Moderation of hyperuricemia in rats via consuming walnut protein hydrolysate diet and identification of new antihyperuricemic peptides†
Qingyong
Li
a,
Xiaoyan
Kang‡
a,
Chuanchao
Shi
a,
Yujuan
Li
a,
Kaustav
Majumder
b,
Zhengxiang
Ning
a and
Jiaoyan
Ren
 *a
*a
aSchool of Food Science and Engineering, South China University of Technology, Guangzhou 510640, China. E-mail: jyren@scut.edu.cn; Fax: +86-20-87113848; Tel: +86-20-87112594
bFood Science & Technology Department, University of Nebraska–Lincoln, NE 68588-6205, USA
First published on 19th September 2017
Abstract
In this study, walnut meal hydrolysates (WMH) and dephenolized walnut meal hydrolysates (DWMH) were found to effectively decrease the serum uric acid level and protect the renal function in potassium oxonate-induced hyperuricemic rats in vivo as well as inhibit xanthine oxidase in vitro. Two novel antihyperuricemic peptides including WPPKN (640.8 Da) and ADIYTE (710.7 Da) were purified from DWMH via Sephadex G-15 gel filtration and reverse-phase high-performance liquid chromatography and identified by LC-ESI-MS/MS. These peptides displayed high in vitro xanthine oxidase inhibition (XOI) activity with IC50 values of 17.75 ± 0.12 mg mL−1 (WPPKN) and 19.01 ± 0.23 mg mL−1 (ADIYTE). Based on the results of molecular simulation, WPPKN entered into the hydrophobic channel and even obstructed the interaction between xanthine and xanthine oxidase (XO), while ADIYTE was positioned on the surface of the B-chain and blocked the entrance of the substrate to the hydrophobic channel. Therefore, the two peptides are partially responsible for the antihyperuricemic properties of DWMH.
Introduction
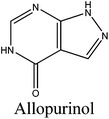
The prevalence of hyperuricemia has been increasing in recent years due to change in eating habits and the intake of high-purine food.1,2 Hyperuricemia refers to the accumulation of excess uric acid in the blood and may lead to the sedimentation of urate crystals in joints and kidneys causing diseases like gout. Earlier studies had shown that gout has a strong link with hypertension, atherosclerosis, insulin resistance, and renal diseases. Uric acid is generated via the oxidation of xanthine under the catalysis of xanthine oxidase (XO).3 Three categories of drugs, including the ones that inhibit XO (e.g. allopurinol, febuxostat) or those that can accelerate urate excretion (e.g. probenecid, RDEA594) or catalyze uric acid into allantoin (e.g. pegloticase), are commonly used to remedy hyperuricemia.4 However, most of them are associated with adverse side effects. Thus, the development of natural and efficient xanthine oxidase inhibitory (XOI) agents has prevailed as a means of relief for hyperuricemia patients. Some flavonoid compounds have been reported to demonstrate potent XOI activities.5–7 Meanwhile, food-derived bioactive peptides with high XOI properties have also attracted much attention in recent years. For example, tryptophan-containing dipeptides from milk protein and shark-cartilage-derived peptides (Tyr-Leu-Asp-Asn-Tyr and Ser-Pro-Pro-Tyr-Trp-Pro-Tyr) had shown XOI activities or even a decreased serum uric acid level in hyperuricemic rats.8,9 Additionally, there is growing interest in discovering other health-promoting functions of bioactive peptides, including antioxidative activity,10–12 anticancer activity,13 ACE inhibitory activity14,15 and neuroprotective activity.16 Besides, bioactive peptides were proved to have the advantage of easy absorption, nontoxicity, and diverse bioactivities.Walnut (Juglans regia L. ) belongs to the family of Juglandaceae and is one of the nuts known globally regularly consumed as a part of regular human diet. Degreased walnut meal is the major by-product of coldly squeezed walnut kernels, containing more than 40% proteins with high contents of nutritive ingredients, which is used as animal feed or fertilizer usually, entailing considerable waste. Therefore, degreased walnut meal is underutilized and its higher value-added product is urgently needed to be created in order to improve the economic value. In the past few years, researchers had found that walnut protein hydrolysates are capable of depressing the amount of reactive oxygen species (ROS)17 and that the presence of Trp residues in its sequences contributes to XOI activity.18 Therefore, walnut protein could be a favorable source to develop antihyperuricemic peptides. So far, to the best of our knowledge, there has been a limited amount of work that is relevant to the XOI properties of walnut protein hydrolysates.
In the present study, the antihyperuricemic properties of walnut (Juglans regia L. ) protein hydrolysates were evaluated in a potassium oxonate-induced rat model in vivo and xanthine oxidase inhibitory effects were evaluated in vitro. In addition, two active peptides isolated and identified from the walnut protein hydrolysates were characterized by their molecular weight and sequence information. Besides, the XOI activities and potential inhibitory mechanisms regarding XO were investigated via in vitro methods and molecular docking simulation, respectively. These results could benefit further understanding of the bioactivity of the walnut protein hydrolysates, which might be conducive to possible commercial applications of the hydrolysates as functional food ingredients with antihyperuricemic properties.
Materials and methods
Chemicals and materials
Allopurinol, xanthine, uric acid, potassium oxonate, and XO (EC 1.17.3.3, 0.8 units per mg protein) from bovine milk, cytochrome C (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 384 Da), aprotinin (6511 Da), zinc bacitracin (1423 Da), oxidized glutathione (612 Da), Gly-Gly-Tyr-Arg (451 Da) and Gly-Gly-Gly (189 Da) were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). Alcalase was purchased from Pangbo Biotech Co., Ltd (Nanning, PR China). Synthetic peptides were bought from GL Biochem Co., Ltd (Shanghai, PR China) using the 9-fluorenylmethyl-oxycarbonyl (Fmoc) strategy. Assay kits of uric acid, creatinine, and urea nitrogen were obtained from Jiancheng Biotech Co., Ltd (Nanjing, PR China). Coldly squeezed degreased walnut meals were obtained from Shangluo City, southeast Shanxi province, China. Methanol (high-performance liquid chromatography (HPLC) grade) was purchased from Yonghua Chemical Technology Co., Ltd (Jiangsu, PR China). And other reagents were of analytical grade or better.
384 Da), aprotinin (6511 Da), zinc bacitracin (1423 Da), oxidized glutathione (612 Da), Gly-Gly-Tyr-Arg (451 Da) and Gly-Gly-Gly (189 Da) were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). Alcalase was purchased from Pangbo Biotech Co., Ltd (Nanning, PR China). Synthetic peptides were bought from GL Biochem Co., Ltd (Shanghai, PR China) using the 9-fluorenylmethyl-oxycarbonyl (Fmoc) strategy. Assay kits of uric acid, creatinine, and urea nitrogen were obtained from Jiancheng Biotech Co., Ltd (Nanjing, PR China). Coldly squeezed degreased walnut meals were obtained from Shangluo City, southeast Shanxi province, China. Methanol (high-performance liquid chromatography (HPLC) grade) was purchased from Yonghua Chemical Technology Co., Ltd (Jiangsu, PR China). And other reagents were of analytical grade or better.
Preparation of the walnut meal hydrolysates and dephenolized walnut meal hydrolysates
The degreased walnut meals were suspended in water to make a final concentration of 10% (w/w) and pre-heated at 85 °C for 15 min. The purpose of pre-heat treatment of this substrate was to partially denature walnut protein and to make it more sensitive to proteases, so as to increase the efficiency of hydrolysis. Then, the mixture was subjected to hydrolysis by using Alcalase 2.4 L protease with an enzyme to substrate ratio (E/S) of 1% (w/w) at 55 °C under agitation in a water bath for 9 h. The hydrolysis was maintained at a constant pH of 7.0 with 1 mol L−1 NaOH. Afterwards, the reaction was terminated by heating to 100 °C for 10 min. The samples were then centrifuged at 7654g for 20 min and the supernatants were lyophilized to obtain walnut meal hydrolysates (WMH) and stored at −20 °C until use. The dephenolized walnut meals were prepared with n-hexane extraction and hydrolyzed in the same way as described above to prepare dephenolized walnut meal hydrolysates (DWMH).In vivo antihyperuricemic effects of WMH and DWMH
Sixty male Sprague-Dawley rats (weight 180–220 g) were bought from the Laboratory Animal Center of Guangzhou University of Chinese Medicine (Guangzhou, PR China). All animals were raised in a comfortable room (temperature, 18–22 °C; humidity, 40–70%) in individual cages in the Laboratory Animal Center of Jinan University (Guangzhou, PR China). The rats were given a standard laboratory diet and filtered water and kept under a 12 h light–dark cycle with minimal suffering. This study was performed in accordance with the standards set out by the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and permitted by the Committee on the Ethics of Animal Experiments of Jinan University (permit number: SCXK2013-0020).The rats were given potassium oxonate by oral gavage once a day 2 g per kg BW at a volume of 1.0 mL per 100 g BW for seven successive days to obtain a high serum uric acid (SUA) level.19,20 The rats with a SUA concentration greater than 110 μmol L−1 were considered as a successful model and selected for future study. Selected rats were weighed and randomly assigned into four subgroups (i.e. n = 12 per group): (i) model control group: rats were administered distilled water for 21 days; (ii) allopurinol group: rats received 25 mg per kg BW allopurinol at a volume of 1.0 mL per 100 g BW via oral gavage once daily; (iii) WMH group: rats were treated with 300 mg per kg BW WMH; (iv) DWMH group: rats were administered 300 mg per kg BW DWMH. The selected rats obtained potassium oxonate treatments after the administration of the test samples to increase their SUA.21,22 The rats in the normal control group received the same volume of distilled water for 28 days (non-hyperuricemic group). The urine volume, health state, and food consumption of all rats were examined daily. The blood samples were drawn from the retroorbital plexus under anaesthesia by injecting 3% sodium pentobarbital (30 mg per kg BW) into the cavum abdominis. The blood was collected and centrifuged at 1530g for 10 min at 4 °C. Then, the obtained supernatant was subjected to the SUA test. It is worth mentioning that the blood samples were taken four times by the 0th, 7th, 14th, and 21st days, respectively. The contents of creatinine and urea nitrogen of all the rats were tested on the 21st day of the treatment. The kidney tissue specimens of the rats were fixed and preserved in a 10% formaldehyde solution for hematoxylin and eosin (H&E) staining and histopathological observation with a fluorescence microscope.
Measurement of XOI activity in vitro
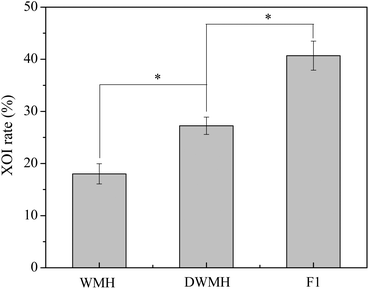
The enzyme activity of XO was detected by quantitating the rate of uric acid produced from xanthine according to the methods described by Wu et al.20 with some modifications. All solutions were dissolved in 50 mM Tris-HCl buffer at pH 8.0. 100 μL of a test sample and 50 μL of XO (0.52 U mL−1) were added to a 10 mL centrifuge tube for pre-incubation at 30 °C for 10 min. Subsequently, an aliquot (2900 μL) of Tris-HCl buffer was added to the mixture followed by the addition of 400 μL of 2 mM xanthine. The final mixed solution was incubated at 30 °C for 20 min. The reaction was terminated by adding 150 μL of 1 M HCl. After that, the reaction solution was filtered via a 0.22 μm PES membrane for HPLC analysis. As for HPLC analysis, a Zorbax Eclipse Plus C18 column (250 × 4.6 mm i.d., 5 μm, Santa Clara, CA, USA) on a Shimadzu LC-20A HPLC system (Shimadzu, Japan) was used, and an isocratic mobile phase (i.e., 95% A in 0–15 min) including 15 mM NH4H2PO4-NH3·H2O buffer (pH 6.5) (A) and methanol (B) was involved. The detection wavelength was 290 nm and the flow rate was 1.0 mL min−1. Tris-HCl buffer (50 mM, pH 8.0) and allopurinol were used as negative and positive controls, respectively.The inhibitory ratio of each sample was calculated according to the following equation:
The IC50 value refers to the required sample concentration (mmol L−1) when the XOI ratio is 50%.
Determination of the molecular weight distribution of DWMH
The molecular weight distribution of DWMH was monitored by using a TSK-Gel G2000 SWxl gel filtration column (7.8 mm × 300 mm, Tosoh Corporation, Tokyo, Japan) attached to a Shimadzu LC-20A HPLC system (Shimadzu, Japan). The samples were dissolved in the mobile phase consisting of acetonitrile/water (1/1, v/v) in the presence of 0.1% (v/v) trifluoroacetic acid and the flow rate was set at 0.5 mL min−1. The calibration curve was obtained using the following standards from Sigma: cytochrome C (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 384 Da), aprotinin (6511 Da), zinc bacitracin (1423 Da), oxidized glutathione (612 Da), Gly-Gly-Tyr-Arg (451 Da) and Gly-Gly-Gly (189 Da).23 Absorbance at 214 nm was measured (data not given).
384 Da), aprotinin (6511 Da), zinc bacitracin (1423 Da), oxidized glutathione (612 Da), Gly-Gly-Tyr-Arg (451 Da) and Gly-Gly-Gly (189 Da).23 Absorbance at 214 nm was measured (data not given).
Separation of DWMH by using gel filtration chromatography
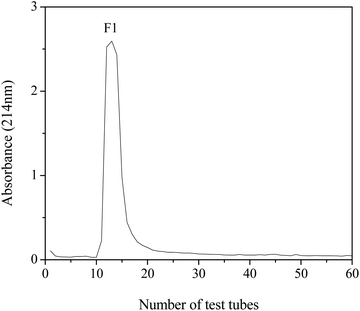
The lyophilized sample of DWMH was dissolved in distilled water (50 mg mL−1) and subjected to separation by using a Sephadex G-15 gel filtration column (1.6 × 50 cm). The column was eluted with distilled water at a flow rate of 0.5 mL min−1. The eluate (3 mL per tube) was collected and detected at 214 nm. The fraction of interest (F1) was collected and lyophilized for further study.Identification of peptides by LC-ESI-MS/MS
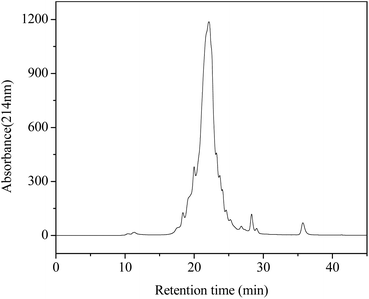
The F1 fraction was dissolved in acetonitrile/water (1/1, v/v) containing 0.1% (v/v) formic acid and then an aliquot (20 μL) of it was subjected to detection by using a C18 column (2.1 × 50 mm, 1.8 μm, Phenomenex, Torrance, CA, USA) on an ultraperformance liquid chromatography-high resolution mass spectrometer (UPLC-HRMS, Agilent 1290/maXis Impact, Bruker Biospin Co., Karlsruhe, Germany). The column was a gradient elution one with a mobile phase including water (A) and acetonitrile (B) in the following procedures: 0–4 min, linear gradient 0–80% B; 4–8 min, 80% B; 8–9 min, linear gradient 80–10% B; 9–10 min, 10% B. Detection was conducted at a flow rate of 0.2 mL min−1 and monitoring at 214 nm. The flow was performed in the positive electrospray ionization mode (ESI+). The scan spectra ranged from 50 to 1500 m/z (mass/charge). In addition, peptide sequencing was performed by processing the MS/MS spectra using Compass DataAnalysis software (Version 4.1, Bruker Daltonik GmbH) as well as manual calculation.Molecular docking studies
Molecular docking of the peptide−XO complex was conducted in order to explore the potential binding details. The molecular docking experiments were conducted by using the MGL tools 1.5.6 with AutoGrid 4.0 and AutoDock 4.0. The X-ray crystal structure of bovine XO in complex with indole-3-aldehyde (PDB ID: 3NVZ) was downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/pdb). The 3D structures of antihyperuricemic peptides were obtained from Chem3D Ultra 12.0. The process was performed as described by Deepthi et al. and Madhuri et al.24,25 The A-chain of the protein and all the water molecules in XO were excluded and polar hydrogen atoms were added before assigning Gasteiger charges to the macromolecule file for the preparation of docking simulation. A grid box with dimensions of 40 Å × 40 Å × 40 Å was defined to enclose the active site with a certain grid spacing of 0.375 Å. Afterwards, the docking simulations were carried out with the default parameters excluding the number of the genetic algorithm runs (100 times). Finally, all the docking models were sorted by the estimated binding energy and the lowest one was selected as its most favorable binding mode.Statistical analysis
All the experimental results were expressed as the mean ± standard deviation. Statistical significance (p < 0.05 and 0.01) was obtained by using one-way ANOVA followed by Tukey's test using SPSS 19.0 (SPSS Inc., Chicago, IL, USA). Statistical analysis was conducted by using Origin 8.5 (OriginLab, Northampton, MA, USA).Results and discussion
In vivo antihyperuricemic effects of WMH and DWMH
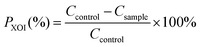
The body weight of all the rats increased gradually over 21 days with no significant difference among five groups (Fig. 1A). Meanwhile, the rats in different groups exhibited shiny hair, good water and food consumption, and normal urine volume, and all the animals were in a healthy state. These results confirmed that WMH and DWMH revealed no side effects on the rats.As shown in Fig. 1B, on the 0th day, the serum uric acid (SUA) levels of the rats treated with potassium oxonate in the four groups (model control group, 225.8 ± 41.7 μM; allopurinol group, 220.0 ± 31.0 μM; WMH group, 211.4 ± 31.0 μM; and DWMH group, 235.2 ± 27.0 μM) were significantly greater (p < 0.01) than those of the rats without potassium oxonate treatment (74.9 ± 28.7 μM). The SUA level of rats in the model control group steadily increased over 21 days under treatment by potassium oxonate in comparison with those of the normal control group. This phenomenon was consistent with the fact that potassium oxonate blocked the function of hepatic uricase and thus drastically increased the SUA levels leading to hyperuricemia in rats.8,26 A single dose of allopurinol (25 mg per kg BW) or DWMH (300 mg per kg BW) could effectively and persistently reduce the serum uric acid contents of the hyperuricemic rats as compared with the model control group (p < 0.01, p < 0.05). A single administration of WMH (300 mg per kg BW) also exhibited significant serum-uric-acid-lowering activity (p < 0.05) in hyperuricemic rats on the 14th day. DWMH demonstrated a similar effective function to that of allopurinol, a traditional drug for gout and hyperuricemia therapy,27 indicating that DWMH possessed the potential to be used as a functional food ingredient for gout and hyperuricemia treatment via reducing the serum uric acid level.
In clinical reports, the serum uric acid level was closely related to kidney disease progression, systemic inflammation or organ dysfunction.28 These diseases were also sensitively associated with clinical and laboratory signs including blood urea nitrogen or creatinine changes.29 Serum creatinine was found to be more specific as a marker of renal function than urea nitrogen, and a change over the baseline might induce acute renal dysfunction.30 As shown in Fig. 1C and D, significant increases were observed for serum urea nitrogen (p < 0.01) and creatinine (p < 0.05) levels of rats after treatment with potassium oxonate for 21 days (model control group vs. normal control group). Allopurinol could also obviously reduce the level of serum creatinine of hyperuricemic rats (p < 0.05) (allopurinol group vs. model control group). A single administration of WMH or DWMH did not change the level of serum urea nitrogen or creatinine as compared with that of the model control group (p > 0.05); nevertheless, DWMH did suppress the levels of serum urea nitrogen and creatinine (DWMH group vs. model control group).
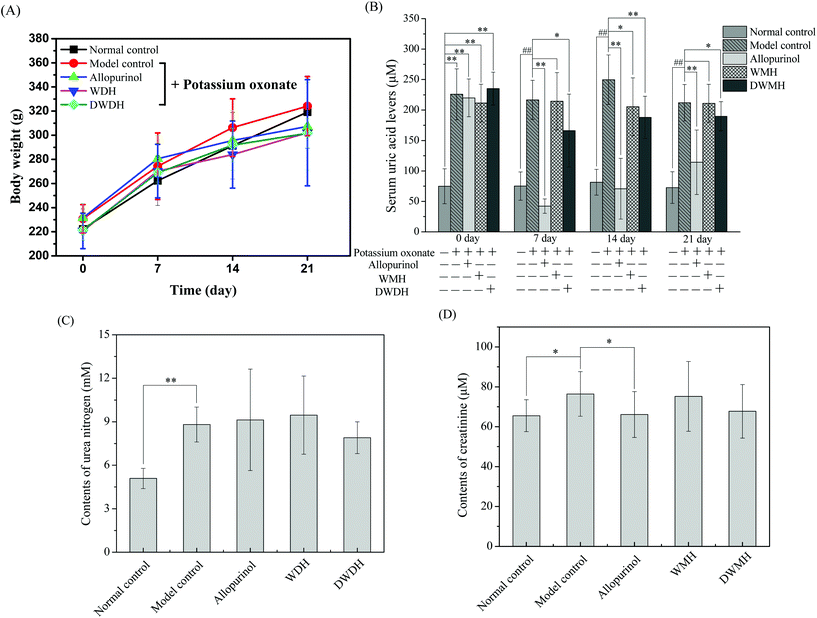
An early study found that potassium oxonate-induced hyperuricemia generated the development of systemic hypertension and kidney dysfunction as induced by intrarenal oxidative stress due to elevated uric acid.31,32 Histopathological observations showed that the kidneys of rats in the model control group (Fig. 2B) were prominently damaged in the interstitial tissues compared to the normal control group (Fig. 2A). The kidneys in Fig. 2B exhibited remarkable detrimental tubule interstitial inflammation, which showed sloughing of the tubular epithelial cells and excessive numbers of the interstitial cellular infiltration. In contrast, rats in the other three groups (allopurinol group, WMH group, DWMH group) retained nearly normal histomorphological structures of the kidney (Fig. 2C–E). These results demonstrated the effective protective function of WMH and DWMH in the improvement of renal failure. Therefore, WMH and DWMH (especially DWMH) could be developed as beneficial antihyperuricemic agents and effective renoprotective agents.
Separation of DWMH based on XOI activity evaluation
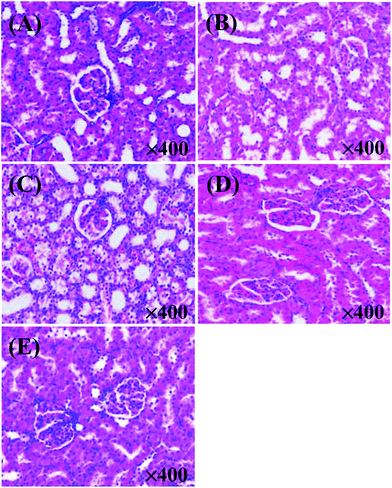
As shown in Fig. 3, the XOI activity of DWMH (27.23 ± 1.67%) was obviously higher than that of WMH (18.02 ± 1.93%) at the same concentration of 20 mg mL−1 (p < 0.05), which was consistent with the results of animal experiments mentioned above, suggesting that the XOI effects of DWMH were not correlated with the phenolic compounds existing in walnut.33 Thus, DWMH was selected for further study.According to a previous report, oligopeptides containing 5–10 amino acids with small molecular weight usually could be more easily absorbed and exhibited specific bioactivity. Besides, they had the advantage of being obtained by enzymatic or chemical synthesis methods.34 The molecular weight distribution of DWMH detected by gel filtration chromatography on a TSK-Gel G2000 SWxl column is displayed in Fig. 4. It was found that almost 68% of the DWMH-derived peptides possessed molecular weights ranging from 180 to 1000 Da, which basically contained less than 10 amino acids and had the potential to interact specifically with biological targets.
The DWMH-derived peptides were then purified by using a Sephadex G-15 gel filtration chromatography column. As shown in Fig. 5, one fraction (F1) was obtained and its XOI activity was 40.70 ± 2.79%, which was better than that of DWMH (27.23 ± 1.67%) before separation at a concentration of 20 mg mL−1 (see Fig. 3).
Identification of XOI peptides by LC-ESI-MS/MS and their bioactivity evaluation
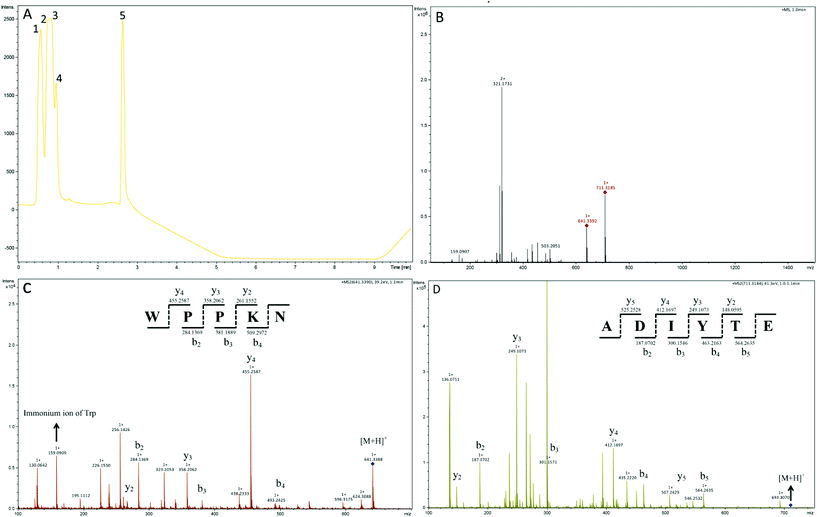
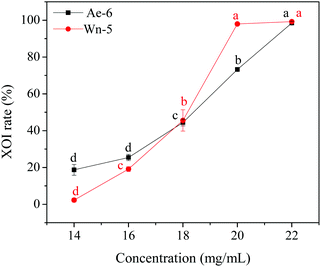
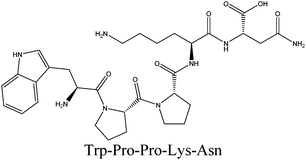
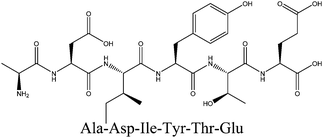
In order to identify the potential active peptides, the F1 fraction was collected and subjected to RP-UPLC coupled on-line to an ion trap mass spectrometer, and the results are shown in Fig. 6. The RP-UPLC-UV chromatographic profile of fraction F1 exhibited five subfractions, as shown in Fig. 6A. Twenty-four compounds were found using the AutoMS(n) function of Compass DataAnalysis software. However, only peak 4 (in Fig. 6A) with retention time 1 min could be confirmed to identify the peptide sequence, and the MS spectrum of peak 4 is shown in Fig. 6B. This spectrum showed that several peptides coeluted at this retention time (1 min). Interestingly, two peptide parent ions with m/z at 641.3392 and 711.3185 were identified automatically by using Compass DataAnalysis software. Therefore, these two peptide ions were naturally selected for fragmentation by the software of the mass spectrometer. The MS/MS spectra of charged ions with m/z at 641.3392 and 711.3185 are illustrated in Fig. 6C and D, respectively. The sequence was determined by the difference in masses of adjacent fragment ions. For example, the difference between the ions with m/z at 641.3392 and 455.2587 is 186.0805, which corresponds to the mass of a tryptophan amino acid residue. In this process, by manual calculation, the Compass DataAnalysis software was a big help to automatically make amino acid annotations. Based on the result of LC-ESI-MS/MS analysis, the sequences of these two peptides were identified to be Trp-Pro-Pro-Lys-Asn (WPPKN) (640.8 Da) and Ala-Asp-Ile-Tyr-Thr-Glu (ADIYTE) (710.7 Da), respectively. Subsequently, the two identified peptides were chemically synthesized and subjected to XOI activity evaluation.As shown in Fig. 7, the addition of Ala-Asp-Ile-Tyr-Thr-Glu (ADIYTE) from 14 to 16 mg mL−1 could slowly reduce XO activity (p > 0.05), and then the suppressing trend of the XO property increased dramatically with increasing contents of the inhibitor (p < 0.05). A similar trend was observed in the results for Trp-Pro-Pro-Lys-Asn (WPPKN) in Fig. 7. Furthermore, as observed in Fig. S1 and S2,† the production of uric acid was apparently decreased with the increasing contents of WPPKN and ADIYTE, which was in accordance with the former results. More detailed information about the comparison between allopurinol and these two synthesized peptides (WPPKN and ADIYTE), including structural formulae, molecular weights and IC50 values, is listed in Table 1. The Trp at the N-terminal in the sequence of WPPKN might contribute to its XOI activity, since Trp has a xanthine-like structure and is analogous to the inhibitors of XO, such as allopurinol.18 However, the sizes of WPPKN and ADIYTE are much greater than that of allopurinol so that it might be harder for them to be well attached to the active center of xanthine oxidase. The IC50 values of WPPKN and ADIYTE were detected to be 17.75 ± 0.12 mg mL−1 and 19.01 ± 0.23 mg mL−1, respectively. Although it seems that the XOI activity of WPPKN or ADIYTE was not as strong as that of allopurinol (IC50 = 10.10 ± 0.29 μg mL−1), as protein-originated natural agents, WPPKN and ADIYTE have the advantage of less side-effects on human health, demonstrating competitive potential use in health care products.
The in vitro XOI properties of two pure synthesized peptides, WPPKN (97.95 ± 0.39%) and ADIYTE (73.21 ± 0.62%), were proved to be higher than that of the fraction F1 (40.70 ± 2.79%), and even more higher than that of the crude DWMH sample (27.23 ± 1.67%), when tested at the same concentration of 20 mg mL−1. This result indicated that WPPKN and ADIYTE played a certain role in depressing the XO activity as effective components of the F1 fraction and DWMH crude sample, which might be responsible for the reason why DWMH demonstrated obvious antihyperuricemic activity in hyperuricemic rats as described at the beginning of this research.
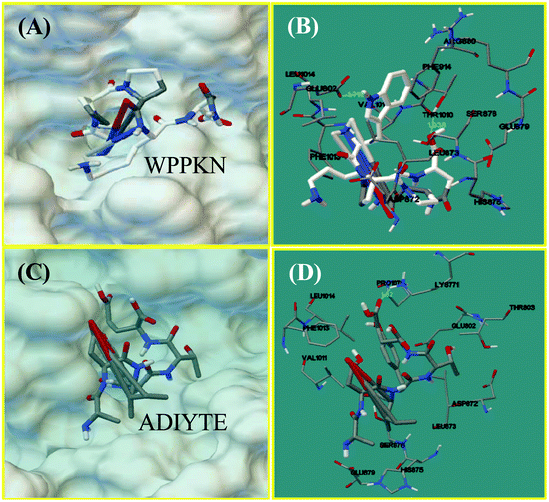
Molecular docking of XO and identified peptides
Molecular docking was designed to improve the understanding of the interaction between the ligand and biomacromolecule. From the docking results of WPPKN, the interaction between WPPKN and XO is illustrated in Fig. 8A and B, and the estimated lowest binding energy was −1.23 kcal mol−1 with an inhibitory constant (Ki) of 125.39 mM. Apparently, the active cavity, a narrow channel leading the substrate to the Mo domain of XO, was partly blocked by the larger inhibitor WPPKN compared to xanthine (Fig. 8A). As a consequence, it was hard for xanthine to be inserted into the Mo domain and more free XO failed to catalyze the substrate. Besides, it has been reported that the Mo domain was embraced by these residues: Phe 649, Asn 768, Glu 802, Leu 873, Arg880 and Phe 914, Phe 1009, Thr 1010, Leu 1014 and Glu 1261, among which Glu 802, Glu 1261 and Arg 880 were demonstrated to play a pivotal role in the catalytic reaction.35,36 As shown in Fig. 8B, WPPKN landed in a cavity formed by these residues: Leu 1014, Glu 802, Phe 914, Arg 880, Val 1011 and Phe 1013, Asp 872, His 875, Leu 873 and Thr 1010, Ser 876 and Glu 879, part of which overlapped with the reported residues in the active cavity. Moreover, two hydrogen bonds were observed, including one formed with the Glu 802 residue with the length of 1.719 Å, and another formed with the Thr 1010 residue (1.938 Å) (Fig. 8B). Thus, it could be concluded that WPPKN entered into the hydrophobic channel and obstructed the protein–ligand interaction between XO and xanthine.As for ADIYTE, the interaction between ADIYTE and XO is illustrated in Fig. 8C and D, and the estimated lowest binding energy was 18.6 kcal mol−1 without giving an inhibitory constant (Ki). In addition, ADIYTE was on the surface of the B-chain and blocked the entrance of the substrate to the hydrophobic channel (Fig. 8C). As observed in Fig. 8D, ADIYTE was positioned in a cavity formed by these residues: Leu 1014, Glu 802, Phe 1013, Glu 879, Val 1011 and Asp 872, His 875, Leu 873 and Thr 803, Ser 876 and Lys771, part of which overlapped with the reported residues in the active cavity. Besides, one hydrogen bond was formed between ADIYTE and the Lys 771 residue with the length of 1.62 Å (Fig. 8D). According to an investigation, Lys754 and Lys771 residues were located next to the surface of the Mo domain,37 which could explain the inhibiting effects of ADIYTE on XO.
Conclusions
In conclusion, WMH and DWMH, especially DWMH, exhibited great potential to act as antihyperuricemic agents and played a critical role in kidney protection as illustrated in the animal test. The DWMH-derived F1 fraction displayed good in vitro XOI activity. According to molecular simulation, the identified peptides both possessed good XOI properties, while ADIYTE was not as effective as WPPKN due to weaker interaction with the Mo domain. This result helped explain why the IC50 value of WPPKN was lower than that of ADIYTE. Moreover, further studies about the in vivo bioavailability and the main driving forces of the interaction between XO and WPPKN or ADIYTE are still in preparation.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors gratefully acknowledge the National Natural Science Foundation of China (No. 31671804), Guangdong Special Funding for Outstanding Young Scholars (2014TQ01N645), the Science and Technology Program of Guangdong (201604020047 & 2015B020230001) and the Fundamental Research Funds for the Central Universities (2017ZD079).References
- D. Conen, V. Wietlisbach, P. Bovet, C. Shamlaye, W. Riesen, F. Paccaud and M. Burnier, Prevalence of hyperuricemia and relation of serum uric acid with cardiovascular risk factors in a developing country, BMC Public Health, 2004, 4, 9 CrossRef CAS PubMed.
- D. Khanna, J. D. Fitzgerald, P. P. Khanna, S. Bae, M. K. Singh, T. Neogi, M. H. Pillinger, J. Merill, S. Lee and S. Prakash, 2012 American College of Rheumatology guidelines for management of gout. Part 1: systematic nonpharmacologic and pharmacologic therapeutic approaches to hyperuricemia, Arthritis Care Res., 2012, 64, 1431–1446 CrossRef CAS PubMed.
- N. Cantu-Medellin and E. E. Kelley, Xanthine oxidoreductase-catalyzed reactive species generation: A process in critical need of reevaluation, Redox Biol., 2013, 1, 353–358 CrossRef CAS PubMed.
- C. M. Burns and R. L. Wortmann, Gout therapeutics: new drugs for an old disease, Lancet, 2011, 377, 165–177 CrossRef CAS.
- Z. Hajdú, A. Martins, O. Orbán-Gyapai, P. Forgo, N. Jedlinszki, I. Máthé and J. Hohmann, Xanthine oxidase-inhibitory activity and antioxidant properties of the methanol extract and flavonoids of Artemisia asiatica, Rec. Nat. Prod., 2014, 8, 299 Search PubMed.
- M. Hamzeh-Mivehroud, S. Rahmani, M.-R. Rashidi, M.-A. Hosseinpour Feizi and S. Dastmalchi, Structure-based investigation of rat aldehyde oxidase inhibition by flavonoids, Xenobiotica, 2013, 43, 661–670 CrossRef CAS PubMed.
- A. Hunyadi, A. Martins, B. Danko, D.-W. Chuang, P. Trouillas, F.-R. Chang, Y.-C. Wu and G. Falkay, Discovery of the first non-planar flavonoid that can strongly inhibit xanthine oxidase: protoapigenone 1′-O-propargyl ether, Tetrahedron Lett., 2013, 54, 6529–6532 CrossRef CAS.
- I. Murota, S. Taguchi, N. Sato, E. Y. Park, Y. Nakamura and K. Sato, Identification of antihyperuricemic peptides in the proteolytic digest of shark cartilage water extract using in vivo activity-guided fractionation, J. Agric. Food Chem., 2014, 62, 2392–2397 CrossRef CAS PubMed.
- A. B. Nongonierma and R. J. FitzGerald, Tryptophan-containing milk protein-derived dipeptides inhibit xanthine oxidase, Peptides, 2012, 37, 263–272 CrossRef CAS PubMed.
- I. Jemil, L. Mora, R. Nasri, O. Abdelhedi, M.-C. Aristoy, M. Hajji, M. Nasri and F. Toldrá, A peptidomic approach for the identification of antioxidant and ACE-inhibitory peptides in sardinelle protein hydrolysates fermented by Bacillus subtilis A26 and Bacillus amyloliquefaciens An6, Food Res. Int., 2016, 89, 347–358 CrossRef CAS PubMed.
- S.-J. Lee, Y.-S. Kim, J.-W. Hwang, E.-K. Kim, S.-H. Moon, B.-T. Jeon, Y.-J. Jeon, J. M. Kim and P.-J. Park, Purification and characterization of a novel antioxidative peptide from duck skin by-products that protects liver against oxidative damage, Food Res. Int., 2012, 49, 285–295 CrossRef CAS.
- C. Wiriyaphan, B. Chitsomboon, S. Roytrakul and J. Yongsawadigul, Isolation and identification of antioxidative peptides from hydrolysate of threadfin bream surimi processing byproduct, J. Funct. Foods, 2013, 5, 1654–1664 CrossRef CAS.
- D. A. Luna-Vital, E. González de Mejía and G. Loarca-Piña, Selective mechanism of action of dietary peptides from common bean on HCT116 human colorectal cancer cells through loss of mitochondrial membrane potential and DNA damage, J. Funct. Foods, 2016, 23, 24–39 CrossRef CAS.
- O. Abdelhedi, M. Jridi, I. Jemil, L. Mora, F. Toldrá, M.-C. Aristoy, A. Boualga, M. Nasri and R. Nasri, Combined biocatalytic conversion of smooth hound viscera: Protein hydrolysates elaboration and assessment of their antioxidant, anti-ACE and antibacterial activities, Food Res. Int., 2016, 86, 9–23 CrossRef CAS.
- M. A. Corrons, C. S. Liggieri, S. A. Trejo and M. A. Bruno, ACE-inhibitory peptides from bovine caseins released with peptidases from Maclura pomifera latex, Food Res. Int., 2017, 93, 8–15 CrossRef CAS PubMed.
- J. Zou, P.-S. Cai, C.-M. Xiong and J.-L. Ruan, Neuroprotective effect of peptides extracted from walnut (Juglans Sigilata Dode) proteins on Aβ25-35-induced memory impairment in mice, J. Huazhong Univ. Sci. Technol., Med. Sci., 2016, 36, 21–30 CrossRef CAS PubMed.
- M.-C. Liu, S.-J. Yang, D. Hong, J.-P. Yang, M. Liu, Y. Lin, C.-H. Huang and C.-J. Wang, A simple and convenient method for the preparation of antioxidant peptides from walnut (Juglans regia L.) protein hydrolysates, Chem. Cent. J., 2016, 10, 1–11 CrossRef PubMed.
- A. B. Nongonierma, C. Mooney, D. C. Shields and R. J. FitzGerald, Inhibition of dipeptidyl peptidase IV and xanthine oxidase by amino acids and dipeptides, Food Chem., 2013, 141, 644–653 CrossRef CAS PubMed.
- V. N. Sarvaiya, K. A. Sadariya, P. G. Pancha, A. M. Thaker, A. C. Patel and A. S. Prajapati, Evaluation of antigout activity of Phyllanthus emblica fruit extracts on potassium oxonate-induced gout rat model, Vet. World, 2015, 8, 1230 CrossRef PubMed.
- X.-H. Wu, J.-L. Ruan, J. Zhang, S.-Q. Wang and Y.-W. Zhang, Pallidifloside D, a saponin glycoside constituent from Smilax riparia, resist to hyperuricemia based on URAT1 and GLUT9 in hyperuricemic mice, J. Ethnopharmacol., 2014, 157, 201–205 CrossRef CAS PubMed.
- I. Murota, T. Tamai, T. Baba, N. Sato, E. Y. Park, Y. Nakamura and K. Sato, Moderation of oxonate-induced hyperuricemia in rats via the ingestion of an ethanol-soluble fraction of a shark cartilage proteolytic digest, J. Funct. Foods, 2012, 4, 459–464 CrossRef CAS.
- X. Wang, J. Zhou, B. Shi, X. Guo and Q. Yan, Hypouricemic and nephroprotective effects of Jianpi Huashi decoction in a rat model of hyperuricemia, Int. J. Clin. Exp. Med., 2016, 9, 455–465 Search PubMed.
- Q.-X. Zhang, Y.-F. Ling, Z. Sun, L. Zhang, H.-X. Yu, S. M. Kamau and R.-R. Lu, Protective effect of whey protein hydrolysates against hydrogen peroxide-induced oxidative stress on PC12 cells, Biotechnol. Lett., 2012, 34, 2001–2006 CrossRef CAS PubMed.
- K. Deepthi, K. Nagoji and A. V. Rao, Computer Aided Molecular Docking Studies on Diarylsulfonylureas as Potential Anticancer Agents, Int. J. Comput. Appl., 2014, 92, 33–36 Search PubMed.
- M. Madhuri, C. Prasad and V. R. Avupati, In Silico Protein-Ligand Docking Studies on Thiazolidinediones as Potential Anticancer Agents, Int. J. Comput. Appl., 2014, 95, 13–16 Search PubMed.
- T. Taniguchi, N. Ashizawa, K. Matsumoto, T. Iwanaga and K. Saitoh, Uricosuric agents decrease the plasma urate level in rats by concomitant treatment with topiroxostat, a novel xanthine oxidoreductase inhibitor, J. Pharm. Pharmacol., 2016, 68, 76–83 CrossRef CAS PubMed.
- Y.-P. Siu, K.-T. Leung, M. K.-H. Tong and T.-H. Kwan, Use of allopurinol in slowing the progression of renal disease through its ability to lower serum uric acid level, Am. J. Kidney Dis., 2006, 47, 51–59 CrossRef CAS PubMed.
- M. Chonchol, M. G. Shlipak, R. Katz, M. J. Sarnak, A. B. Newman, D. S. Siscovick, B. Kestenbaum, J. K. Carney and L. F. Fried, Relationship of Uric Acid With Progression of Kidney Disease, Am. J. Kidney Dis., 2007, 50, 239–247 CrossRef CAS PubMed.
- M. Gao, L. Zhang, Y. Liu, M. Yang, N. Wang, K. Wang, D. Ou, M. Liu, G. Chen, K. Liu and X. Xiao, Use of blood urea nitrogen, creatinine, interleukin-6, granulocyte–macrophage colony stimulating factor in combination to predict the severity and outcome of abdominal sepsis in rats, Inflammation Res., 2012, 61, 889–897 CrossRef CAS PubMed.
- R. Bellomo, C. Ronco, J. A. Kellum, R. L. Mehta and P. Palevsky, Acute renal failure – definition, outcome measures, animal models, fluid therapy and information technology needs: the Second International Consensus Conference of the Acute Dialysis Quality Initiative (ADQI) Group, Crit. Care, 2004, 8, 1–9 CrossRef PubMed.
- L. G. Sánchez-Lozada, V. Soto, E. Tapia, C. Avila-Casado, Y. Y. Sautin, T. Nakagawa, M. Franco, B. Rodríguez-Iturbe and R. J. Johnson, Role of oxidative stress in the renal abnormalities induced by experimental hyperuricemia, Am. J. Physiol.: Renal, Fluid Electrolyte Physiol., 2008, 295, F1134–F1141 CrossRef PubMed.
- O. A. Kensara, Protective effect of vitamin C supplementation on oxonate-induced hyperuricemia and renal injury in rats, Int. J. Nutr. Metab., 2013, 5, 61–68 CAS.
- X. Wang, M. Zhao, G. Su, M. Cai, D. Sun-Waterhouse, C. Zhou and L. Lin, Antihyperuricemic activities of an ethanolic and aqueous extract of Walnut (Juglans regia L.) shell and a new aldehyde xanthine oxidase inhibitor, Int. J. Food Sci. Technol., 2016, 51, 453–460 CrossRef CAS.
- D. J. Craik, D. P. Fairlie, S. Liras and D. Price, The Future of Peptide-based Drugs, Chem. Biol. Drug Des., 2013, 81, 136–147 CAS.
- T. Nishino, K. Okamoto, B. T. Eger, E. F. Pai and T. Nishino, Mammalian xanthine oxidoreductase–mechanism of transition from xanthine dehydrogenase to xanthine oxidase, FEBS J., 2008, 275, 3278–3289 CrossRef CAS PubMed.
- J. M. Pauff, H. Cao and R. Hille, Substrate orientation and catalysis at the molybdenum site in xanthine oxidase crystal structures in complex with xanthine and lumazine, J. Biol. Chem., 2009, 284, 8760–8767 CrossRef CAS PubMed.
- K. Ichida, Y. Amaya, K. Okamoto and T. Nishino, Mutations associated with functional disorder of xanthine oxidoreductase and hereditary xanthinuria in humans, Int. J. Mol. Sci., 2012, 13, 15475–15495 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7fo01174a |
| ‡ Co-first author. |
| This journal is © The Royal Society of Chemistry 2018 |