Musa paradisiaca inflorescence induces human colon cancer cell death by modulating cascades of transcriptional events†
Arun
K. B.
 a,
Aravind
Madhavan
b,
Reshmitha
T. R.
ac,
Sithara
Thomas
ac and
P.
Nisha
a,
Aravind
Madhavan
b,
Reshmitha
T. R.
ac,
Sithara
Thomas
ac and
P.
Nisha
 *ac
*ac
aAgro Processing and Technology Division, National Institute for Interdisciplinary Science and Technology (CSIR-NIIST), Thiruvananthapuram-695019, Kerala, India. E-mail: bp.nisha@yahoo.com; Tel: +91 471 2515348
bMicrobial Processes and Technology Division, National Institute for Interdisciplinary Science and Technology (CSIR-NIIST), Thiruvananthapuram-695019, Kerala, India
cAcademy of Scientific and Innovative Research (AcSIR), New Delhi 110001, India
First published on 29th November 2017
Abstract
Colorectal cancer (CRC) is one of the leading causes of cancer death, and diet plays an important role in the etiology of CRC. Traditional medical practitioners in many South Asian countries use plantain inflorescence to treat various gastro-intestinal ailments. The aim of the present study was to investigate the anticancer effects of extracts of inflorescence of Musa paradisiaca against HT29 human colon cancer cells and elucidate the mechanism of these effects by studying the modulation of cascades of transcriptional events. In vitro assays depicted that methanol extract of Musa paradisiaca inflorescence (PIMET) was cytotoxic to HT29 cells. PIMET induced DNA damage and arrested the cell cycle at the G2/M phase. Expression studies showed that PIMET pretreatment upregulates pro-apoptotic Bcl2 and downregulates anti-apoptotic Bax proteins. Different assays showed that the deregulation of pro/antiapoptotic proteins reduces the mitochondrial membrane potential and ATP production; moreover, it enhances cytochrome c release, which triggers the apoptotic pathway, and further cleaves caspase 3 and PARP proteins, resulting in apoptosis. Changes in the protein expression profile of HT29 cells after PIMET treatment were analyzed using mass-spectrometry-based proteomics. PIMET treatment significantly altered the expression of HT29 protein; interestingly, X-linked inhibitor of apoptosis protein was also downregulated. Alteration in the expression of this protein has significant effects, leading to HT29 cell death.
1. Introduction
Epidemiological studies suggest that diets rich in bioactive phytochemicals can play a major role in the prevention and management of non-communicable diseases such as cancer.1 Consumption of fruits high in polyphenolic compounds is currently increasing due to their chemotherapeutic potential.2 There are many convincing reports indicating that the consumption of fruits and vegetables may reduce the risk of colorectal cancer by approximately 50%.3 Colorectal cancer (CRC) is the third most common type of cancer worldwide and a major cause of death by cancer.4 Torre et al. (2015) reported that 693![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 deaths occurred due to CRC in 2012.5 The onset of CRC has been reported to be associated with dietary and lifestyle factors, wherein 90% of CRC mortality is attributed to dietary factors.6,7 The anticancer properties of fruits and vegetables are reported to be associated with constituents such as carotenoids, vitamins (C and E), dietary fibre, folic acid, selenium, and polyphenols; this may be due to the radical scavenging activities of the bioactive phytochemicals or their interactions with metabolic and molecular events.8,9 At present, 5-fluorouracil is the primary option for colon cancer treatment;10 it inhibits DNA synthesis.11 Moreover, most anticancer drugs have severe side effects. Hence, nutraceuticals and phytochemicals with fewer side effects have been investigated extensively for colon cancer therapeutics; this has been reviewed in many studies.12,13
900 deaths occurred due to CRC in 2012.5 The onset of CRC has been reported to be associated with dietary and lifestyle factors, wherein 90% of CRC mortality is attributed to dietary factors.6,7 The anticancer properties of fruits and vegetables are reported to be associated with constituents such as carotenoids, vitamins (C and E), dietary fibre, folic acid, selenium, and polyphenols; this may be due to the radical scavenging activities of the bioactive phytochemicals or their interactions with metabolic and molecular events.8,9 At present, 5-fluorouracil is the primary option for colon cancer treatment;10 it inhibits DNA synthesis.11 Moreover, most anticancer drugs have severe side effects. Hence, nutraceuticals and phytochemicals with fewer side effects have been investigated extensively for colon cancer therapeutics; this has been reviewed in many studies.12,13
Musa paradisiaca (plantain) is an herbaceous plant that is cultivated for fruit mainly in southern parts of Asia. Plantain inflorescence (PI) is widely used as a vegetable in southern India, Malaysia, Thaiwan, Srilnaka and some African countries.14 Additionally, inflorescence is traditionally used for the treatment of ailments such as dysentery, ulcers, menorrhagia and diabetes.15,16 Different parts of the plantain, especially its inflorescence, have been traditionally used for maintenance of gastrointestinal health.16 It is reported that PI is a rich source of phenolic compounds with potential radical scavenging activities.17 Our previous study showed that PI is a very good source of dietary fibre (12.54%) and that the extracts are rich in polyphenols. We identified and quantified twelve polyphenols in the methanol extract of PI and demonstrated their potential antidiabetic and cardiovascular protection properties.18 Dietary fibre is known to act as a matrix which holds biologically active molecules.19 These molecules, which are released during digestion as well as fermentation by probiotic bacteria, exert beneficial effects on gastrointestinal health.20,21 The dietary fibre and polyphenols present in inflorescence can play an important role in the maintenance of gastrointestinal health and in reducing the risk of developing colon cancer. Although it is a rich source of dietary fibre with bioactive compounds, the effects of plantain inflorescence on cancer, in particular colorectal carcinoma cell growth, have not been studied in detail. In the present study, we selected the Nendran variety of Musa paradisiaca because it is one of the most important plantain varieties grown in India, particularly in Kerala. The study was designed to assess the anticancer potential of extracts of Musa paradisiaca inflorescence against HT29 colon cancer cells and to decode the underlying mechanisms which impart its specific effects.
2. Materials and methods
2.1 Chemicals
Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), trypsin, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), dimethyl sulfoxide (DMSO), acridine orange (AO), ethidium bromide (EtBr), propidium iodide and rhodamine 123 (Rh123) were purchased from Sigma Aldrich Chemicals, St Louis, USA. Penicillin and streptomycin were obtained from Himedia Laboratories, Mumbai, India. Antibodies were purchased from Santa Cruz Biotechnology, Inc., USA.2.2 Sample
Plantain inflorescence (PI) from the Nendran variety, identified as Musa paradisiaca, was collected from a local banana farm located in the Thiruvananthapuram district of Kerala, India. A voucher specimen of Musa paradisiaca (TBGT 81481) has been deposited in the herbarium (TBGT) of the Jawaharlal Nehru Tropical Botanic Garden and Research Institute, Thiruvananthapuram, Kerala, India. The inflorescence was washed, drained and sliced into small pieces. It was then freeze-dried using a lyophilizer (VirTis genesis, USA). The dried sample was ground (Ultra centrifugal millZM200, Retsch, Germany) and sieved (20 mm mesh – Vibro Sifter-PVS30, Prism Pharma Machinery, India) to obtain a fine powder.2.3 Preparation of PI extracts
The PI powder was defatted with hexane and extracted with ethyl acetate (PIETH) and methanol (PIMET) at room temperature. The extracts were filtered through Whatman No. 1 filter paper, and the solvent was removed under reduced pressure (BUCHI R215, Switzerland) followed by lyophilization. Prior to the cell-based assays, the extracts were reconstituted in 50% DMSO and diluted further so that the final concentration of DMSO was less than 0.1% in the media.2.4 Cell line and culture medium
HT29 colon cancer cell lines were obtained from the National Centre for Cell Science, Pune, India. Cells were cultured in DMEM supplemented with 10% FBS, penicillin (100 U mL−1) and streptomycin (100 μg mL−1) and were maintained at 5% CO2 and 37 °C.2.5 Determination of cytotoxicity
The cytotoxicity of the PI extract against the HT29 cell line was determined by MTT and LDH release assays. The MTT assay was performed as described by Mosmann,22 and the LDH assay was performed using an assay kit (Cayman Chemical Company, USA). 5-Fluorouracil (5-FU) and dimethyl sulfoxide (DMSO) were used as positive controls for the MTT and LDH assays, respectively.2.6 Detection of cell death by AO/EtBr staining
AO/EtBr dual staining was performed to determine the effects of PI on cell death.232.7 Hoechst 33342 staining
The apoptotic activity of the extract on HT29 cells was determined by Hoechst 33342 staining as described by Harada et al.242.8 Analysis of cell cycle distribution profile
The effects of the extract on HT29 cell cycle distribution were assessed by flow cytometry after propidium iodide staining.25 1 × 104 cells per well were treated with different concentrations of PIMET for 24 h. Cells in the control group received only media containing 0.1% DMSO, and Taxol (50 nm) was used as a positive control. Cells were harvested, washed with PBS, fixed with ice-cold 70% ethanol and maintained at 4 °C for 12 h. The cells were washed again with cold PBS, stained with propidium iodide (5 μg mL−1) containing 0.1 mg mL−1 RNAse, and incubated in the dark for 30 min. The cellular DNA content was analyzed using Fluorescence Activated Cell Sorting (BD FACS Aria II, USA) and the percentages of cells in the G0/G1, S, and G2/M phases of the cell cycle were determined using BD FACSDiva™ Software v6.1.2.2.9 Apoptosis assay by flow cytometry
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide staining was used to detect the effects of PI extract on apoptosis in HT29 cells using an annexin V-FITC/propidium iodide apoptosis detection kit (Cayman Chemical Company, USA). Briefly, 1 × 104 cells were treated with different concentrations of PI extract and incubated for 24 h. Control group cells were treated with media containing 0.1% DMSO, and camptothecin (50 μM) was used as a positive control. Following 24 h incubation, the cells were harvested, washed with cold PBS and resuspended in binding buffer. The cells were treated with annexin V-FITC conjugate and incubated for 15 min at room temperature in the dark. The cells were then stained with propidium iodide (5 μg mL−1) and analyzed by FACS.2.10 Detection of mitochondrial membrane potential in HT29 cells
The effects of PI extract on mitochondrial membrane potential were identified by staining with Rh123, a green fluorescent cationic dye that binds to the polarized mitochondrial membrane and accumulates as aggregates in the mitochondria of normal cells. When the mitochondrial membrane potential is reduced, the dye washes out, resulting in reduction of green fluorescence. To analyze changes in their mitochondrial trans-membrane potential, HT29 cells were treated with different concentrations of PI extract for 24 h.2.11 Effects on ATP production
The ATP levels in HT29 cells after treatment with extract were determined using HPLC.262.12 Detection of cytochrome C release
Cytochrome c release due to mitochondrial damage was assessed according to Radhakrishnan et al.27 with slight modifications. Detailed methods are provided in the ESI.†2.13 Western blot analysis
Cells (1 × 105) were seeded in a 24-well culture plate for 1 day before treatment with the PI extract. After 24 h incubation, the cells were collected and lysed for western blot analysis. Antibodies specific against β-actin, PARP, cleaved PARP, caspase 3, cleaved caspase 3, BCL2, and BAX were used. After cell lysis, the protein concentration was determined with the Bradford method, and 40 μg of proteins were separated on 10% SDS polyacrylamide gel and transferred to a PVDF transfer membrane (Immobilon P™, Millipore®, USA). For immunoblotting, primary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and IgG-HRP secondary antibody (1
1000) and IgG-HRP secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) were used. The blots were later washed with wash buffer and were detected using DAB-Peroxidase substrate solution (0.05% DAB, 0.015% H2O2, 0.01 M PBS, pH 7.2). The pixel densities of specific protein bands (ImageJ software) were compared to that of the housekeeping gene β actin and were plotted to obtain graphs.
2000) were used. The blots were later washed with wash buffer and were detected using DAB-Peroxidase substrate solution (0.05% DAB, 0.015% H2O2, 0.01 M PBS, pH 7.2). The pixel densities of specific protein bands (ImageJ software) were compared to that of the housekeeping gene β actin and were plotted to obtain graphs.
2.14 Two-dimensional electrophoresis
In order to prepare the cell lysates, 5 × 105 HT29 cells per well were plated on 6-well plates. After incubation with PIMET extract, the cells were scraped out and washed with PBS; then, lysis buffer was added, and the cells were incubated for 30 min on ice followed by centrifugation at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g (Kubota-7780, Japan) for 20 min at 4 °C to collect the supernatant. Proteins were precipitated by 10% TCA in acetone, and the pelleted proteins were solubilized in buffer containing 7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 50 mM DTT, 0.2% 100× BioLyte 3/10 ampholyte, and 0.002% (w/v) bromophenol blue. The total soluble protein concentration was determined by Bradford's method.
000g (Kubota-7780, Japan) for 20 min at 4 °C to collect the supernatant. Proteins were precipitated by 10% TCA in acetone, and the pelleted proteins were solubilized in buffer containing 7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 50 mM DTT, 0.2% 100× BioLyte 3/10 ampholyte, and 0.002% (w/v) bromophenol blue. The total soluble protein concentration was determined by Bradford's method.
The first dimension based on the isoelectric point of the proteins was run using a strip-based Isoelectric Focusing System (Protean IEF®, Bio-Rad, USA) at 20 °C with a current of 50 μA per strip. Protein sample (160 μg PIMET-treated protein) was loaded onto IPG Dry Strip™ gels with pH 3 to 10 (Bio-Rad, USA) by in-gel rehydration. IEF was performed under the following conditions: 500 V × 20 min, 4000 V × 2.5 h, and 8000 V for 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 V h. Prior to the second dimension (SDSPAGE), the IPG strips were equilibrated for 15 min in 6 M urea, 30% glycerol (v/v), 2% SDS (w/v), and 0.375 M Tris-HCl (pH 8.8) containing 2% (w/v) DTT, and then for 15 min in the same buffer additionally containing 2.5% (w/v) iodoacetamide. Equilibrated strips were transferred onto SDS-polyacrylamide gels for the second dimension. The proteins were separated on 12% gels. Subsequently, the gels were stained using Coomassie Blue R-250. The gels were scanned with a GS-800 calibrated imaging densitometer (Bio-Rad Laboratories GmbH, Munich, Germany) using QuantityOne software. For each sample, three replicate 2D-gels were comparatively analyzed using PDQuest 8.0 software (Bio-Rad Laboratories GmbH, Munich, Germany), which allowed automatic detection with manual correction and quantification of protein spots (based on pixel density). The significance of the differences between the protein spots was evaluated; a p value lower than 0.05 was considered to be significant. An additional selection criterion was a fold change value (based on the ratio of the band intensity of the sample protein to that of the corresponding untreated control protein) of a protein spot which maintained consistency in repeated experiments.
000 V h. Prior to the second dimension (SDSPAGE), the IPG strips were equilibrated for 15 min in 6 M urea, 30% glycerol (v/v), 2% SDS (w/v), and 0.375 M Tris-HCl (pH 8.8) containing 2% (w/v) DTT, and then for 15 min in the same buffer additionally containing 2.5% (w/v) iodoacetamide. Equilibrated strips were transferred onto SDS-polyacrylamide gels for the second dimension. The proteins were separated on 12% gels. Subsequently, the gels were stained using Coomassie Blue R-250. The gels were scanned with a GS-800 calibrated imaging densitometer (Bio-Rad Laboratories GmbH, Munich, Germany) using QuantityOne software. For each sample, three replicate 2D-gels were comparatively analyzed using PDQuest 8.0 software (Bio-Rad Laboratories GmbH, Munich, Germany), which allowed automatic detection with manual correction and quantification of protein spots (based on pixel density). The significance of the differences between the protein spots was evaluated; a p value lower than 0.05 was considered to be significant. An additional selection criterion was a fold change value (based on the ratio of the band intensity of the sample protein to that of the corresponding untreated control protein) of a protein spot which maintained consistency in repeated experiments.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with 10 to 20 mg mL−1 DAHC. The analysis was performed using an ultrafleXtreme MALDI-TOF/TOF instrument from Bruker Daltronik. The mass spectra were imported into a database search engine (BioTools v2.2 connected to Mascot, version 2.2.04; Matrix Science). Mascot searches were performed using the NCBI non-redundant database (Human Genome).
1 ratio with 10 to 20 mg mL−1 DAHC. The analysis was performed using an ultrafleXtreme MALDI-TOF/TOF instrument from Bruker Daltronik. The mass spectra were imported into a database search engine (BioTools v2.2 connected to Mascot, version 2.2.04; Matrix Science). Mascot searches were performed using the NCBI non-redundant database (Human Genome).
2.15 Statistical analysis
The experimental results were expressed as mean ± SD (standard deviation) of triplicate measurements. The data were subjected to one way analysis of variance (ANOVA), and the significance of the differences between means was calculated by Duncan's multiple range test using SPSS for Windows, standard version 7.5.1, SPSS (SPSS Inc., USA); significance was accepted at p ≤ 0.05.3. Results and discussion
The risk of developing colorectal cancer is usually associated with an unhealthy diet that primarily lacks dietary fibre and antioxidants. The concept of antioxidant dietary fibre is gaining increasing attention in the prevention and management of colon cancer.28 Based on this background, as a preliminary study, we screened various agro-industrial residues and discovered that plantain (Musa paradisiaca) inflorescence can be utilized as a good source of antioxidant dietary fibre. HPLC analysis of a methanol extract of plantain inflorescence showed the presence of twelve polyphenols: gallic acid (1.29 mg mL−1), catechol (0.323 mg mL−1), chlorogenic acid (0.115 mg mL−1), caffeic acid (0.068 mg mL−1), syringic acid (0.233 mg mL−1), p-coumaric acid (0.108 mg mL−1), ferulic acid (0.019 mg mL−1), ellagic acid (0.028 mg mL−1), myricetin (0.072 mg mL−1), cinnamic acid (0.086 mg mL−1), kaempferol (0.056 mg mL−1) and apigenin (0.025 mg mL−1).21 In the present study, we will validate the anticancer efficacy of the methanol extract of plantain inflorescence (PIMET) against HT29 colon cancer cells and attempt to delineate the mechanistic pathway through which these anticancer properties are executed.3.1 Cytotoxic effect of PI extracts
The cytotoxic effects of ethyl acetate and methanol extracts were determined by MTT assay and LDH release assay. The cellular reduction of MTT occurs in mitochondria by the action of the enzyme succinate dehydrogenase and outside the mitochondria by the action of NADH and NADPH. Therefore, only live cells in a population can reduce MTT. The LDH release assay is based on the fact that cytosolic LDH leaks out of cells with damaged membranes.The MTT assay (ESI Fig. S1[A]†) indicated that among the extracts of PI, methanol extract showed the best cytotoxic effects against HT29 colon cancer cells. The IC50 values were found to be 259.84 and 130.26 μg mL−1, respectively, for the PIETH and PIMET extracts. The cytotoxic effects of the extracts increased in a concentration-dependent manner. PIMET exhibited better cytotoxic activity than PIETH. 5-FU (positive control) inhibited 50% cell growth at 51.95 μM (≈ 6.794 μg mL−1, Fig. S1[B]†).
The cytotoxic effects of the PI extracts were again confirmed by LDH release assay (ESI Fig. S1[B]†). The results indicated that the viability of HT29 cells decreased significantly with increasing concentration of PI extract. These results were in accordance with the MTT assay; also, PIMET exhibited better activity than PIETH. The IC50 values were found to be 268.62 and 195.31 μg mL−1, respectively, for the PIETH and PIMET extracts. The activities of the extracts were better than those of the positive control (DMSO) used for the assay (Fig. S1[D]†). The IC50 value for DMSO was found to be 0.431 M (≈ 33.674 mg mL−1).
Our preliminary studies showed that PIMET is rich in polyphenols, especially gallic acid, syringic acid, catechol and ferulic acid.18 The cytotoxic effects of the PI extracts may be related to the higher phenolic content of PIMET, as phenolic compounds are reported to have anticancer activity. Gallic acid, syringic acid, catechol and ferulic acid are reported to have anticancer activities on various cancer cell lines.29–32 The extracts may affect the membrane integrity of HT29 cells, which acts as a trigger for inducing cell death. Because PIMET exhibited better activity with the MTT and LDH release assays, for further anticancer assays, two concentrations of PIMET (1 to 50 μg mL−1 and 2 to 100 μg mL−1) were selected.
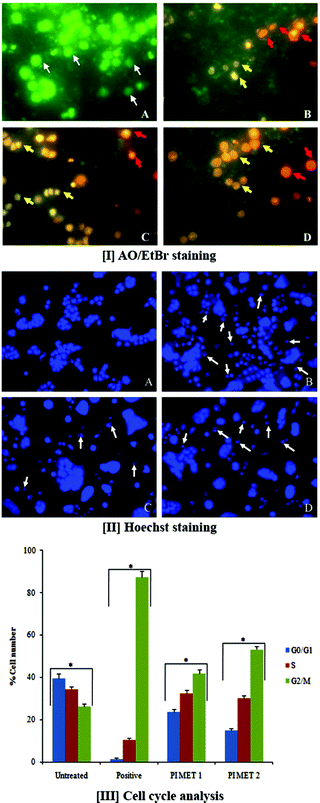
3.2 Changes in cell morphology upon treatment with a methanol extract of PI using AO/EtBr double staining
AO/EtBr double staining allows rapid and easy recognition of live and dead cells when visualized by fluorescence microscopy. AO is a crucial dye which can stain cells that have lost membrane integrity. Under fluorescent emission, AO is taken up by both viable and non-viable cells and emits a green fluorescence, whereas EtBr is taken up only by non-viable cells and emits red fluorescence when intercalated into DNA strands. Hence, live cells have normal green nuclei, whereas early apoptotic cells have bright green-yellow nuclei with condensed or fragmented chromatin and late apoptotic cells show condensed or fragmented orange-red chromatin.The morphological observations of HT29 cells treated with PIMET showed various morphological changes compared to the untreated control cells (Fig. 1[I]). The control cells appeared to be intact oval shapes, and the nuclei were stained uniformly green due to binding with AO dye. However, PIMET-treated HT29 cells showed signs of apoptosis, such as cell shrinkage, formation of apoptotic bodies and membrane blebbing. The percentages of early apoptotic cells for PIMET (50 and 100 μg mL−1) and for the standard H2O2 (250 μM) were 27.33 ± 1.57%, 31.5 ± 1.65% and 61.5 ± 1.78%, respectively, whereas the percentages of late apoptotic cells were 3.67 ± 0.2%, 6.13 ± 0.49% and 1.57 ± 0.21%, respectively (ESI Fig. S2†). An increase in the percentage of apoptotic cells with increasing concentration was noted.
3.3 Methanol extract of PI induces DNA damage in HT29 cells
The DNA damage induced by PIMET extract in HT29 cells was determined by Hoechst staining. Hoechst 33342 binds to adenine–thymine-rich regions of DNA in the minor groove. On binding to DNA, the fluorescence greatly increases, which helps identify characteristic changes. After cells were treated with PIMET extract for 24 h, marked morphological changes related to cell apoptosis, such as condensation of chromatin and nuclear fragmentation, were clearly observed using Hoechst 33342 staining. These morphological changes related to cell apoptosis, such as condensation of chromatin and nuclear fragmentation, were clearly observed in Fig. 1[II]. The morphological analysis indicated the ability of PIMET to induce apoptosis in HT29 cells. Apoptotic cells gradually increased in a concentration-dependent manner when cells were treated with PIMET extract. The cells with normal nuclei after treatment with PIMET/H2O2 were quantified; it was found that the percentage of cells with normal nuclei after PIMET treatment at 100 μg mL−1 concentration was only 56.17 ± 4.53%, and that for the positive control was 41.35 ± 3.21% (ESI Fig. S3†).3.4 Effects of PI extract on cell cycle
Normal cells usually transform into cancerous cells when the proteins involved in regulating cell division events fail to appropriately drive progression from one cell cycle stage to the next. Cancer cells reproduce at a rate far beyond the normal tightly regulated limits of the cell cycle. To determine the cellular mechanism of growth inhibition of PIMET in HT29 cells, we investigated cell cycle progression after PIMET treatment. The distribution of cells in all three phases of HT29 changed significantly as the PIMET concentration increased from 50 μg mL−1 to 100 μg mL−1 (Fig. 1[III]). It was noted that the number of G2/M phase cells increased significantly, whereas the number of G0/G1 phase cells decreased. However, the activity of PIMET was less than that of the standard taxol (50 nM) used in the assay.Previous studies have confirmed that grape seed, Longon seed, and Longon flower extract increase S phase cells in colorectal cancer.33–35 The cell cycle is controlled by a group of regulatory proteins named cyclins. Reports showed that cyclin B1 expression is decreased when G2/M phase arrest occurs, which prevents cells from entering mitosis.36 Gallic acid and kaempferol are reported to arrest the cell cycle at G2/M phase by decreasing cyclin B1 expression.37,38 In the present study, HT29 cells exhibited significant increases in the number of G2/M phase cells following treatment with PIMET. G2/M phase cells increased from 26.2 ± 1.2% (untreated control) to 53.2 ± 1.6% (PIMET 100 μg mL−1). These findings suggested that the anti-proliferative effects induced by polyphenol-rich naturally occurring products involve different cell cycle controlling mechanisms. The different composition of polyphenols in each natural product may induce different expressions of cyclin proteins to control the cell cycle in colorectal cancer cells. The molecular mechanism responsible for the perturbation of the cells from M to G1 phase of the cell cycle in HT29 cells requires further investigation. Additionally, we strongly believe that this extract may contain biologically active compounds other than polyphenols which may play significant roles in the anticancer potential of PIMET. Further chemical analysis is required to validate this statement.
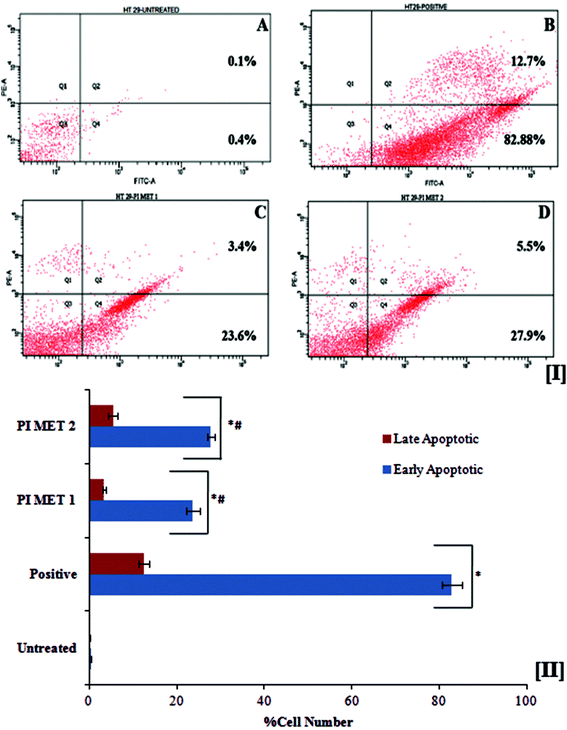
3.5 Assessment of apoptosis
The death of aged cells in the human body is programmed by a sequence of events known as apoptosis. Cancer research has led to great developments in exploring in the role of oncogenic mutations which alter the apoptotic pathway, resulting in tumor initiation, progression or metastasis. Polyphenols are known to overcome this oncogenic mutation and prompt apoptosis in cancer cells. In apoptotic cells, the membrane phosphatidylserine is translocated from the inner to the outer plasma membrane, thereby exposing phosphatidylserine to the external cellular environment. Hence, the effects of PIMET extract on apoptosis were investigated by the flow cytometric detection of phosphatidylserine expression using the annexin V-FITC and propidium iodide combination staining method. Annexin V has high affinity for phosphatidylserine and binds to exposed phosphatidylserine on the surface of apoptotic cells. Viable cells with intact membranes exclude propidium iodide, whereas it can permeate the membranes of dead and damaged cells. Therefore, viable cells are both annexin V and propidium iodide negative, while cells that are in early apoptosis are annexin V positive and propidium iodide negative and cells that are in late apoptosis or already dead are both annexin V and propidium iodide positive.The flow cytometry results (Fig. 2[I]) indicated that PIMET extracts can induce apoptosis in HT29 colon cancer cells. The untreated HT29 cells showed only 0.4 ± 0.02% early apoptotic cells and 0.11 ± 0.01% late apoptotic cells. Treatment with PIMET significantly increased the percentages of cells in both stages. About 23.7 ± 1.54% of cells were in the early stage of apoptosis after treatment with 50 μg mL−1 PIMET; this percentage increased to 27.8 ± 0.78% after treatment with 100 μg mL−1 PIMET (Fig. 2[II]). However, the activity of PIMET is less than that of the standard camptothecin used in the assay (82.91 ± 2.21% (early stage) and 12.5 ± 1.19% (late stage)).
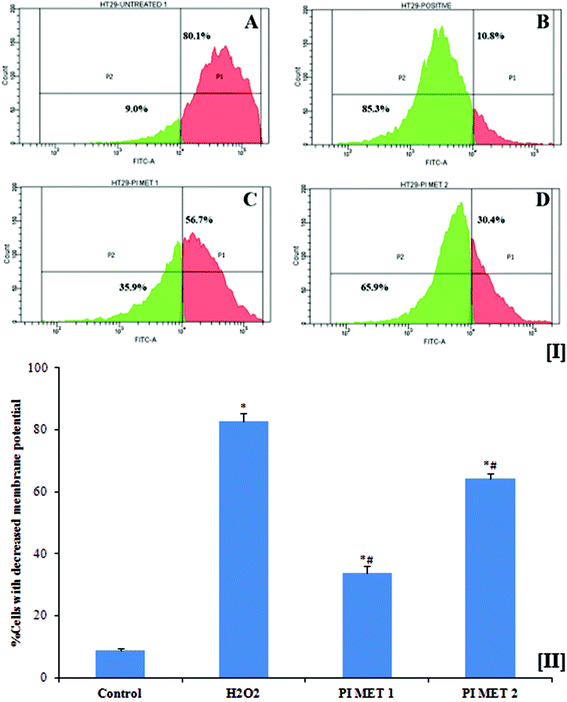
3.6 Assessment of mitochondrial membrane potential
Mitochondria are vital to life, mainly due to their key roles in processes such as ATP generation and apoptotic cell death. Mitochondrial membrane potential plays an important role in inducing apoptosis. This membrane potential is relevant because a decrease in the mitochondrial membrane potential triggers the release of cytochrome c and other pro-apoptotic proteins. Another important factor is that cancer cells exhibit higher mitochondrial membrane potential, which in turn enables them to inhibit apoptosis. General elevations in mitochondrial membrane potential have been linked to colonic carcinoma cells. The disintegration of the mitochondrial membrane potential is accompanied by the opening of the mitochondrial membrane transition pores, leading to the release of cytochrome c into the cytosol, which in turn activates subsequent events in the apoptotic cascade. Therefore, depletion of mitochondrial membrane potential is considered to be an important event in the induction of apoptosis with respect to prevention, management and treatment of cancer.The mitochondrial membrane potentials of HT29 colon cells were determined using rhodamine 123 dye (Fig. 3[I]). Rh123 selectively enters mitochondria with intact membrane potentials and is retained in the mitochondria. Once the membrane potential is lost, the dye is washed out of the mitochondria, resulting in reduction of Rh123 fluorescence. The reduced fluorescence was measured using flow cytometry and correlated with the mitochondrial membrane potential. As is evident from the results, the mitochondrial membrane potential of HT29 colon cancer cells decreased when the cells were pre-treated with PIMET. The percentage of cells with decreased mitochondrial membrane potential was found to be 8.9 ± 0.458% in the untreated group. The percentages of cells with decreased membrane potential increased substantially to 33.73 ± 2.254% and 64.13 ± 1.75%, respectively, when treated with 50 and 100 μg mL−1 PIMET (Fig. 3[II]). Meanwhile, 82.7 ± 2.551% of HT29 cells lost their mitochondrial membrane potential upon treatment with 250 μM H2O2 (positive control). These results are promising, as the loss of mitochondrial membrane potential in cancer cells affects the production of ATP and may facilitate the initial steps of apoptosis, e.g., release of cytochrome c. To further confirm this, ATP production and cytochrome c release by cells upon treatment with PIMET were studied.
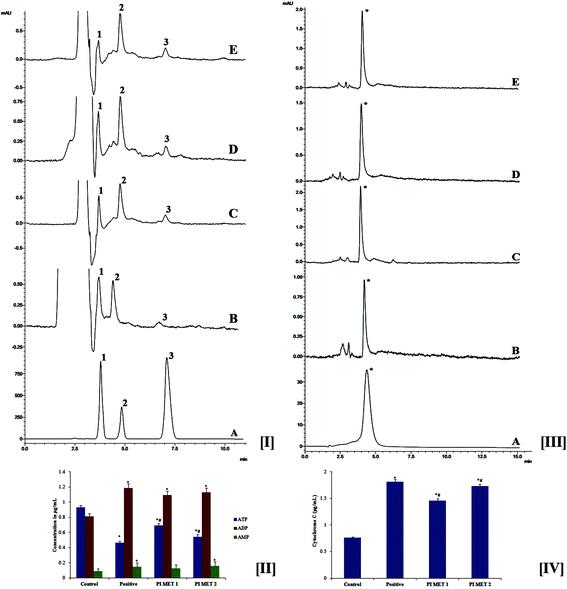
3.7 Determination of ATP production
During late-stage apoptosis, ATP levels sharply decrease, mostly due to loss of mitochondrial function and consumption by ATP-dependent proteases. Thus, ATP production plays an important role in the survival of cancer cells. Cancer cells are reported to rely mostly on the glycolytic pathway rather than oxidative phosphorylation for ATP production.39 Cancer cells exhibit this altered metabolism to meet energy needs during tumor progression; this is known as the Warburg effect.40 Hence, cancer research has focused on targeting ATP production. The effects of PIMET extract on ATP production in HT29 cells were analyzed by HPLC at 259 nm (Fig. 4[I]). The retention times were found to be 3.808, 4.858 and 7.119 min, respectively, for ATP, ADP and AMP. These results clearly indicate that ATP production in HT29 colon cancer cells decreased significantly after treatment with PIMET extract. The ATP content in the control group was found to be 0.931 ± 0.024 μg mL−1; this was reduced to 0.465 ± 0.017 μg mL−1, 0.69 ± 0.029 μg mL−1 and 0.543 ± 0.03 μg mL−1, respectively, for cells treated with H2O2 (250 μM), PIMET-50 μg mL−1 and PIMET-100 μg mL−1 (Fig. 4[II]). Thus, these results confirm that the decreased mitochondrial potential affected the production of ATP in cancer cells after treatment with PIMET extract. The extract may also have affected enzymes of the glycolytic pathway, consecutively reducing ATP production. Further studies are required to confirm this.3.8 Determination of cytochrome c release
The effects of PIMET on the release of cytochrome c were analyzed by HPLC at 393 nm. The retention time was found to be 4.492 min (Fig. 4[III]). It was observed that the release of cytochrome c increased after treatment with PIMET. The cytochrome c release increased from 0.761 ± 0.016 μg mL−1 (control) to 1.461 ± 0.031 and 1.734 ± 0.037 μg mL−1, respectively, for 50 and 100 μg mL−1 PIMET (Fig. 4[IV]). The activity of PIMET was comparable to that of the positive control H2O2 (1.815 ± 0.039 μg mL−1) used in the assay. Many pro-apoptotic signals trigger mitochondrial cytochrome c release, leading to caspase activation and ultimately to cellular breakdown. Mitochondrial cytochrome c release and caspase activation are often impaired in tumors with BCL2 overexpression or BAX-defective status.413.9 Western blot analysis
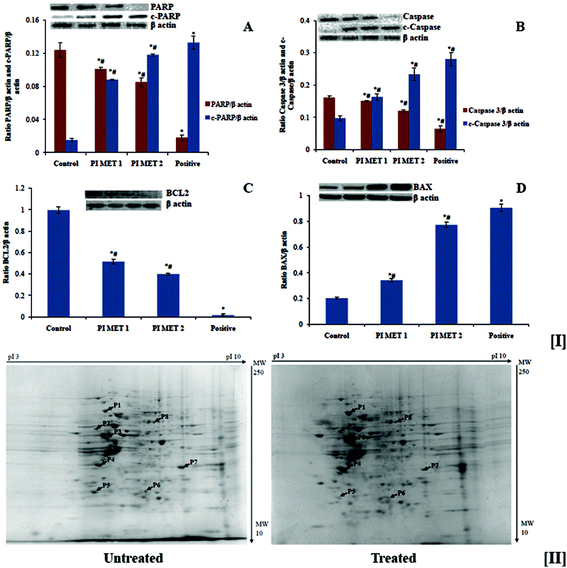
The effects of PI methanol extract on the expression of various proteins involved in apoptosis were analyzed by western blot analysis (Fig. 6). Anti-apoptotic protein BCL2, pro-apoptotic protein BAX, caspase 3, c-caspase 3, PARP and c-PARP were analyzed. There was a dose-dependent reduction in the expression of anti-apoptotic protein BCL2 and a dose-dependent increase in the expression of pro-apoptotic protein BAX (Fig. 5[I(A)] and [I(B)]). The PIMET extract also induced activation of cleavage of caspase 3 and PARP (Fig. 5[I(C)] and [I(D)]). The changes in expression of the abovementioned proteins may be the reason for the apoptotic effects of methanol extract of PI. The activity of PIMET is almost comparable to that of 5-fluorouracil (50 μM) in the activation of c-caspase 3 and c-PARP.Apoptosis is moderated by intrinsic and extrinsic pathways. The intrinsic pathway is initiated when the mitochondrial membrane potential decreases and cytochrome c is released from the mitochondria into the cytoplasm. Cytochrome c induces assemblage of the apoptosome, which contains the Apaf-1 protein and caspase-9. The extrinsic pathway receives signals through the binding of extracellular ligands to proapoptotic death receptors (such as Fas ligand and TRAIL) situated on the cell surface; this results in formation of the death-inducing signaling complex, which is composed of the death receptor, Fas-associated protein, with death domain and caspase-8. These two pathways lead to the activation of effector caspases, caspase 3 and caspase 7; the cleaved caspase 3 activates the cleavage of PARP, resulting in apoptosis. BCL2 and the BAX family of proteins are involved in maintaining mitochondrial membrane potential. BCL2 is antiapoptotic and prevents the release of cytochrome c, whereas BAX is proapoptotic and facilitates release of cytochrome c. PARP in the active stage enhances repair of DNA damage; thus, cleavage of PARP inactivates it and thereby ceases repair of DNA damage, which in turn triggers cells to shift to apoptosis. The western blot analysis clearly showed that treatment of HT29 colon cancer cells with PIMET enhances apoptosis, particularly through the intrinsic pathway. Because PIMET activates cleavage of caspase 3 and PARP, it is possible that components of PIMET bind the death receptors, which then encourage the cascade of apoptosis (extrinsic pathway) which leads downstream to the cleavage of caspase 3 and PARP.
The relationship between natural polyphenols, apoptosis and cancer has been evaluated by different studies, and the capacity of these compounds to perform as cancer chemopreventive and/or chemotherapeutic agents has also been investigated.42 We previously determined that PIMET is rich in polyphenols, especially gallic acid and p-coumaric acid.18 Gallic acid and p-coumaric acid have been revealed to induce apoptosis in colon cancer cells.43,44 All these reports suggests that the ability of PIMET extract to induce activation and deactivation of pro- and anti-apoptotic proteins can be attributed to the polyphenols present in the extract.
Gallic acid (1.6 to 29 mM), syringic acid (4 to 6 mM) and catechol (319 mM) have been reported to exhibit anticancer potential individually against colon cancer cells.30,31 These polyphenols, i.e. gallic acid, syringic acid, and catechol, are abundant in PIMET;18 moreover, the IC50 values of these individual polyphenols in the present study are within the concentration limits that have been reported to induce anticancer efficacy. However, in the present study, we strongly believe that the anticancer potential arises more from a synergistic effect of all the polyphenols and other unidentified compounds present in PIMET than from the effects of gallic acid, syringic acid or catechol alone. Hence, further studies should identify the specific compounds in PIMET responsible for its anticancer effects other than polyphenols, if any.
3.10 Changes in the protein expression profile of HT-29 human colon cancer cells after treatment with methanol extract of PI
The above studies confirmed the anticancer potential of PIMET by its ability to induce cell cycle arrest and apoptosis in HT29 colon cancer cells. To better understand the anticancer potential of PIMET, it is important to know its mechanisms of action as well as the cellular response towards the extract. Hence, we performed proteomic profiling using 2D-PAGE and peptide mass fingerprinting for the identification of differentially expressed proteins after treating HT-29 cells with PIMET extract.The gels with stained protein spots were scanned, and computer-assisted image analysis was performed using PDQuest 8.0 software (Bio-Rad Laboratories GmbH, Munich, Germany). Selected differentially expressed proteins were excised manually from the gels and subjected to peptide mass fingerprinting for identification of proteins. The two-dimensional map of protein expression of HT29 cells after treatment with PIMET is shown in Fig. 5[II]. Eight proteins were identified in this experiment. Among these eight proteins, four were upregulated and another four were downregulated after treatment with PIMET. The identified proteins with their theoretical molecular weights (Mw), isoelectric points (pI) and fold changes with respect to an untreated control (ratio of band intensity of the sample to that of the untreated control) are summarized in Table 1.
| Proteins | Molecular weight (kDa) | Isoelectric point (pI) | Fold change (treated/control) | |
|---|---|---|---|---|
| Spot | Upregulated proteins identified | |||
| P1 | 78 kDa glucose-regulated protein precursor | 72.33 | 5.07 | 2.417 ± 0.204 |
| P2 | Calreticulin precursor | 48.14 | 4.29 | 1.346 ± 0.105 |
| P3 | COP9 signalosome complex subunit 4 | 46.26 | 5.57 | 2.832 ± 0.141 |
| P4 | Pyruvate dehydrogenase (lipoamide) beta | 37.19 | 5.64 | 2.723 ± 0.108 |
| Spot | Downregulated proteins identified | |||
| P5 | Polymerase (RNA) II subunit C | 31.44 | 4.79 | 0.831 ± 0.031 |
| P6 | Peroxiredoxin 6 | 25.03 | 6.00 | 0.849 ± 0.019 |
| P7 | Solute carrier family 25 member 35 | 32.43 | 9.21 | 0.534 ± 0.125 |
| P8 | X-linked inhibitor of apoptosis | 56.68 | 6.22 | 0.965 ± 0.067 |
78 kDa glucose-regulated protein precursor, calreticulin precursor, COP9 signalosome complex subunit 4 and pyruvate dehydrogenase (lipoamide) beta are the upregulated proteins. Among the selected upregulated proteins, COP9 signalosome complex subunit 4 had the highest fold difference (2.832 ± 0.141). The downregulated proteins are polymerase (RNA) II subunit C, peroxiredoxin 6, solute carrier family 25 member 35 and X-linked inhibitor of apoptosis. Among the selected downregulated proteins, solute carrier family 25 member 35 had the most significant fold change (0.534 ± 0.125).
Glucose-regulated protein 78 (GRP78) is a key chaperone and stress response protein that resides primarily in the endoplasmic reticulum and plays a role in facilitating the assembly of multimeric protein complexes inside the endoplasmic reticulum. Increased expression of the ER stress protein GRP78 has been found in several types of cancer, including colon cancer.45 However, the role of GRP78 expression in the prognosis of colon cancer is controversial. GRP78 silencing has been found to enhance apoptosis in colon cancer cells and suppress colon cancer growth through downregulation of the VEGF/VEGFR2 signaling pathway.46 In another study, a low level of GRP78 was found to increase metastasis ability in colon cancer cells by altering E-cadherin and vimentin expression and activating the NRF-2/HO-1 signaling pathway.47 It was found in the present study that 78 kDa glucose-regulated protein precursors increased after treatment with PIMET. By the upregulation of GRP78 after PIMET treatment, we were unable to reach a conclusion whether PIMET enhanced the formation of GRP78 or inhibited the conversion of GRP78 precursors to the active molecule. Further studies are required to confirm this.
Calreticulin is a calcium-binding chaperone that promotes folding, oligomeric assembly and quality control in the endoplasmic reticulum via the calreticulin/calnexin cycle. It is also found in the nucleus and can act as an important modulator of the regulation of gene transcription by nuclear hormone receptors. Proteomic analyses of calreticulin in human colon adenocarcinomas show conflicting results. Toquet et al.48 reported that calreticulin expression was downregulated in 51.7% of human colon adenocarcinomas. Our results showed that calreticulin precursor was upregulated after treatment, which indicates that the extract may inhibit the conversion of precursor molecule to active calreticulin.
COP9 signalosome complex (CSN), a complex protein involved in various cellular and developmental processes, is a potential player in tumorigenesis, and its subunits are often overexpressed in tumors.49 Recently, it was reported that reduced expression of different CSN subunits in HT29 may lead to increased levels of the CDK inhibitor p27 and the tumor suppressor p53 in HT29 cells.50 Our results agreed with the latter results, as the COP9 signalosome complex subunit 4 was found to be upregulated after treatment with PIMET. The relevance of this upregulation requires further study.
The pyruvate dehydrogenase (PDH) complex is a nuclear-encoded mitochondrial multienzyme complex that catalyzes the overall conversion of pyruvate to acetyl-CoA and carbon dioxide; it provides the primary link between glycolysis and the tricarboxylic acid cycle. The PDH complex is composed of three enzymatic components: pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2) and lipoamide dehydrogenase (E3). The E1 enzyme is a heterotetramer of two alpha and two beta subunits. From the proteomics data, we found that the pyruvate dehydrogenase (lipoamide) beta subunit is upregulated after PIMET treatment. The relevance of this upregulation is unclear; no studies have been reported to date. It is possible that PIMET inhibits formation of the PDH complex (hence increasing the lipoamide subunit), which hinders the conversion of pyruvate to acetyl CoA, thereby affecting the normal functioning of cells. Additional data are required to validate this statement.
Polymerase (RNA) II subunit C (POLR2C) is a subunit of RNA polymerase II, which is involved in the synthesis of mRNA. This protein was found to be downregulated after PIMET treatment. By downregulating POLR2C, PIMET extract may inhibit the transcription process in cancer cells, thereby affecting the functioning of cancer cells.
Peroxiredoxin 6 (PRDX6) is a member of the peroxidise family and has glutathione peroxidase and calcium-independent phospholipase A2 activities. Because peroxiredoxins are antioxidants, they support survival and tumor maintenance by protecting cells from oxidative stress-induced apoptosis. In a recent study, overexpression of PRDX 6 attenuated cisplatin-induced apoptosis in human ovarian cancer cells.51 In contrast, reduction of PRDX6 expression increased peroxide-induced cell death in liver cancer cells.52 Peroxiredoxin 6 was found to be downregulated in the present study after treatment with PIMET, which may initiate cell death.
Solute carrier family 25 member 35 (SLC25A35) belongs to the SLC25 family of mitochondrial carrier proteins. The mitochondrion relies on compartmentalization of certain enzymes, ions and metabolites for the sake of efficient metabolism. In order to perform these activities, numerous carriers are expressed, targeted and folded in the inner mitochondrial membrane. Among these carriers, the six-transmembrane-helix mitochondrial SLC25 (solute carrier family 25) proteins facilitate transport of solutes with disparate chemical identities across the inner mitochondrial membrane. The decrease in mitochondrial membrane potential after treatment with PIMET may have resulted in downregulated SLC25A35, as evident from proteomics analysis. This downregulation will negatively affect the proper functioning of mitochondria and, thereby, the functioning of HT29 cells.
X-linked inhibitor of apoptosis belongs to a family of apoptotic suppressor proteins. This protein functions by binding to tumor necrosis factor receptor-associated factors TRAF1 and TRAF2 and inhibiting apoptosis. This protein is upregulated in many human colon carcinomas; it also inhibits Caspase 3, 6 and 7 by blocking the activity of Caspase 9.53 However, in our study, this apoptotic inhibitor protein was downregulated after the treatment of HT29 cells with PIMET extract; this supports our earlier observation that PIMET can induce apoptosis in HT29 colon cancer cells.
Thus proteomic analysis of proteins from HT29 showed that exposure to methanol extract of PI altered the expression of different proteins of HT29 cells. The downregulation of peroxiredoxin 6 negatively affected the ability of cells to withstand stress, and the downregulation of solute carrier proteins interfered with the normal functioning of the mitochondria of HT29 cells. The downregulation of X-linked inhibitor of apoptosis in particular may have forced the HT29 cells to enter apoptosis. Further identification of the remaining protein spots is necssary; however, this is not in the scope of this study.
4. Conclusions
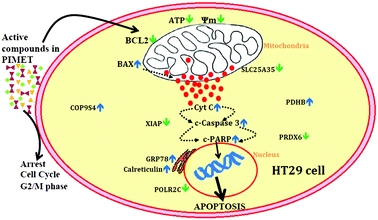
To summarize this study (Fig. 6), a methanol extract of PI was found to be cytotoxic against HT29 colon cancer cells. The extract could arrest the cell cycle at G2/M phase and reduce mitochondrial membrane potential, resulting in decreased ATP production and increased cytochrome C release and inducing apoptosis in HT29 cells. Additionally, western blotting showed that the extract has significant effects on apoptosis, and 2D electrophoresis analysis showed that expression of some significant proteins was altered in favour of inducing apoptosis; especially, the key apoptotic inhibitor protein XIAP was downregulated. Thus, we conclude that plantain infloresence is a good source of biologically active compounds with significant anticancer potential against HT29 colon cancer cells. Further studies are required to isolate the compound or compounds other than polyphenols, if any, which are potentially related to the anticancer effects of the methanol extract of plantain inflorescence.Conflicts of interest
The authors declare no competing interests.Acknowledgements
The first author appreciates the financial support (junior research fellowship) provided by the Indian Council of Medical Research (ICMR), India. The authors are also thankful to the Council of Scientific and Industrial Research (CSIR), India for providing financial support for this research activity. The authors gratefully acknowledge Dr K. B. Ramesh kumar, Jawaharlal Nehru Tropical Botanic Garden and Research Institute (JNTBGRI), India, for his support in plant identification. We also thank Dr Rajeev K Sukumaran of the MPTD, CSIR-NIIST for help provided in conducting 2D gel electrophoresis. The authors would like to thank Mr Saravanakumar, Rajiv Gandhi Centre for Biotechnology for his assistance with mass spectrometry.References
- J. L. Roberts and R. Moreau, Functional properties of spinach (Spinaciaoleracea L.) phytochemicals and bioactives, Food Funct., 2016, 7, 3337–3353 CAS.
- E. Turrini, L. Ferruzzi and C. Fimognari, Potential effects of pomegranate polyphenols in cancer prevention and therapy, Oxid. Med. Cell. Longevity, 2015, 2015, 938475 Search PubMed.
- T. J. Key, Fruit and vegetables and cancer risk, Br. J. Cancer, 2011, 104(1), 6–11 CrossRef CAS PubMed.
- American Cancer Society, Cancer Facts & Fig. 2017, American Cancer Society, Atlanta, Ga, 2017 Search PubMed.
- L. A. Torre, F. Bray, R. L. Siegel, J. Ferlay, J. Lortet-Tieulent and A. Jemal, Global cancer statistics, 2012, CA-Cancer J. Clin., 2015, 65(2), 87–108 CrossRef PubMed.
- M. Pericleous, D. Mandair and M. E. Caplin, Diet and supplements and their impact on colorectal cancer, J. Gastrointest. Oncol., 2013, 4(4), 409–423 CAS.
- F. M. F. Roleira, E. J. Tavares-da-Silva, C. L. Varela, S. C. Costa, T. Silva, J. Garrido and F. Borges, Plant derived and dietary phenolic antioxidants: Anticancer properties, Food Chem., 2015, 183, 235–258 CrossRef CAS PubMed.
- H. Wang, X. Guo, X. Hu, T. Li, X. Fu and R. H. Liu, Comparison of phytochemical profiles, antioxidant and cellular antioxidant activities of different varieties of blueberry (Vaccinium spp.), Food Chem., 2017, 217, 773–781 CrossRef CAS PubMed.
- M. S. Donaldson, Nutrition and cancer: A review of the evidence for an anti-cancer diet, Nutr. J., 2004, 3, 19 CrossRef PubMed.
- A. Gonzalez-Sarrias, J. Tome-Camerio, A. Bellesia, F. A. Tomas-Barberian and J. C. Espin, Theellagic acid-derived gut microbiota metabolite, urolothin A, potentiates the anticancer effects of 5-fluorouracil chemotherapy on human colon cancer cells, Food Funct., 2015, 6, 1460–1469 CAS.
- M. J. Wargovich, J. Morris, V. Brown, J. Ellis, B. Logothetis and R. Weber, Nutraceutical use in late-stage cancer, Cancer Metastasis Rev., 2010, 29, 503–510 CrossRef CAS PubMed.
- P. Kuppusamy, M. M. Yusoff, G. P. Maniam, S. J. Arieflchwan, I. Soundharrajan and N. Govindan, Nutraceuticals as potential therapeutic agents for colon cancer: A review, Acta Pharm. Sin. B, 2014, 4(3), 173–181 CrossRef PubMed.
- E. Ranzato, S. Martinotti, C. M. Calabrese and G. Calabrese, Role of nutraceuticals in cancer therapy, J. Food Res., 2014, 3(4), 35882 Search PubMed.
- H. P. Singh, Harnessing the potential of banana and plantain in asia and the pacific for inclusive growth, The Proceedings of the International ISHS-ProMusa Symposium on Global Perspectives on Asian Challenges, 2011, Acta Horticulture 897 ISHS 2011. Available from http://www.musalit.org Search PubMed.
- M. Z. Imam and S. Akter, Musa paradisiaca L. and Musa sapientumL.: A Phytochemical and Pharmacological Review, J. Appl. Pharm. Sci., 2011, 01(05), 14–20 Search PubMed.
- K. P. S. Kumar, D. Bhowmik, S. Duraivel and M. Umadevi, Traditional and medicinal uses of banana, J. Pharmacogn. Phytochem., 2012, 1(3), 51–63 Search PubMed.
- P. Jayamurthy, B. Aparna, G. Gayathri and P. Nisha, Evaluation of antioxidant potential of inflorescence and stalk of plantain (Musa Sapientum), J. Food Biochem., 2011, 32, 2–7 Search PubMed.
- K. B. Arun, T. Sithara, T. R. Reshmitha, G. C. Akhil and P. Nisha, Dietary fibre and phenolic-rich extracts from Musa paradisiaca inflorescence ameliorates type 2 diabetes and associated cardiovascular risks, J. Funct. Foods, 2017, 31, 198–207 CrossRef CAS.
- H. Palafox-Carlos, J. F. Ayala-Zavala and G. A. González-Aguilar, The role of dietary fiber in the bioaccessibility and bioavailability of fruit and vegetable antioxidants, J. Food Sci., 2011, 76(1), R6–R15 CrossRef CAS PubMed.
- D. Tagliazucchi, E. Verzelloni, D. Bertolini and A. Conte, In vitro bio-accessibility and antioxidant activity of grape polyphenols, Food Chem., 2009, 120(2), 599–606 CrossRef.
- F. Saura-Calixto and M. E. Díaz-Rubio, Polyphenols associated with dietary fibre in wine: a wine polyphenols gap?, Food Res. Int., 2007, 40(5), 613–619 CrossRef CAS.
- T. Mosmann, Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays, J. Immunol. Methods, 1983, 65(1–2), 55–63 CrossRef CAS PubMed.
- D. Pitchai, A. Roy and C. Ignatius, In vitro evaluation of anticancer potentials of lupeol isolated from Elephantopus scaber L. on MCF-7 cell line, J. Adv. Pharm. Technol. Res., 2014, 5(4), 179–184 CrossRef PubMed.
- K. Harada, S. Kawaguchi, Supriatno, Y. Kawashima, H. Yoshida and S. Mitsunobu, S-1, an oral fluoropyrimidine anti-cancer agent, enhanced radio sensitivity in a human oral cancer cell line in vivo and in vitro: involvement possibility of inhibition of survival signal, Akt/PKB, Cancer Lett., 2005, 226, 161–168 CrossRef CAS PubMed.
- M. S. Lin, W. C. Chen, X. Bai and Y. D. Wang, Activation of peroxisome proliferator-activated receptor γ inhibits cell growth via apoptosis and arrest of the cell cycle in human colorectal cancer, J. Dig. Dis., 2007, 8, 82–88 CrossRef CAS PubMed.
- A. Hahn-Windgassen, V. Nogueira, C. C. Chen, J. E. Skeen, N. Sonenberg and N. Hay, Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity, J. Biol. Chem., 2005, 280, 32081–32089 CrossRef CAS PubMed.
- J. Radhakrishnan, S. Wang, I. M. Ayoub, J. D. Kolarova, R. F. Levine and R. J. Gazmuri, Circulating levels of cytochrome c after resuscitation from cardiac arrest: A marker of mitochondrial injury and predictor of survival. American Journal of Physiology, Am. J. Physiol., 2007, 292(2), H767–H775 CrossRef CAS PubMed.
- L. Lahouar, F. Ghrairi, A. E. Arem, W. Sghaeir, M. E. Felah, H. B. Salem, B. Sriha and L. Achour, Attenuation of histopathological alterations of colon, liver and lung by dietary fibre of barley Rihane in azoxymethane-treated rats, Food Chem., 2014, 149, 271–276 CrossRef CAS PubMed.
- M. Glei and S. Klenow, New insight into the influence of carob extract and gallic acid on hemin induced modulation of HT29 cell growth parameters, Toxicol. in Vitro, 2009, 23(6), 1055–1061 CrossRef PubMed.
- M. S. Abaza, R. Al-Attiyah, R. Bharadwaj, G. Abbadi, M. Koyippally and M. Afzal, Syringic acid from Tamarixaucheriana possesses antimitogenic and chemo-sensitizing activities in human colorectal cancer cells, Pharm. Biol., 2013, 51(9), 1110–1124 CrossRef CAS PubMed.
- H. Deng and Y. Fang, The three catecholics benserazide, catechol and pyrogallol are gpr35 agonists, Pharmaceuticals, 2013, 6(4), 500–509 CrossRef CAS PubMed.
- M. Whent, H. Huang, Z. Xie, H. Lutterodt, L. Yu, E. P. Fuerst, C. F. Morris, L. L. Yu and D. Luthria, Phytochemical composition, anti-inflammatory, and anti proliferative activity of whole wheat flour, J. Agric. Food Chem., 2012, 60(9), 2129–2135 CrossRef CAS PubMed.
- C. P. Hsu, Y. H. Lin, S. P. Zhou, Y. C. Chung, C. C. Lin and S. C. Wang, Longan flower extract inhibits the growth of colorectal carcinoma, Nutr. Cancer, 2010, 62(2), 229–236 CrossRef PubMed.
- Y. C. Chung, C. C. Lin, C. C. Chou and C. P. Hsu, The effect of Longan seed polyphenols on colorectal carcinoma cells, Eur. J. Clin. Invest., 2010, 40(8), 713–721 CrossRef CAS PubMed.
- Y. C. Chung, C. C. Huang, C. H. Chen, H. C. Chiang, K. B. Chen, Y. J. Chen, C. L. Liu, L. T. Chuang, M. Liu and C. P. Hsu, Grape-seed procyanidins inhibit the in vitro growth and invasion of pancreatic carcinoma cells, Pancreas, 2012, 41(3), 447–454 CrossRef CAS PubMed.
- A. Lindqvist, V. Rodríguez-Bravo and R. H. Medema, The decision to enter mitosis: feedback and redundancy in the mitotic entry network, J. Cell Biol., 2009, 185(2), 193–202 CrossRef CAS PubMed.
- J. D. Hsu, S. H. Kao, T. T. Ou, Y. J. Chen, Y. J. Li and C. J. Wang, Gallic acid induces G2/M phase arrest of breast cancer cell MCF-7 through stabilization of p27Kip1 attributed to disruption of p27Kip1/Skp2 complex, J. Agric. Food Chem., 2011, 59(5), 1996–2003 CrossRef CAS PubMed.
- H. J. Cho and J. H. Y. Park, Kaempferol induces cell cycle arrest in HT-29 human colon cancer cells, J. Cancer Prev., 2013, 18(3), 257–263 CrossRef PubMed.
- Y. Zhao, E. B. Butler and M. Tan, Targeting cellular metabolism to improve cancer therapeutics, Cell Death Dis., 2013, 4, e532, DOI:10.1038/cddis.2013.60.
- M. Jang, S. S. Kim and J. Lee, Cancer cell metabolism: implications for therapeutic targets, Exp. Mol. Med., 2013, 45(10), e45, DOI:10.1038/emm.2013.85.
- M. Seervi, J. Joseph, P. K. Sobhan, B. C. Bhavya and T. R. Santhoshkumar, Essential requirement of Cytochrome C release for caspase activation by procaspase-activating compound defined by cellular models, Cell Death Dis., 2011, 2, e207, DOI:10.1038/cddis.2011.90.
- G. D. Stoner and H. Mukhtar, Polyphenols as cancer chemopreventive agents, J. Cell. Biochem. Suppl., 1995, 22, 169–180 CrossRef CAS PubMed.
- A. P. Subramanian, S. K. Jaganathan, M. Mandal, E. Supriyanto and I. I. Muhamad, Gallic acid induced apoptotic events in HCT-15 colon cancer cells, World J. Gastroenterol., 2016, 22(15), 3952–3961 CrossRef CAS PubMed.
- S. K. Jaganathan, E. Supriyanto and M. Mandal, Events associated with apoptotic effect of p-Coumaric acid in HCT-15 colon cancer cells, World J. Gastroenterol., 2013, 19(43), 7726–7734 CrossRef CAS PubMed.
- X. Xing, M. Lai, Y. Wang, E. Xu and Q. Huang, Over expression of glucose regulated protein 78 in colon cancer, Clin. Chim. Acta, 2006, 364(1–2), 308–315 CrossRef CAS PubMed.
- L. J. Kuo, C. S. Hung, W. Y. Chen, Y. J. Chang and P. L. Wei, Glucose-regulated protein 78 silencing down-regulates vascular endothelial growth factor/vascular endothelial growth factor receptor 2 pathway to suppress human colon cancer tumor growth, J. Surg. Res., 2013, 185(1), 264–272 CrossRef CAS PubMed.
- Y. J. Chang, W. Y. Chen, C. Y. Huang, H. H. Liu and P. L. Wei, Glucose-regulated protein 78 (GRP78) regulates colon cancer metastasis through EMT biomarkers and the NRF-2/HO-1 pathway, Tumor Biol., 2015, 36(3), 1859–1869 CrossRef CAS PubMed.
- C. Toquet, A. Jarry, C. Bou-Hanna, K. Bach, M. G. Denis, J. F. Mosnier and C. L. Laboisse, Altered calreticulin expression in human colon cancer: maintenance of calreticulin expression is associated with mucinous differentiation, Oncol. Rep., 2007, 17(5), 1101–1107 CAS.
- M. A. Kouvaraki, G. Z. Rassidakis, L. Tian, R. Kumar, C. Kittas and F. X. Claret, Jun activation domain-binding protein 1 expression in breast cancer inversely correlates with the cell cycle inhibitor p27 (Kip1), Cancer Res., 2003, 63, 2977–2981 CAS.
- M. Feist, X. Huang, J. M. Müller, B. Rau and W. Dubiel, Canhyperthermic intraperitoneal chemotherapy efficiency be improved by blocking the DNA repair factor COP9 signalosome?, Int. J. Colorectal Dis., 2014, 29, 673–680 CrossRef PubMed.
- J. H. Pak, W. H. Choi, H. M. Lee, W. D. Joo, J. H. Kim, Y. T. Kim, Y. M. Kim and J. H. Nam, Peroxiredoxin 6 overexpression attenuates cisplatin-induced apoptosis in human ovarian cancer cells, Cancer Invest., 2011, 29, 21–28 CrossRef CAS PubMed.
- B. Walsh, A. Pearl, S. Suchy, J. Tartaglio, K. Visco and S. A. Phelan, Overexpression of Prdx6 and resistance to peroxide-induced death in Hepa1-6 cells: Prdx suppression increases apoptosis, Redox Rep., 2009, 14, 275–284 CrossRef CAS PubMed.
- L. Yang, Z. Cao, H. Yan and W. C. Wood, Coexistence of high levels of apoptotic signaling and inhibitor of apoptosis proteins in human tumor cells: implication for cancer specific therapy, Cancer Res., 2003, 63, 6815–6824 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7fo01454f |
| This journal is © The Royal Society of Chemistry 2018 |