Open Access Article
Open Access ArticleImpact of upstream chlorination on filter performance and microbial community structure of GAC and anthracite biofilters†
Glen Andrew
de Vera
 a,
Daniel
Gerrity
a,
Daniel
Gerrity
 b,
Mitchell
Stoker
a,
Wilbur
Frehner
a and
Eric C.
Wert
b,
Mitchell
Stoker
a,
Wilbur
Frehner
a and
Eric C.
Wert
 *a
*a
aApplied Research and Development Center, Southern Nevada Water Authority, P.O. Box 99954, Las Vegas, NV 89193, USA. E-mail: eric.wert@snwa.com; Tel: +1(702) 856 3669
bDepartment of Civil and Environmental Engineering and Construction, University of Nevada, Las Vegas, 4505 S. Maryland Parkway, Las Vegas, NV 89154-4015, USA
First published on 11th June 2018
Abstract
Drinking water filters may be operated to promote or deter biological activity through upstream oxidant addition. While there are several water quality benefits from biofiltration, microbial growth in biofilters warrants further investigation. In this study, routine monitoring detected target DNA sequences for Naegleria fowleri in source water and Acanthamoeba spp. in source water and biofilter effluent, triggering further microbial community characterization. Full-scale anthracite and granular activated carbon (GAC) filters receiving chlorinated waters were compared in terms of effluent water quality (i.e., turbidity and particle counts), biological activity (i.e., adenosine triphosphate (ATP)), and the composition of the microbial community (i.e., 16S/18S rRNA gene sequencing, free-living amoeba). Because of rapid chlorine quenching by GAC, greater biomass development was observed in the GAC biofilter (ATP = 5 × 103–5 × 104 pg cm−3 media) than the anthracite filter (ATP = 4 × 102–1 × 103 pg cm−3 media). Due to possible sloughed biomass, GAC effluent also had consistently greater turbidity, particle counts, and cellular ATP than the anthracite filter. 16S rRNA gene sequencing revealed distinct taxonomic differences between the anthracite and GAC filters, and GAC also hosted a more diverse population (Shannon index: GAC = 3.7–5.0 and anthracite = 2.4–2.9). At the genus level, the anthracite filter contained mostly Undibacterium-like taxon (45–68%), while the GAC biofilter was dominated by Massilia (8–36%), Herbaspirillum (2–44%), and unknown Comamonadaceae (9–18%), among others. The presence of viable free-living amoebas was also detected in the GAC biofilter media. Further characterization of the eukaryotic 18S rRNA gene showed that anthracite predominantly harbored copepod Leptodiaptomus (67%), while a majority of the sequences in GAC are unknown.
Water impactThis study emphasizes the need to weigh the benefits of biofiltration (e.g., DOC removal) against potential microbial risks associated with enhanced biomass development within filtration systems. Because of the reactivity of granular activated carbon (GAC) towards oxidants, GAC biofilters may allow for accumulation and release of unwanted organisms. In contrast, filters that maintain a disinfectant residual (e.g., chlorinated anthracite) may mitigate these risks. |
1. Introduction
Biofiltration provides water quality benefits such as improved distribution system biostability and biodegradation of organic carbon, disinfection byproduct precursors, and other contaminants,1–4 while meeting regulatory filter effluent turbidity guidelines.5 Monitoring biofilter efficiency may include water quality parameters such as biological activity (i.e., adenosine triphosphate (ATP)), dissolved oxygen consumption, or various organic carbon measurements (e.g., biodegradable/assimilable organic carbon, fluorescence excitation–emission matrices, size-exclusion chromatography, etc.).6,7 Strategies such as nutrient addition and upstream oxidant usage have also emerged to improve biofilter performance with respect to a variety of operational challenges (i.e., head loss development, reduced filter time, increased turbidity and particle counts).8 With these advances in monitoring and operating strategies, water utilities must develop greater understanding of the microbial structure within biofiltration systems to fully evaluate the benefits and possible risks of biofiltration for a specific utility. Current microbiological methods now allow for greater opportunity to explore and optimize biofilters to meet treatment objectives.Different organisms have different catabolic abilities for certain substrates. Hence, depending on the influent water quality, certain microbes can be enriched in the filter media, thereby impacting the overall composition of the microbial community. For example, granular activated carbon (GAC) biofilters respond to changes in the carbon![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nitrogen
nitrogen![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phosphorous ratios of the influent. Under nutrient-limited conditions, Bradyrhizobium may proliferate, produce excessive amounts of extracellular polymeric substances (EPS), and cause rapid filter clogging.9 The addition of phosphorous has also been shown to cause dramatic changes in the microbial community of activated carbon bioreactors.10 Preceding treatment processes such as oxidation also impact characteristics of the filter microbiome. Greater eukaryotic markers were found in biofilters receiving non-ozonated waters compared to ozonated waters.11 Some organisms may have also developed tolerance against chlorination.12 Microbial community structure could also differ with respect to filter depth. The top layer of the filter can be composed of microbes adapted for efficient consumption of readily biodegradable dissolved organic carbon (DOC),11 which can shift with the backwashing procedure.13 Filter-specific occurrence patterns have also been observed in a recent study (e.g., Bradyrhizobium in GAC and Nitrospira in sand-associated filters).14
phosphorous ratios of the influent. Under nutrient-limited conditions, Bradyrhizobium may proliferate, produce excessive amounts of extracellular polymeric substances (EPS), and cause rapid filter clogging.9 The addition of phosphorous has also been shown to cause dramatic changes in the microbial community of activated carbon bioreactors.10 Preceding treatment processes such as oxidation also impact characteristics of the filter microbiome. Greater eukaryotic markers were found in biofilters receiving non-ozonated waters compared to ozonated waters.11 Some organisms may have also developed tolerance against chlorination.12 Microbial community structure could also differ with respect to filter depth. The top layer of the filter can be composed of microbes adapted for efficient consumption of readily biodegradable dissolved organic carbon (DOC),11 which can shift with the backwashing procedure.13 Filter-specific occurrence patterns have also been observed in a recent study (e.g., Bradyrhizobium in GAC and Nitrospira in sand-associated filters).14
It is essential to understand changes in the microbial composition of the filter media, as they dictate the microbiological quality of the filtered water. Pinto et al. (2012) found that ‘leaky colonizers’ (e.g., Hydrogenophaga, Acidovorax and Denitratisoma) can persist in GAC/sand filters and transfer to distributed waters.15 Lautenschlager et al. (2014) also reported similarities in microbial taxa in biofilters and their respective effluents, but the corresponding abundances differed by filter type (e.g., relative abundance of proteobacteria: GAC > rapid sand filter > slow sand filter).16 Various invertebrates that colonize GAC filters can also be transferred in the filter effluent.17,18 These observations are important especially in the context of microbial risk. Previous studies observed harboring of possible pathogenic species (e.g., Acinetobacter spp., Pseudomonas spp., Klebsiella pneumoniae, Aeromonas, among others) by the GAC media, unintentionally releasing them in the distribution waters through filter fines shed from the biofilters.19,20
Free-living amoebas (FLA) comprise an emerging class of organisms which has not been thoroughly studied in drinking water biofilters. These organisms have generated considerable interest in drinking water distribution systems due to the presence of Naegleria and Acanthamoeba species. Each of these organisms has been listed on the U.S. Environmental Protection Agency's Contaminant Candidate List (Naegleria on CCL4 and Acanthamoeba on CCL1) and both can result in disease (i.e., amoebic meningoencephalitis, keratitis) or even death.21 Because of its status as a CCL4 contaminant, Naegleria fowleri may ultimately be regulated under the Safe Drinking Water Act.22 While regarded as a distribution system phenomenon,23 greater surveillance is needed in drinking water biofilters to determine if they provide habitat for these organisms to proliferate.
In addition to direct health effects, FLA can also have indirect health implications by hosting pathogenic bacteria within its vacuoles.7,24,25 Under these situations, the amoeba may live as trophozoites in suitable environments (such as a biofilter) or as cysts when under stress conditions (such as oxidation).26,27 If present as cysts, FLA pose a risk to drinking water quality during distribution because the cyst form is highly resistant to chlorine, chloramine, and chlorine dioxide,28–31 which are commonly used secondary oxidants in distribution systems. Thus, greater monitoring of FLA is warranted in drinking water treatment plants. To date, there is limited information available that explores the role of biofilters in harboring/releasing FLA.
This study investigates the filter dynamics in parallel full-scale drinking water biofilters by integrating the conventional engineering aspects of filtration (e.g., 10 year turbidity and particle counts data) with microbial characterization. This work was prompted by qPCR detection of Naegleria fowleri and Acanthamoeba spp. target DNA sequences in the raw water, followed by an increase in Acanthamoeba DNA in GAC filter effluent. Various microbial characterization tools were used to meet the objectives of the study: (1) to quantify the amount of biomass (via ATP measurements) in chlorinated GAC and anthracite filters, (2) to determine the effect of biological activity on the effluent turbidity and particle counts, (3) to identify the presence and viability of FLA in the different filters, and (4) to determine differences in microbial diversity and relative abundance via sequencing of the 16S rRNA gene of bacteria and 18S rRNA gene of eukaryotes/invertebrates in the different filters.
2. Experimental section
2.1. Sampling site
Media and water samples were obtained from a full-scale drinking water treatment plant that has been operating for more than 10 years (River Mountains Water Treatment Facility (RMWTF), Henderson, NV, USA). Raw water quality characteristics (±standard deviation) are included in Table S1 in ESI† (DOC = 2.58 ± 0.08 mg L−1; turbidity = 0.38 ± 0.10 NTU; pH = 8.1 ± 0.2; temperature = 16.1 ± 1.8 °C). The treatment process begins with ozonation (O3 = 1.5 mg L−1 and O3/DOC = 0.6 to target 1–2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log inactivation of Cryptosporidium) followed by coagulation [0.6 mg L−1 FeCl3 (as 40% product)], flocculation, chlorination (free Cl2 dose = 1.8 mg L−1), and filtration. The applied chlorine dose before the filters was optimized based on the plant CT requirement. The filtration process is designed with 20 filters operated in parallel: 18 anthracite/sand (filters 1–18) and 2 GAC/sand filters (filters 19–20) (Fig. S1†). The filter bed is composed of 1.8 m of anthracite [effective size (ES) = 1.2 mm; uniformity coefficient (UC) < 1.4; particle density (ρ) = 1.65 g cm−3] or GAC (ES = 1.1 mm; UC < 1.4; ρ = 1.38 g cm−3) followed by 0.2 m of sand (ES = 0.60 mm; UC < 1.4; ρ = 2.62 g cm−3) on a support plate (type “S”, Leopold, Zelionople, PA, USA). The average empty bed contact time is 15 min, the hydraulic loading rate is 7 m h−1, and filter run times are 200 and 150 h for the anthracite and GAC filters, respectively. The anthracite filter allows passage of chlorine into the filter effluent (free Cl2 residual = 1.0 ± 0.3 mg L−1). The GAC filters quench the free chlorine residual within the initial 0.5 m of media depth, allowing the remaining portion of the filter bed (depth > 0.5 m) to have enhanced biological activity. The backwash process uses chlorinated (∼1.8 mg Cl2 L−1) finished water with surface wash, air scour, low rate water backwash, high rate water backwash, and low rate water backwash.
log inactivation of Cryptosporidium) followed by coagulation [0.6 mg L−1 FeCl3 (as 40% product)], flocculation, chlorination (free Cl2 dose = 1.8 mg L−1), and filtration. The applied chlorine dose before the filters was optimized based on the plant CT requirement. The filtration process is designed with 20 filters operated in parallel: 18 anthracite/sand (filters 1–18) and 2 GAC/sand filters (filters 19–20) (Fig. S1†). The filter bed is composed of 1.8 m of anthracite [effective size (ES) = 1.2 mm; uniformity coefficient (UC) < 1.4; particle density (ρ) = 1.65 g cm−3] or GAC (ES = 1.1 mm; UC < 1.4; ρ = 1.38 g cm−3) followed by 0.2 m of sand (ES = 0.60 mm; UC < 1.4; ρ = 2.62 g cm−3) on a support plate (type “S”, Leopold, Zelionople, PA, USA). The average empty bed contact time is 15 min, the hydraulic loading rate is 7 m h−1, and filter run times are 200 and 150 h for the anthracite and GAC filters, respectively. The anthracite filter allows passage of chlorine into the filter effluent (free Cl2 residual = 1.0 ± 0.3 mg L−1). The GAC filters quench the free chlorine residual within the initial 0.5 m of media depth, allowing the remaining portion of the filter bed (depth > 0.5 m) to have enhanced biological activity. The backwash process uses chlorinated (∼1.8 mg Cl2 L−1) finished water with surface wash, air scour, low rate water backwash, high rate water backwash, and low rate water backwash.
2.2. Monitoring plan
Routine monthly monitoring of Acanthamoeba spp., Naegleria fowleri, and Balamuthia mandrillaris target DNA was initiated at the Las Vegas Wash (a wastewater-influenced tributary of Lake Mead), the raw water supply from Lake Mead, and filter effluents from RMWTF due to concerns of pathogenic FLA proliferation induced by drought conditions (i.e., increasing temperature, decreasing surface water level). In May 2017, filter media sampling was conducted in Filter 18 (anthracite/sand; F18) and Filters 19–20 (GAC/sand; F19–F20) at different depths (0–2 m). Media samples were collected to assess the biological activity on the filter media, identify viable FLA, and profile microbial communities present in the anthracite and GAC filters. Filter 18 (anthracite/sand) serves as the control filter (chlorinated) to determine the effect of chlorine quenching on filter performance and microbial structure of GAC/sand filters (n = 2, each for top and bottom layers, before and after backwash). The filter effluents were also equipped with turbidity (1720E, Hach, USA) and particle count (IBR, USA) sensors.Biological activity within the filters was investigated using filter cores obtained using a wheat thief sampling device (Fig. S2†). Compartments are located every ∼0.25 m along the sampling device allowing for complete profiles of the filter media. Once the media was collected, it was stored wet until analysis within 24 hours. To quantify biomass, attached ATP was measured using a Deposit & Surface Analysis test kit (LuminUltra Technologies Ltd., Canada).32,33 Attached ATP values of GAC and anthracite were reported in mass of ATP per cm3 of media particle to account for differences in media density. A similar method was employed to monitor sloughed biomass in the effluent (Quench-Gone Aqueous test kit, LuminUltra Technologies Ltd., Canada). Chlorine residuals were also measured at various filter depths using the N,N-diethyl-p-phenylenediamine free chlorine colorimetric method (Hach, USA).
2.3. Molecular methods
The analysis of water samples for Naegleria fowleri, Acanthamoeba spp., and Balamuthia mandrillaris target DNA sequences was performed using real time quantitative polymerase chain reaction (qPCR). At each sampling location, a 10 liter sample was collected and subjected to tangential flow ultrafiltration (TFU) to concentrate the samples to about 200 mL. The TFU setup (Fig. S3†) is composed of a Rexeed-25S hollow fiber filter (Asahi Kasei Medical Co., Ltd, U.S.) fed with the water sample at a flow rate of 0.6–1.2 mL min−1. Microorganisms attached to the ultrafilter were eluted using a 500 mL buffer solution (Tween 80 (Fisher Scientific) + Antifoam A (Sigma Aldrich) + 10% sodium polyphosphate) until a 150 mL concentrate was collected. The sample retentate and filter eluate were combined (total volume = 350 mL) and centrifuged for 15 min at 800 × g and 24 °C. The resulting pellet was resuspended in 0.40 mL of sterile reagent water and stored at −80 °C. Nucleic acid extraction and purification followed using a MagAttract Virus Mini M48 Kit on a BioRobot M48 workstation (Qiagen, Germany). Further details on sample preparation and DNA extraction are described in Text S1.† Using the extracts, qPCR (Applied Biosystems, CA, USA) was performed to quantify the pathogenic FLA using the standards, primers, and probes presented in Text S1.†34 The qPCR reaction mixture contained 5 μL extracted DNA, 12.5 μL TaqMan Environmental Mix 2.0 (Life Technologies, USA), 0.3 μL of forward and reverse primer (final concentration = 0.24 μM of each primer), 0.25 μL probe (final concentration = 0.20 μM), and 6.65 μL PCR grade water (for a total reaction volume of 25 μL). The thermal cycling parameters were as follows: a single cycle at 95 °C for 10 min, 45 cycles at 95 °C for 15 s, and final extension at 63 °C for 1 min.35 This assay showed no cross-reactivity among different amoebas and had a detection limit of 10 cells per L. An example amplification plot is shown in Fig. S4.†Media samples found to have the greatest attached ATP concentration were selected to assess the presence of viable amoeba within the filters. Approximately 100 mL of core sample was transferred to a 250 mL centrifuge tube containing 200 mL of Page's amoeba saline (PAS). This was mixed for 1 min, and the core samples were allowed to settle for another min. The liquid was removed and pipetted into a separate 250 mL centrifuge tube. This sample was centrifuged at 800 × g and 24 °C for 15 min, the supernatant was decanted, and the resulting pellet was resuspended in 1 mL of PAS. The sample was analyzed by cell culture analysis by adding 500 μL of each sample to two 25 cm2 tissue culture flasks that contained 8.0 mL of PAS and approximately 108 cells of E. coli. The culture flasks were then incubated for 3 days at 24 °C. After incubation, microscopic examination for FLA was conducted using an Eclipse TS100 inverted microscope equipped with a DS-Fi2 color camera (Nikon Instruments, Japan). Following cell culturing, DNA from 400 μL of the sample from the culture flask was extracted using a MagAttract Virus Mini M48 Kit on a BioRobot M48 workstation (Qiagen, Germany) and analyzed by qPCR. A similar approach was conducted using raw water samples.
Microbial community profiling was performed by RTLGenomics (Lubbock, TX, USA) for 16S (bacteria assay) and 18S (eukaryote assay) rRNA gene sequencing. DNA from 50 g media samples was extracted by RTLGenomics using the following procedure. First, 100 mL of warm molecular grade water was added to each media sample. The mixture was agitated, and the water was decanted into two-50 mL centrifuge tubes. The water was then vacuum filtered through a 0.2 μm polyethersulfone membrane, and 1/6th of the filter membrane was processed for DNA extraction using a Qiagen MagAttract Powersoil DNA KF Kit. The extracts were amplified using primers targeting the 16S (515yF: 5′-GTGYCAGCMGCCGCGGTAA-3′ and 926pfR: 5′-CCGYCAATTYMTTTRAGTTT–3′) and 18S (TAReukF: 5′-CCAGCASCYGCGGTAATTCC-3′ and TAReukR: 5′-ACTTTCGTTCTTGATYRA-3′) rRNA genes. After Illumina MiSeq sequencing, data analysis was performed using the standard pipeline of RTLGenomics.36 Denoising and chimera detection were performed using UCHIME37 and UPARSE38 and in-house scripts. Operational taxonomic units (OTUs) selection was performed at 97% (subsequence) identity using the UPARSE algorithm.38 USEARCH_global39 was used to classify the OTU centroid with 90% identity cutoff against an in-house database of RTLGenomics (#2016-02-10) created from respective gene fragments from nucleotide (nr/nt) and genome databases found in NCBI (National Center for Biotechnology Information). Database sequences were at least 500 base pairs, annotated to be partial rRNA genes, and had taxonomic classification of at least the phylum level. The assigned NCBI taxonomy for each sequence was used and screened to contain seven levels, with missing information filled in with ‘Unclassified’ terms. Post-classification scripts were applied to alter the called taxonomy to reflect confidence in the call of <50% agreement for the top six hits at each taxonomic level. For confirmation, assignment of taxonomy was also performed using a naïve Bayesian classifier trained on the 97% majority SILVA (release 128) database.40 Quantitative Insights into Microbial Ecology (QIIME 2)41 was used to evaluate alpha and beta diversity after rarefaction at sequencing depths of 6816 and 6358 sequences (to include all samples) for the 16S and 18S rRNA genes, respectively. The OTU sequences were deposited in GenBank under the following accession numbers: 16S rRNA gene: MG430521-MG430799; 18S rRNA gene: MG418682-MG418825. All raw sequence data were also deposited in the Sequence Read Archive (SRA) under accession number SRP140670.
3. Results and discussion
3.1. Filter performance and biomass development
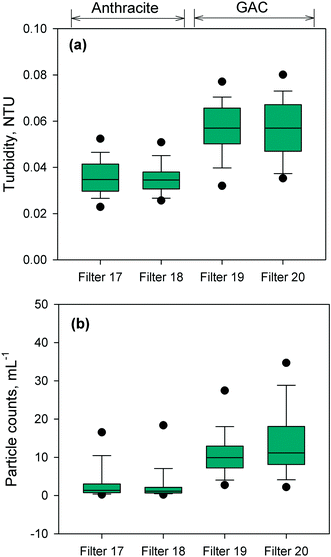
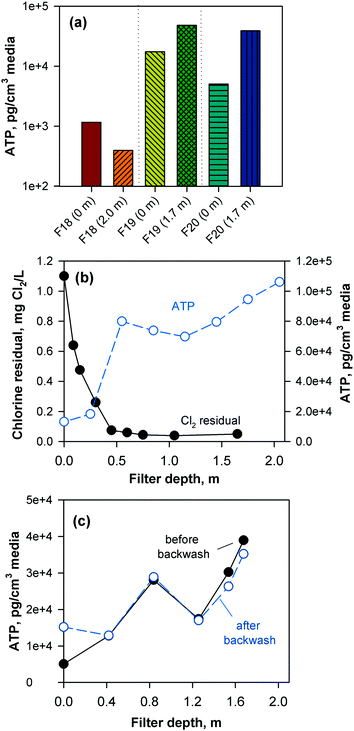
The performance of the anthracite and GAC filters was assessed in terms of turbidity and particle removal. Effluent turbidity and particle counts data (monthly averages) from 2007–2017 are compiled in Fig. 1. Based on the raw water turbidity (range of monthly medians = 0.3–0.8 NTU, Fig. S5†), the filters were found to be effective in removing turbidity-causing particles. Both were able to achieve effluent turbidity <0.15 NTU, in accordance with the Long Term 2 Enhanced Surface Water Treatment (LT2) Rule.5 However, significant differences in turbidity (p < 0.001, Mann–Whitney rank sum test) were observed between the effluent of the chlorinated anthracite and GAC filters (Fig. 1a). The anthracite filters (filters 17–18) had lower median effluent turbidities (<0.04 NTU) than GAC (∼0.06 NTU) over the monitoring period. For the GAC filters (filters 19–20), the effluent turbidities appeared to increase gradually through the years (Fig. S6†). This may be attributed to progressively greater amounts of steady-state biological activity over years of operation. The removal of raw water particles, for which median counts ranged from 1500–3500 particles per mL (Fig. S7†), also differed between the two media types (Fig. 1b) (p < 0.001, Mann–Whitney rank sum test). Consistently, particle counts in GAC effluent were higher than those from the anthracite filter (Fig. S8†). The median particle counts in effluents of anthracite and GAC filters were about 1 and 10 particles per mL, respectively (Fig. 1b). The results for turbidity and particle counts seem to agree with the amount of sloughed biomass in the filter effluents, which was measured by cellular ATP (Fig. S9†). For anthracite, minimal biomass was present in the filter effluent (i.e., ATP < 1 pg mL−1), with only a few samples with detectable ATP. For the biological GAC filters, the presence of sloughed biomass in the effluent translated to consistent ATP detection, with concentrations ranging from of 2–44 pg mL−1.To confirm biomass growth in the media, attached ATP was also measured at various filter depths (Fig. 2). The chlorinated anthracite filters had ATP values of 4 × 102–1 × 103 pg cm−3 media, while GAC biofilters had significantly greater ATP concentrations ranging from 5 × 103–5 × 104 pg cm−3 media. The GAC biofilters were found to contain more biomass toward the bottom of the filter (i.e., attached ATP > 7 × 104 pg cm−3), which was attributed to the absence of a free chlorine residual (i.e., <0.08 mg L−1 Cl2) at media depths >0.5 m (Fig. 2b). In contrast, the anthracite filter effluent contained a free chlorine residual of approximately 1 mg L−1 Cl2 (Fig. S9†), which likely inhibited colonization of the media. Also, backwashing only achieved slight changes in biomass in the GAC (Fig. 2c). Although the backwash water contained chlorine, dechlorination also occurred as the backwash water contacted the GAC in the fluidized upflow media bed [i.e., at the GAC/sand interface (d = 1.3–1.8 m)]. This creates an intermediate zone (d = 0.5–1.3 m) favorable for the proliferation of microorganisms due to the availability of substrate and absence of oxidant exposure at all times.
3.2. Monitoring of amoeba in filter effluent
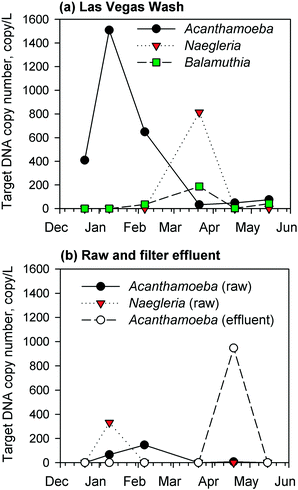
Due to a lack of chlorine residual and subsequent biomass development in the GAC filters, the corresponding filter effluents were monitored by qPCR for the presence of target DNA sequences of pathogenic Acanthamoeba spp., Naegleria fowleri, and Balamuthia mandrillaris. Complementary measurements were conducted for samples from the Las Vegas Wash (a wastewater-impacted tributary to Lake Mead) and the raw water intake for the drinking water facility. The results showed regular occurrence of Acanthamoeba spp., Naegleria fowleri, and/or Balamuthia mandrillaris target DNA in the environmental samples from the Las Vegas Wash (Fig. 3a), presumably due to the high organic and bacterial content of the Wash, but at the raw water intake, there were fewer detections and lower target DNA concentrations of Naegleria fowleri and Acanthamoeba spp. (Fig. 3b). For the GAC filter effluent, there were consistent non-detects by qPCR until April 2017 (Fig. 3b), at which point target Acanthamoeba spp. DNA was detected at a concentration of 950 copies per L, which was 120 times higher than those measured in the corresponding raw water (8 copies per L) and wastewater-impacted Las Vegas Wash (i.e., 48 copies per L) samples. This unexpected spike warranted further investigation into the potential presence of viable FLA in the GAC biofilters, which might serve as a source of Acanthamoeba during sloughing events. Naegleria fowleri and Balamuthia mandrillaris target DNA sequences were not detected in the GAC effluent. These target microbes were absent in the effluent of the anthracite filter sampled in May 2017.3.3. Growth tests for viable amoeba
The filter core samples containing the greatest amount of biomass (i.e., ATP) were used to assess the presence of viable amoeba within the GAC and anthracite filters. After a 72 h growth/incubation period in the presence of E. coli, viable, fan-shaped FLA were found in samples originating from the GAC media (Fig. S10†) but not from the anthracite. Similar organisms were also found in the raw water samples (Fig. S10†). The observed amoebas had diameters of 20–30 μm. Comparison with literature suggests that these viable amoebas exhibit similar morphology to the genus Vannella, which consists of flabellate amoebas found in freshwater, soil, and seawater (typical diameter = 9–80 μm),42 as well as in swamps,43 and moist sanitary areas.44 They have also been identified in drinking water treatment plants, drinking water biofilms, hot water tanks, and residential and hospital tap waters.25,45,46 While this genus is considered non-pathogenic, there have been a few studies showing that it can host several bacterial species and human pathogens such as Microsporidia parasites.47,48 No target pathogenic amoebas were detected by qPCR after this test.3.4. Microbial taxonomic profile of the filters
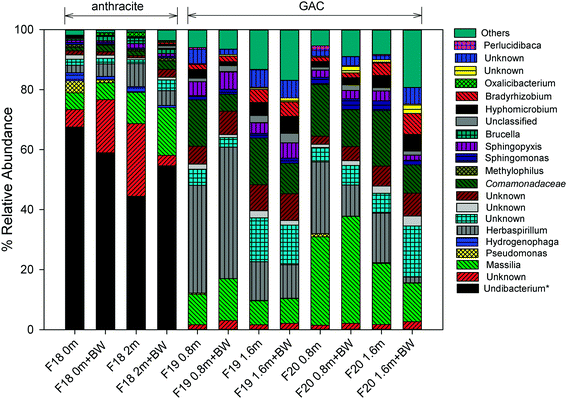
For more than 10 years, the microbial community structure of each filter was shaped by selective pressures caused by processes occurring at the filter media surface. The results of the 16S and 18S rRNA gene sequencing indicated that these processes have resulted in distinct communities for the anthracite and GAC. GAC hosts a richer, more diverse bacterial community than anthracite (Table S2†), which is likely attributable to greater availability of substrate (e.g., adsorbed organic matter) and absence of a free chlorine residual in the GAC filter bed. In all samples collected (i.e., at different filter depths, before and after backwash), the Shannon index, which accounts for both species richness and evenness, was greater in GAC (3.7–5.0) than in anthracite (2.4–2.9). Higher Simpson and Chao1 indices were also observed for the 16S rRNA gene sequences originating from GAC (Table S2†). Using the 18S rRNA gene sequence data, Simpson and Shannon indices were also found greater in GAC than anthracite (Table S3†), although Chao1 values (i.e., estimate of total OTU richness) for both media were relatively similar. Beta diversity metrics such as unweighted/weighted UniFrac distances and Bray Curtis dissimilarities were also evaluated (see Fig. S11 and S12† for the principal coordinate analyses (PCoA)). These diversity metrics indicated that filter media type had a significant impact on beta diversity of the bacterial communities (p < 0.05 when comparing anthracite versus the two GAC filters; Table S4†). For eukaryotes, filter media appeared to be an insignificant factor for beta diversity (Table S5†), although this result could be an artifact due to the small sample size possibly affecting statistical significance (anthracite: n = 1; other anthracite samples had insufficient DNA in the extract, presumably due to the free chlorine residual in the anthracite filter bed). Nevertheless, the distant positioning of anthracite from GAC in the PCoA plots (Fig. S12†) suggests differences in invertebrate community structure between the filters.Among the proteobacteria, betaproteobacteria (almost entirely Burkholderiales) dominated both filters, but alphaproteobacteria were also abundant in the GAC, which is consistent with Lautenschlager et al. (2014).16 The presence of Burkholderiales is not surprising as they have been detected previously in biofilms associated with conventional treatment (i.e., coagulation, flocculation, sand/anthracite filtration), conventional + GAC filtration, and membrane filtration systems.15,54
In the anthracite filter in the current study (i.e., filter 18), the free chlorine residual was expected to cause selective pressures that might limit the observed genera to those that can tolerate disinfection. Consistent with this theory, an unknown taxon that is Undibacterium-like (<51% confidence) was the most abundant at 45–68%. Taxonomic assignments using the SILVA database also confirmed Undibacterium as the dominant genus. This genus commonly occurs in drinking water and membrane biofilms55,56 and has also been reported to have high relative abundance after chlorination.12,57 On the other hand, Undibacterium had low relative abundance (<0.1%) on the GAC media, possibly due to being outcompeted by other genera (e.g., Massilia, Herbaspirillum, unknown Comamonadaceae) in the absence of free chlorine.
Pseudomonas and Hydrogenophaga are known to exhibit resistance to disinfection,12,58 and they were also observed on the anthracite media. Increases in relative abundance of Pseudomonas have been observed in water samples after chlorination,12 and Hydrogenophaga are known to colonize chlorinated distribution pipes.58 Although there are many non-pathogenic Pseudomonas species, the presence of Pseudomonas may be concerning for water utilities due to clinical relevance of some of its species like P. aeruginosa.59Pseudomonas was found at the top of the anthracite bed with a relative abundance of 4% but became less abundant deeper in the filter (0.3% at the bottom) and after backwashing (0%). A similar trend was observed for Hydrogenophaga. These observations suggest possible influence of nutrient availability on microbial abundance across the filter bed.
In GAC, the abundance of certain organisms was also affected by filter depth and backwashing. Massilia, a common soil/root colonizing bacterium,60 had higher abundance at 0.8 m (filter 19: 10.1%, filter 20: 29.5%) than at 1.6 m (filter 19: 8.0%, filter 20: 20.3%). Backwashing also appeared to select for this genus, as demonstrated by its 21–38% increase in relative abundance after backwashing. Unlike Massilia, backwashing reduced the levels of an unknown Comamonadaceae (e.g., filter 19 at a depth 0.8 m: 15.5% before backwash and 5.7% after backwash). Herbaspirillum, another soil bacterium, also showed higher abundance at a depth of 0.8 m (24–36%) than 1.6 m (12.9–16.5%). These bacteria may colonize the upper layers of the filter bed to gain access to readily biodegradable, lower molecular weight carbon, which is expected to be present in the ozonated feed water. In contrast, Bradyrhizobium and Hyphomicrobium (members of alphaproteobacteria) were more abundant deeper in the filter bed (1.4–2.9% at 0.8 m vs. 4.0–4.4% at 1.6 m), and their relative abundance increased further after backwashing. The occurrence of Bradyrhizobium and Hyphomicrobium in the deep zones of the filter bed can be ascribed to the inherent property of alphaproteobacteria to thrive under low nutrient conditions and degrade complex organic materials,16 among others. This agreed well with a previous study in which Bradyrhizobium was shown to dominate a nutrient-limited biofilter, leading to the release of EPS and head loss development.9 The quenching of chlorine by GAC might have also allowed growth of Bradyrhizobium, consistent with Oh et al. (2018) who reported dominance of Bradyrhizobium in a GAC filter receiving ozonated, non-chlorinated waters.14 Other than these organisms, Nitrospira (0.4–0.6%), known for nitrite oxidation, was also detected at a depth of 1.6 m. The presence of bacteria involved in nitrogen cycling (including Bradyrhizobium for nitrogen fixation) indicates that nitrification occurs in GAC, despite prior oxidation of inorganic and organic nitrogen by ozone.61 The occurrence of N-metabolizing bacteria suggests secondary production of ammonia and amino acids by protozoan activity within the GAC.62,63
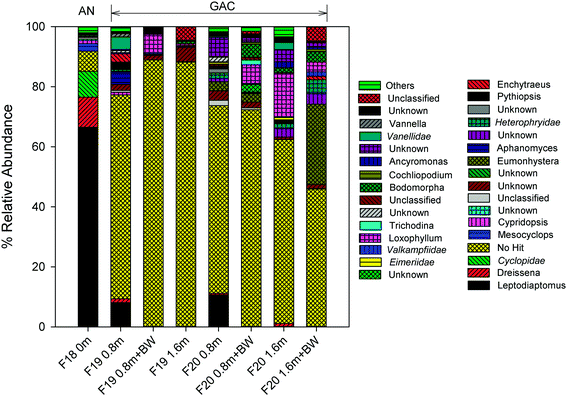
Calanoid copepods (Leptodiaptomus), which are common in fresh water bodies (e.g., the Great Lakes) and sediments,65 dominated the eukaryote community within the upper layer of the anthracite (relative abundance of 66.5%; Table S7†). Their high abundance in the chlorinated anthracite can likely be attributed to their tolerance for oxidants (e.g., chlorinated cooling towers; other studies have reported copepod resistance to UV irradiation as well).66,67 Furthermore, the data in the current study are consistent with the copepod chlorine tolerance hierarchy described by Ershath et al. (2008): calanoids > cyclopoida > harpacticoids > naupliar stages.68 The relative abundance of cyclopoida (e.g., Mesocyclops) was <10% in the chlorinated anthracite filter.
In contrast with anthracite, Leptodiaptomus was observed in the upper layer (i.e., depth = 0.8 m) of the GAC but only at a relative abundance of 8.0–10.7%. Other eukaryotes, including Eumonhystera (up to 26.8%) and Loxophyllum (up to 14.5%), exhibited high relative abundance in the GAC but not the anthracite, possibly due to the absence of a disinfectant residual in the GAC. The nematode Eumonhystera was also detected in a GAC filtrate in a previous study.18 While 18S rRNA gene amplification was successful in many of the GAC samples, a majority of the OTUs remained unknown as many sequences did not match the applied taxonomy classifier or were not yet assigned to any taxa.36 Taxonomic assignment using the SILVA database40 confirmed the presence of predominantly unknown eukaryotic sequences (>33%). Consistent with the amoeba viability test results, members of the family Vannellidae (e.g., Vannella) were observed in the GAC but not the anthracite, further supporting the hypothesis that bioactive GAC offers favorable conditions for amoeba colonization.
3.5. Implications
Upon completion of this study, the GAC biofilters (filters 19–20) and the chlorinated anthracite filters (filters 1–18) were evaluated in the context of treatment performance and microbiological water quality. One of the justifications for employing biofiltration is stabilization of ozonated water, which typically contains significant quantities of assimilable organic carbon. However, there have been no historical concerns about the biostability of finished water from RMWTF or microbial regrowth in the distribution system, thereby negating the need for biofiltration. In addition, only marginal reductions (<10%) in DOC and total trihalomethane (TTHM) formation potential have been achieved by the GAC biofilters at RMWTF (Fig. S13†). The biomass accumulation in the GAC has even reduced filter run times (from 200 to 150 hours) and increased turbidity and particle counts (Fig. 1), presumably due to biomass sloughing. Therefore, considering the negligible effect on biostability, the adverse impacts on effluent water quality, and the potential for pathogen harboring and release, RMWTF decided to suspend operation of the GAC biofilters and initiate media replacement with anthracite and sand. As a result, the full-scale filters at RMWTF are now exclusively chlorinated anthracite filters with a sand base.This study demonstrates the importance of integrating both the engineering and microbial aspects of biofiltration to investigate the overall effect of upstream chlorine addition on filter performance and microbial community structure in the filter media. It is among the first to report the detection of viable FLA occurrence in drinking water biofilters, presumably linked to the lack of residual disinfectant and an abundant food source (i.e., bacteria). The occurrence of pathogenic and non-pathogenic FLA as well as oxidant-resistant microorganisms requires further investigation into the microbial communities of drinking water biofilters and the microbiological water quality of the finished drinking water. Should additional occurrence data highlight the role of drinking water biofilters as a significant source of potential pathogens, then the water quality benefits of biofiltration must be weighed against the associated microbial risks.
4. Conclusions
Full-scale chlorinated GAC and anthracite filters were investigated for their biological activity, effluent quality, and microbial community characteristics. The following conclusions can be drawn from this study:• GAC rapidly quenches residual chlorine, which leads to greater biomass development and biological activity in GAC biofilters than chlorinated anthracite filters. This increase in biological activity can be advantageous for stabilization of ozonated water but can also lead to greater biomass sloughing, turbidity, and particle counts.
• GAC biofilters provide favorable conditions for a variety of prokaryotic and eukaryotic organisms, including free-living amoebas. For example, environmental Acanthamoeba spp. can reach drinking water intakes, accumulate in biofilters, and get released into the filter effluent during sloughing events, as documented by the qPCR assays in this study. Other types of viable free-living amoebas, including Vannella, may also be found in biofilters according to the culture-based assays in the current study.
• Based on 16S and 18S rRNA gene sequencing, chlorinated anthracite filters host a less diverse bacterial community and are dominated by microbes that are possibly chlorine-resistant (e.g., Undibacterium-like taxon). In contrast, the absence of residual disinfectant in GAC filters allows for colonization by Massilia, Herbaspirillum, unknown Comamonadaceae, Bradyrhizobium, and Hyphomicrobium, among others. Filter depth and backwashing dynamics appeared to affect the relative abundance of these taxa. With respect to eukaryotes, Calanoid copepods (e.g., Leptodiaptomus) dominated the chlorinated anthracite filter, while a variety of invertebrates (e.g., nematodes) and unknown taxa were detected in the GAC. Further studies are needed to identify the unknown taxa that dominate the eukaryotic fraction of GAC's microbial community.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors would like to thank the following SNWA staff members: Mei Mei Dong, Julia Lew, Katherine Greenstein, and Mary Murphy for their assistance with filter core sampling and ATP measurements; Kevin Fisher, David Rexing, Stan van Wagenen, and Richard Giltner for helpful discussions regarding future direction; Jeff Waufle and other SNWA staff for their help with data collection and sampling at River Mountains WTF. The authors are also grateful to the anonymous reviewers of this paper for all their constructive comments. Lars Koenig, Sarah Mangum, and Rocio Navarro of RTLGenomics are also acknowledged for their assistance with 16S/18S rRNA gene sequencing.References
- C. Liu, C. I. Olivares, A. J. Pinto, C. V. Lauderdale, J. Brown, M. Selbes and T. Karanfil, The control of disinfection byproducts and their precursors in biologically active filtration processes, Water Res., 2017, 124, 630–653 CrossRef PubMed.
- D. Urfer, P. M. Huck, S. D. J. Booth and B. M. Coffey, Biological filtration for BOM and particle removal: a critical review, J. - Am. Water Works Assoc., 1997, 89, 83–98 CrossRef.
- T. L. Zearley and R. S. Summers, Removal of trace organic micropollutants by drinking water biological filters, Environ. Sci. Technol., 2012, 46, 9412–9419 CrossRef PubMed.
- G. A. de Vera, J. Keller, W. Gernjak, H. Weinberg and M. J. Farre, Biodegradability of DBP precursors after drinking water ozonation, Water Res., 2016, 106, 550–561 CrossRef PubMed.
- US EPA, Long Term 2 Enhanced Surface Water Treatment Rule Toolbox Guidance Manual. Office of Water (4606). EPA 815-R-09-016, 2010.
- P. J. Evans, J. L. Smith, M. W. LeChevallier, O. D. Schneider, L. A. Weinrich and P. K. Jjemba, Biological filtration monitoring and control toolbox: Guidance manual, Water Research Foundation, CO, USA., 2013, ISBN 978-1-60573-194-0 Search PubMed.
- D. van der Kooij and P. W. J. J. van der Wielen, Microbial growth in drinking-water supplies: Problems, causes, control and research needs, IWA Publishing, UK., 2014, ISBN 9781780400402 Search PubMed.
- C. V. Lauderdale, P. Chadik, M. J. Kirisits and J. Brown, Engineered biofiltration: Enhanced biofilter performance through nutrient and peroxide addition, J. - Am. Water Works Assoc., 2012, 104, E298–E309 CrossRef.
- C. V. Lauderdale, Engineered biofiltration for enhanced hydraulic and water treatment performance, PhD dissertation, University of Florida, 2011 Search PubMed.
- X. Li, G. Upadhyaya, W. Yuen, J. Brown, E. Morgenroth and L. Raskin, Changes in the structure and function of microbial communities in drinking water treatment bioreactors upon addition of phosphorus, Appl. Environ. Microbiol., 2010, 76, 7473–7481 CrossRef PubMed.
- D. M. Moll, R. S. Summers and A. Breen, Microbial characterization of biological filters used for drinking water treatment, Appl. Environ. Microbiol., 1998, 64, 2755–2759 Search PubMed.
- S. Jia, P. Shi, Q. Hu, B. Li, T. Zhang and X.-X. Zhang, Bacterial community shift drives antibiotic resistance promotion during drinking water chlorination, Environ. Sci. Technol., 2015, 49, 12271–12279 CrossRef PubMed.
- I. Kasuga, D. Shimazaki and S. Kunikane, Influence of backwashing on the microbial community in a biofilm developed on biological activated carbon used in a drinking water treatment plant, Water Sci. Technol., 2007, 55, 173–180 CrossRef PubMed.
- S. Oh, F. Hammes and W.-T. Liu, Metagenomic characterization of biofilter microbial communities in a full-scale drinking water treatment plant, Water Res., 2018, 128, 278–285 CrossRef PubMed.
- A. J. Pinto, C. Xi and L. Raskin, Bacterial community structure in the drinking water microbiome is governed by filtration processes, Environ. Sci. Technol., 2012, 46, 8851–8859 CrossRef PubMed.
- K. Lautenschlager, C. Hwang, F. Ling, W.-T. Liu, N. Boon, O. Koster, T. Egli and F. Hammes, Abundance and composition of indigenous bacterial communities in a multi-step biofiltration-based drinking water treatment plant, Water Res., 2014, 62, 40–52 CrossRef PubMed.
- Q. Wang, W. You, X. Li, Y. Yang and L. Liu, Seasonal changes in the invertebrate community of granular activated carbon filters and control technologies, Water Res., 2014, 51, 216–227 CrossRef PubMed.
- H. Schreiber, D. Schoenen and W. Traunspurger, Invertebrate colonization of granular activated carbon filters, Water Res., 1997, 31, 743–748 CrossRef.
- A. K. Camper, M. W. LeChevallier, S. C. Broadway and G. A. McFeters, Bacteria associated with granular activated carbon particles in drinking water, Appl. Environ. Microbiol., 1986, 52, 434–438 Search PubMed.
- M. Burr, A. K. Camper, R. DeLeon and P. Hacker, Colonization of biologically active filter media with pathogens, AwwaRF and AWWA, CO, USA., 2000, ISBN 1-58321-036-9 Search PubMed.
- A. J. Martinez, Free-living amebas: Naegleria, Acanthamoeba, and Balamuthia, Medical Microbiology, ed. S. Baron, 4th edn, University of Texas Medical Branch, Galveston (TX), 1996, ch. 81 Search PubMed.
- US EPA, Contaminant Candidate List 4 - CCL4. https://www.epa.gov/ccl/microbial-contaminants-ccl-4 (accessed February 2018).
- J. M. Thomas and N. J. Ashbolt, Do free-living amoebae in treated drinking watersystems present an emerging health risk?, Environ. Sci. Technol., 2011, 45, 860–869 CrossRef PubMed.
- H. Y. Buse and N. J. Ashbolt, Counting Legionella cells within single amoeba host cells, Appl. Environ. Microbiol., 2012, 78, 2070–2072 CrossRef PubMed.
- V. Thomas, G. McDonnell, S. P. Denyer and J.-Y. Maillard, Free-living amoebae and their intracellular pathogenic microorganisms: risk for water quality, FEMS Microbiol. Rev., 2010, 34, 231–259 CrossRef PubMed.
- S. Kilvington and J. Price, Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure, J. Appl. Microbiol., 1990, 68, 519–525 CrossRef PubMed.
- M. Taylor, K. Ross and R. Bentham, Legionella, Protozoa, and Biofilms: Interactions within complex microbial systems, Microb. Ecol., 2009, 58, 538–547 CrossRef PubMed.
- J. De Johnckheere and H. van de Voorde, Differences in destruction of cysts of pathogenic and nonpathogenic Naegleria and Acanthamoeba by chlorine, Appl. Environ. Microbiol., 1976, 31, 294–297 Search PubMed.
- V. Thomas, T. Bouchez, V. Nicolas, S. Robert, J. F. Loret and Y. Levi, Amoeba in domestic water systems: resistance to disinfection treatments and implication in Legionella persistence, J. Appl. Microbiol., 2004, 97, 950–963 CrossRef PubMed.
- S. Cervero-Arago, S. Rodriguez-martinez, A. Puertas-Bennasar and R. M. Araujo, Effect of common drinking water disinfectants, chlorine and heat, on free Legionella and amoeba-associated Legionella, PLoS One, 2015, e0134726, DOI:10.1371/journal.pone.0134726.
- M. Dupuy, F. Berne, P. Herbelin, M. Binet, N. Berthelot, M.-H. Rodier, S. Soreau and Y. Hechard, Sensitivity of free-living amoeba trophozoites and cysts to water disinfectants, Int. J. Hyg. Environ. Health, 2014, 217, 335–339 CrossRef PubMed.
- S. Velten, M. Boller, O. Koster, J. Helbing, H.-U. Weilenmann and F. Hammes, Development of biomass in drinking water granular active carbon (GAC) filter, Water Res., 2011, 45, 6347–6354 CrossRef PubMed.
- A. Magic-Knezev and D. van der Kooij, Optimisation and significance of ATP analysis for measuring active biomass in granular activated carbon filters used in water treatment, Water Res., 2004, 38, 3971–3979 CrossRef PubMed.
- Y. Qvarnstrom, G. S. Visvesvara, R. Sriram and A. J. da Silva, Multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri, J. Clin. Microbiol., 2006, 44, 3589–3595 CrossRef PubMed.
- D. Sullivan, B. Fahey and D. Titus, Fast PCR: General considerations for minimizing run times and maximizing throughput, Bio-Rad Laboratories, Inc. Technical Note, 5362, 2006 Search PubMed.
- RTLGenomics, Data Analysis Methodology, Version 2.3.1, http://www.rtlgenomics.com/docs/Data_Analysis_Methodology.pdf (accessed February 2018).
- R. C. Edgar, B. J. Haas, J. C. Clemente, C. Quince and R. Knight, UCHIME improves sensitivity and speed of chimera detection, Bioinformatics, 2011, 27, 2194–2200 CrossRef PubMed.
- R. C. Edgar, UPARSE: highly accurate OTU sequences from microbial amplicon reads, Nat. Methods, 2013, 10, 996–998 CrossRef PubMed.
- R. C. Edgar, Search and clustering orders of magnitude faster than BLAST, Bioinformatics, 2010, 26, 2460–2461 CrossRef PubMed.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, J. Peplies and F. O. Glockner, The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools, Nucleic Acids Res., 2013, 41, D590–D596 CrossRef PubMed.
- J. G. Caporaso, Quantitative insights into microbial ecology (QIIME) 2, https://qiime2.org. (Accessed April 2018).
- S. Maciver, Vannella. Biomedical Sciences, University of Edinburgh, http://www.bms.ed.ac.uk/research/others/smaciver/vannella.htm (accessed February 2018).
- A. Garcia, P. Goni, J. Cieloszyk, M. T. Fernandez, L. Calvo-Begueria, E. Rubio, M. F. Fillat, M. L. Peleato and A. Clavel, Identification of free-living amoeba and amoeba-associated bacteria from reservoirs and water treatment plants by molecular techniques, Environ. Sci. Technol., 2013, 47, 3132–3140 CrossRef PubMed.
- U. Rohr, S. Weber, R. Michel, F. Selenka and M. Wilhelm, Comparison of free-living amoebae in hot water systems of hospitals with isolates from moist sanitary areas by identifying genera and determining temperature tolerance, Appl. Environ. Microbiol., 1998, 64, 1822–1824 Search PubMed.
- A. Edagawa, A. Kimura, T. Kawabuchi-Kurata, Y. Kusuhara and P. Karanis, Isolation and genotyping of potentially pathogenic Acanthamoeba and Naegleria species from tap-water sources in Osaka, Japan, Parasitol. Res., 2009, 105, 1109–1117 CrossRef PubMed.
- H. Y. Buse, J. Lu, I. T. Struewing and N. J. Ashbolt, Eukaryotic diversity in premise drinking water using 18S rDNA sequencing: implications for health risks, Environ. Sci. Pollut. Res., 2013, 20, 6351–6366 CrossRef PubMed.
- R. Hoffmann, R. Michel, E. N. Schmid and K.-D. Muller, Natural infection with microsporidian organisms (KW19) in Vannella spp. (Gymnamoebia) isolated from a domestic tap-water supply, Parasitol. Res., 1998, 84, 164–166 CrossRef PubMed.
- P. Scheid, Mechanism of intrusion of a microspordian-like organism into the nucleus of host amoebae (Vannella sp.) isolated from a keratitis patient, Parasitol. Res., 2007, 101, 1097–1102 CrossRef PubMed.
- R. J. Newton, S. J. Jones, A. Eiler, K. D. McMahon and S. Bertilsson, A guide to the natural history of freshwater lake bacteria, Microbiol. Mol. Biol. Rev., 2011, 75, 14–49 CrossRef PubMed.
- X. Liao, C. Chen, Z. Wang, R. Wan, C.-H. Chang, X. Zhang and S. Xie, Changes in biomass and bacterial communities in biological activated carbon filters for drinking water treatment, Process Biochem., 2013, 48, 312–316 CrossRef.
- W. Lin, Z. Yu, H. Zhang and I. P. Thompson, Diversity and dynamics of microbial communities at each step of treatment plant for potable water generation, Water Res., 2014, 52, 218–230 CrossRef PubMed.
- D. Gerrity, M. Arnold, E. Dickenson, D. Moser, J. D. Sackett and E. C. Wert, Microbial community characterization of ozone-biofiltration systems in drinking water and potable reuse applications, Water Res., 2018, 135, 207–219 CrossRef PubMed.
- J. Tian, J. Lu, Y. Zhang, J.-C. Li and L.-C. Sun, Z.-L. Hu, Microbial community structures and dynamics in the O3/BAC drinking water treatment process, Int. J. Environ. Res. Public Health, 2014, 11, 6281–6290 CrossRef PubMed.
- J. L. Shaw, P. Monis, R. Fabris, L. Ho, K. Braun, M. Drikas and A. Cooper, Assessing the impact of water treatment on bacterial biofilms in drinking water distribution systems using high-throughput DNA sequencing, Chemosphere, 2014, 117, 185–192 CrossRef PubMed.
- S. Kwon, E. Moon, T.-S. Kim, S. Hong and H.-D. Park, Pyrosequencing demonstrated complex microbial communities in a membrane filtration system for a drinking water treatment plant, Microbes Environ., 2011, 26, 149–155 CrossRef PubMed.
- P. Kampfer, R. Rossello-Mora, M. Hermansson, F. Persson, B. Huber, E. Falsen and H.-J. Busse, Undibacterium pigrum gen. nov., sp. nov., isolated from drinking water, Int. J. Syst. Evol. Microbiol., 2007, 57, 1510–1515 CrossRef PubMed.
- Y. Zhang, Microbial community dynamics and assembly: Drinking water treatment and distribution, PhD diss., University of Tennessee, 2012 Search PubMed.
- M. W. LeChevallier, C. D. Norton, A. Camper, P. Morin, B. Ellis, W. Jones, A. Rompre, M. Prevost, J. Coallier, P. Servais, D. Holt, A. Delanoue and J. Colbourne, Microbial impact of biological filtration, AWWA Res. F., Am. Water Works Assoc., USA, 1998, ISBN 0-89867-939-7 Search PubMed.
- K. D. Mena and C. P. Gerba, Risk assessment of Pseudomonas aeruginosa in Water, Reviews of Environmental Contamination and Toxicology, ed. D. Whitacre, Reviews of Environmental Contamination and Toxicology (Continuation of Residue Reviews), vol 201, Springer, Boston, MA., 2009, vol. 201 Search PubMed.
- M. Ofek, Y. Hadar and D. Minz, Ecology of root colonizing Massilia (Oxalobacteraceae), PLoS One, 2012, 7, e40117 Search PubMed.
- G. A. de Vera, W. Gernjak, H. Weinberg, M. J. Farre, J. Keller and U. von Gunten, Kinetics and mechanisms of nitrate and ammonium formation during ozonation of dissolved organic nitrogen, Water Res., 2017, 108, 451–461 CrossRef PubMed.
- A. Knezev, Microbial activity in granular activated carbon filters in drinking water treatment, PhD dissertation, University of Wageningen, 2015, ISBN 978-94-6257-244-7 Search PubMed.
- E. B. Sherr and B. F. Sherr, Significance of predation by protists in aquatic microbial food webs, Antonie van Leeuwenhoek, 2002, 81, 293–308 CrossRef PubMed.
- J. H. van Lieverloo, D. W. Bosboom, G. L. Bakker, A. J. Brouwer, R. Voogt and J. E. De Roos, Sampling and quantifying invertebrates from drinking water distribution mains, Water Res., 2004, 38, 1101–1112 CrossRef PubMed.
- USGS, Key to the species of Great Lakes calanoids. https://www.glsc.usgs.gov/greatlakescopepods/Introduction.php?GROUP=Calanoid (accessed February 2018).
- Z.-B. Jiang, J.-N. Zeng, Q.-Z. Chen, Y.-B. Liao, X.-Q. Xu, L. Shou, J.-J. Liu, A.-G. Gao, Y.-Q. Zhao and Y.-J. Huang, Toxicity of residual chlorine on copepods under temperature increase in different seasons, Shuisheng Shengwu Xuebao, 2009, 33, 896–904 Search PubMed.
- S. L. Cooke and C. E. Williamson, Positive effects of UV radiation on a calanoid copepod in a transparent lake: do competition, predation or food availability play a role?, J. Plankton Res., 2005, 28, 171–179 CrossRef.
- M. Ershath, K. Altaff, P. Sriyutha Murthy and V. P. Venugopalan, Chlorine toxicity to copepods: implications in the context of zooplankton entrainment in power plants, India: Department of Atomic Energy, 2008, vol. 40, p. 40041575 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ew00115d |
| This journal is © The Royal Society of Chemistry 2018 |