Compr. Rev. Food Sci. Food Saf., 2013, 12, 662–678 CrossRef.
- L. Vercruysse, G. Smagghe, G. Herregods and J. Van Camp, J. Agric. Food Chem., 2005, 53, 5207–5211 CrossRef CAS PubMed.
- Q. Wu, J. Jia, H. Yan, J. Du and Z. Gui, Peptides, 2015, 68, 17–24 CrossRef CAS PubMed.
- M. Tao, C. Wang, D. Liao, H. Liu, Z. Zhao and Z. Zhao, Process Biochem., 2017, 54, 172–179 CrossRef CAS.
- E. Zielińska, B. Baraniak, M. Karaś, K. Rybczyńska and A. Jakubczyk, Food Res. Int., 2015, 77(Part 3), 460–466 CrossRef.
- D. Vandeweyer, S. Crauwels, B. Lievens and L. Van Campenhout, Int. J. Food Microbiol., 2017, 242, 13–18 CrossRef CAS PubMed.
- R. Caparros Megido, S. Desmedt, C. Blecker, F. Béra, É. Haubruge, T. Alabi and F. Francis, Insects, 2017, 8, 12 CrossRef PubMed.
- F. G. Hall, O. G. Jones, M. E. O'Haire and A. M. Liceaga, Food Chem., 2017, 224, 414–422 CrossRef CAS PubMed.
- E. Zielińska, B. Baraniak and M. Karaś, Nutrients, 2017, 9, 970 CrossRef PubMed.
- A. B. Nongonierma, S. Le Maux, C. Dubrulle, C. Barre and R. J. FitzGerald, J. Cereal Sci., 2015, 65, 112–118 CrossRef CAS.
- K. Kilcawley, M. Wilkinson and P. Fox, Enzyme Microb. Technol., 2002, 31, 310–320 CrossRef CAS.
- A. B. Nongonierma, C. Mazzocchi, S. Paolella and R. J. FitzGerald, Food Res. Int., 2017, 94, 79–89 CrossRef CAS PubMed.
- S. Le Maux, A. B. Nongonierma, C. Barre and R. J. FitzGerald, Food Chem., 2016, 199, 246–251 CrossRef CAS PubMed.
- C. van der Ven, H. Gruppen, D. B. A. de Bont and A. G. J. Voragen, Int. Dairy J., 2002, 12, 813–820 CrossRef CAS.
- A. B. Nongonierma, S. Le Maux, C. Esteveny and R. J. FitzGerald, J. Sci. Food Agric., 2017, 97, 1093–1101 CrossRef CAS PubMed.
- A. B. Nongonierma and R. J. FitzGerald, RSC Adv., 2016, 6, 75400–75413 RSC.
- D. J. Walsh, H. Bernard, B. A. Murray, J. MacDonald, A. K. Pentzien, G. A. Wright, J. M. Wal, A. D. Struthers, H. Meisel and R. J. FitzGerald, J. Dairy Sci., 2004, 87, 3845–3857 CrossRef CAS PubMed.
- G. R. Lankas, B. Leiting, R. S. Roy, G. J. Eiermann, M. G. Beconi, T. Biftu, C.-C. Chan, S. Edmondson, W. P. Feeney and H. He, Diabetes, 2005, 54, 2988–2994 CrossRef CAS PubMed.
- A. B. Nongonierma and R. J. FitzGerald, Peptides, 2012, 37, 263–272 CrossRef CAS PubMed.
- D. Spellman, G. O'Cuinn and R. J. FitzGerald, Food Chem., 2009, 114, 440–446 CrossRef CAS.
- T. Nakashima, K. Tokuyasu and M. Funatsu, Agric. Biol. Chem., 1965, 29, 307–314 CrossRef CAS.
- M. A. Khan, Entomol. Exp. Appl., 1963, 6, 181–193 CrossRef CAS.
- F. J. Clissold, B. J. Tedder, A. D. Conigrave and S. J. Simpson, Proc. R. Soc. B, 2010, 277, 1751–1759 CrossRef PubMed.
- W. Lam, G. M. Coast and R. C. Rayne, Insect Biochem. Mol. Biol., 2000, 30, 85–94 CrossRef CAS PubMed.
- J. Spit, S. Zels, S. Dillen, M. Holtof, N. Wynant and J. Vanden Broeck, Insect Biochem. Mol. Biol., 2014, 48, 100–109 CrossRef CAS PubMed.
- A. B. Nongonierma and R. J. FitzGerald, J. Food Biochem., 2017 DOI:10.1111/jfbc.12451 , in press.
- M. Smyth and R. J. FitzGerald, Int. Dairy J., 1998, 8, 819–827 CrossRef CAS.
- A. B. Nongonierma and R. J. FitzGerald, J. Funct. Foods, 2013, 5, 1909–1917 CrossRef CAS.
- K.-C. Hsu, Y.-S. Tung, S.-L. Huang and C.-L. Jao, in Bioactive Food Peptides in Health and Disease, ed. B. Hernández-Ledesma, In Tech, http://www.intechopen.com/books/bioactive-food-peptides-in-health-and-disease/dipeptidyl-peptidase-iv-inhibitory-activity-of-peptides-in-porcine-skin-gelatin-hydrolysates, 2013, pp. 205–218, DOI:10.5772/51264.
- G. Tulipano, L. Faggi, A. Nardone, D. Cocchi and A. M. Caroli, Int. Dairy J., 2015, 48, 62–72 CrossRef.
- M. Engel, T. Hoffmann, L. Wagner, M. Wermann, U. Heiser, R. Kiefersauer, R. Huber, W. Bode, H.-U. Demuth and H. Brandstetter, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 5063–5068 CrossRef CAS PubMed.
- A. B. Nongonierma, C. Mooney, D. C. Shields and R. J. FitzGerald, Food Chem., 2013, 141, 644–653 CrossRef CAS PubMed.
- A. B. Nongonierma and R. J. FitzGerald, Food Dig., 2015, 6, 19–29 CAS.
- S. Le Maux, A. B. Nongonierma, C. Lardeux and R. J. FitzGerald, Int. J. Food Sci. Technol., 2017 DOI:10.1111/ijfs.13576 , in press.
- A. Sila, O. Martinez-Alvarez, A. Haddar, M. C. Gómez-Guillén, M. Nasri, M. P. Montero and A. Bougatef, Food Chem., 2015, 168, 478–486 CrossRef CAS PubMed.
- R. H. Oliver, A. N. J. Albury and T. A. Mousseau, J. Insect Physiol., 2007, 53, 30–39 CrossRef CAS PubMed.
- L. Vercruysse, J. Van Camp and G. Smagghe, J. Agric. Food Chem., 2005, 53, 8106–8115 CrossRef CAS PubMed.
- T. Hatanaka, Y. Inoue, J. Arima, Y. Kumagai, H. Usuki, K. Kawakami, M. Kimura and T. Mukaihara, Food Chem., 2012, 134, 797–802 CrossRef CAS PubMed.
- T. Hatanaka, K. Kawakami and M. Uraji, J. Enzyme Inhib. Med. Chem., 2014, 29, 823–828 CrossRef CAS PubMed.
- E. C. Y. Li-Chan, S.-L. Hunag, C.-L. Jao, K.-P. Ho and K.-C. Hsu, J. Agric. Food Chem., 2012, 60, 973–978 CrossRef CAS PubMed.
- A. B. Nongonierma, M. Lalmahomed, S. Paolella and R. J. FitzGerald, Food Chem., 2017, 231, 202–211 CrossRef CAS PubMed.
- A. B. Nongonierma, S. Paolella, P. Mudgil, S. Maqsood and R. J. FitzGerald, J. Funct. Foods, 2017, 34, 49–58 CrossRef CAS.
- J. Sanchón, S. Fernández-Tomé, B. Miralles, B. Hernández-Ledesma, D. Tomé, C. Gaudichon and I. Recio, Food Chem., 2018, 239, 486–494 CrossRef PubMed.
- L. Egger, P. Schlegel, C. Baumann, H. Stoffers, D. Guggisberg, C. Brügger, D. Dürr, P. Stoll, G. Vergères and R. Portmann, Food Res. Int., 2017 DOI:10.1016/j.foodres.2017.09.047 , in press.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7fo01568b |
| This journal is © The Royal Society of Chemistry 2018 |