Open Access Article
Open Access ArticleChlorinated cobalt alkyne complexes derived from acetylsalicylic acid as new specific antitumor agents†
Victoria
Obermoser‡
a,
Daniel
Baecker‡
a,
Carina
Schuster
a,
Valentin
Braun
a,
Brigitte
Kircher
bc and
Ronald
Gust
 *a
*a
aDepartment of Pharmaceutical Chemistry, Institute of Pharmacy, Center for Molecular Biosciences Innsbruck, University of Innsbruck, CCB – Centrum for Chemistry and Biomedicine, Innrain 80-82, 6020 Innsbruck, Austria. E-mail: ronald.gust@uibk.ac.at; Tel: +43-512-507-58200
bImmunobiology and Stem Cell Laboratory, Department of Internal Medicine V (Hematology and Oncology), Innsbruck Medical University, Anichstrasse 35, 6020 Innsbruck, Austria
cTyrolean Cancer Research Institute, Innrain 66, 6020 Innsbruck, Austria
First published on 21st February 2018
Abstract
[(Prop-2-ynyl)-2-acetoxybenzoate]dicobalthexacarbonyl (Co-ASS), an organometallic derivative of the irreversible cyclooxygenase-1/2 (COX-1/2) inhibitor acetylsalicylic acid (ASS), demonstrated high growth-inhibitory potential against various tumor cell lines and inhibition of both COX isoenzymes. With the objective of increasing the selectivity for COX-2, we introduced a chlorine substituent in position 3, 4, 5, or 6 of the ASS moiety, respectively. Increased COX-2 selectivity is desirable as this isoenzyme is predominantly related to the development of cancer and abnormal tissue growth. The new compounds were investigated in comprehensive cellular biological assays to identify the impact of the chlorine substitution at the complex on COX-1/2 inhibition, antiproliferative activity, apoptosis, metabolic activity, cell-based COX inhibition, and cellular uptake. Chlorination distinctly reduced the effects at isolated COX-1 (about 25% inhibition at 10 μM; Co-ASS: 82.7%), while those at COX-2 remained almost unchanged (about 65% inhibition at 10 μM; Co-ASS: 78.5%). In cellular systems, with exception of the 6-Cl derivative, all compounds showed notable antitumor activity in COX-1/2 expressing tumor cells (HT-29 (IC50 = 1.5–2.7 μM), MDA-MB-231 (IC50 = 5.2–8.0 μM)), but were distinctly less active in the COX-1/2-negative MCF-7 breast cancer cell line (IC50 = 15.2–22.9 μM). All complexes possess high selectivity for tumor cells, because they did not influence the growth of the non-tumorigenic, human bone marrow stromal cell line HS-5. These findings clearly demonstrate that the interference with the COX-1/2 cascade contributes to the mode of anticancer action of the cobalt alkyne complexes.
Introduction
Cyclooxygenases are homodimeric enzymes catalyzing the conversion of arachidonic acid to the cyclic prostaglandins (PGs) G2 and H2, which are further converted to PGE2, PGD2, PGI2, and thromboxane via their respective synthases. The constitutively active isoform COX-1 is responsible for basal PG synthesis in nearly all human tissues. Besides their functions in platelet activity, reproduction, and regulation of peripheral vascular resistance, COX-1-derived PGs are particularly important in the maintenance of the protecting gastric mucosa.1,2 The second short-living isoform COX-2 is basally expressed only in the central nervous system, the kidney and in female reproduction organs. However, it is inducible upon stimulation by cytokines, endotoxins, and mitogens in the context of pathophysiological processes and contributes via specific PG production to inflammation and carcinogenesis.3–6 COX-2 is upregulated in various types of cancer, such as colon and breast cancer, osteosarcoma, chronic myeloid leukemia, melanoma, ovarian, and gastric cancer.7–10 Its downstream product PGE2 acts as harmful mediator in cancer tissue. PGE2 inhibits apoptosis, increases proliferation, initiates angiogenesis, supports invasiveness and suppresses immune answer via interaction with its specific receptors EP1–4, leading overall to accelerated tumor growth and increased tumor aggressiveness.11–14The cancer chemopreventative and adjuvant chemotherapeutic properties of COX inhibitory nonsteroidal anti-inflammatory drugs (NSAIDs) have therefore been extensively explored in cancers expressing the enzyme. Acetylsalicylic acid (ASS) exhibited good results in the prevention of both breast and colon cancers or as adjuvant in combination therapy.15–18 But to the best of our knowledge it does not cause any cytotoxic effect in vitro or solely administered in vivo.
In our group, we tried to increase the antitumor activity of ASS by coordinating it to an organometallic moiety. For this purpose, ASS was esterified with propargyl alcohol and bound to dicobalthexacarbonyl.19 The resulting [(prop-2-ynyl)-2-acetoxybenzoate]dicobalthexacarbonyl (Co-ASS) showed considerable growth inhibitory potential in colon (HT-29) and breast (MDA-MB-231) cancer cell lines with IC50 values in the low micromolar range. Compared to ASS, the potency to inhibit COX isoenzymes 10-fold increased for both COX-1 and COX-2, suggesting an interference with the arachidonic acid cascade as a probable mode of action.20 However, the high inhibitory effect on COX-1 points to therapy-limiting side effects, such as ulcers, which can be reduced by shifting the COX isoenzyme selectivity towards COX-2.21
The amino acids Ile523, His513, and Ile434 in COX-1 lead to a smaller binding cave compared to COX-2 (Val523, Arg513, and Val434). Especially, Ile523 in COX-1 blocks the access to a small hydrophilic side pocket. The larger substrate channel in COX-2 was already utilized to design COX-2 selective NSAIDs.1,22–24
Therefore, we tried to use these differences in the volume of the binding pocket to reduce the COX-1 inhibitory effects and to maintain the activity at COX-2 by introduction of a bulkier chlorine substituent into the aromatic moiety of Co-ASS. The derivatives 3-Cl-Co-ASS, 4-Cl-Co-ASS, 5-Cl-Co-ASS, and 6-Cl-Co-ASS were investigated for COX inhibitory effects at isolated or human recombinant isoenzymes and in cell culture experiments against HT-29, MDA-MB-231, and MCF-7 cells regarding their influence on proliferation, metabolic activity, and caspase-3/7 induction. Furthermore, we studied their potential to inhibit intracellular PGE2 production in HT-29 cells and the time-dependent cellular uptake was monitored to complete the characterization of their pharmacological profiles. Finally, the selectivity of the compounds towards cancer cells was investigated using the healthy (non-cancerous) human bone marrow stromal cell line HS-5 for comparison.
Results and discussion
Synthesis
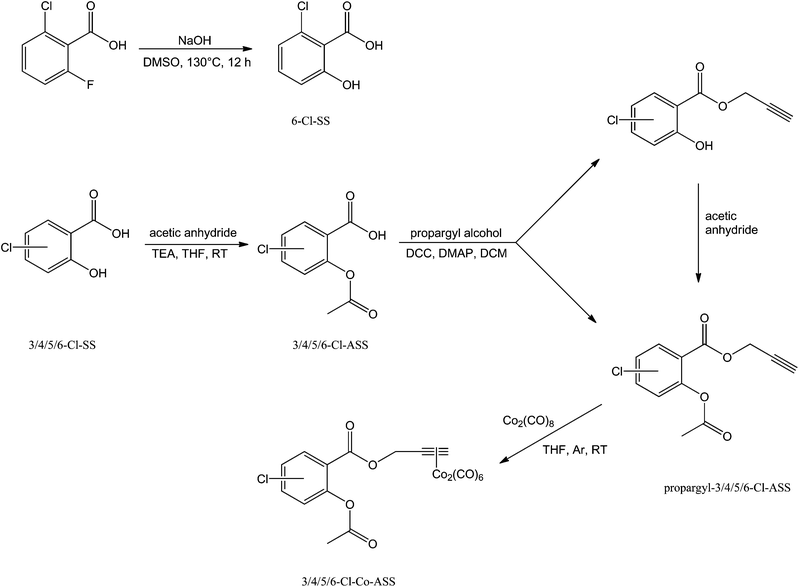
6-Chlorosalicylic acid (6-Cl-SS) was obtained by reacting 2-chloro-6-fluorobenzoic acid with powdered sodium hydroxide in dimethyl sulfoxide (DMSO). Thereby, the fluorine substituent was selectively exchanged by a hydroxyl group (Scheme 1).25 3-Chlorosalicylic acid (3-Cl-SS), 4-chlorosalicylic acid (4-Cl-SS), and 5-chlorosalicylic acid (5-Cl-SS) were commercially available.Acetylation of the chlorine-substituted salicylic acids with acetic anhydride and triethylamine (TEA) in tetrahydrofuran (THF) resulted in the respective chlorine-substituted acetylsalicylic acids (3/4/5/6-Cl-ASS), which were subsequently esterified with propargyl alcohol (leading to propargyl-3/4/5/6-Cl-ASS) according to a method described by Steglich et al. using N,N′-dicyclohexylcarbodiimide (DCC) and the Steglich-catalyst 4-(dimethylamino)pyridine (DMAP).26 Propargylchlorosalicylates, which were built as by-products in this reaction were reacetylated as described above without separation from the respective ASS ester (Scheme 1).
Finally, reaction of the propargyl-3/4/5/6-chloroacetylsalicylates with dicobaltoctacarbonyl in dry THF under an argon-modified atmosphere yielded the respective dicobalthexacarbonyl complexes (Scheme 1).
Biological evaluation
| Compound | COX-1 inhibitiona [%] | COX-2 inhibitiona [%] |
|---|---|---|
a Values are represented as % COX inhibition calculated from initial activity of vehicle treated enzyme. Each experiment was conducted ≥![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 times independently and the values are presented as mean ± SD. 3 times independently and the values are presented as mean ± SD.
|
||
| ASS [10 μM] | 28.8 ± 3.7 | 0.6 ± 2.8 |
| ASS [100 μM] | 73.3 ± 8.7 | 59.4 ± 8.7 |
| Propargyl-ASS [100 μM] | 25.4 ± 4.4 | 34.5 ± 4.2 |
| 3-Cl-ASS [100 μM] | 27.0 ± 6.1 | 29.5 ± 3.5 |
| Propargyl-3-Cl-ASS [100 μM] | 32.5 ± 1.9 | 52.2 ± 5.7 |
| Co-ASS [10 μM] | 82.7 ± 3.2 | 78.5 ± 6.1 |
| 3-Cl-Co-ASS [10 μM] | 24.9 ± 2.6 | 71.4 ± 8.3 |
| 4-Cl-Co-ASS [10 μM] | 22.9 ± 1.7 | 69.9 ± 2.7 |
| 5-Cl-Co-ASS [10 μM] | 25.0 ± 1.0 | 64.0 ± 7.5 |
| 6-Cl-Co-ASS [10 μM] | 24.4 ± 3.0 | 70.5 ± 3.5 |
A chlorine substituent at position 3 or 6 reduced the COX inhibition of ASS to about 30% (100 μM) at both isoenzymes, while substitution at position 4 or 5 maintained the inhibitory effects nearly unchanged (see Table S1 in ESI†). After esterification of the Cl-ASS moiety with propargyl alcohol, the activity was about 30% at COX-1 and about 50% at COX-2, independent of the position of the chlorine substituent at ASS. The COX-2 inhibition induced by the unsubstituted propargyl-ASS was slightly weaker (34.5%). Binding of propargyl-ASS to dicobalthexacarbonyl, resulting in Co-ASS, distinctly increased the COX inhibitory effects. An inhibition of about 80% was determined at both isoenzymes at 10 μM. Interestingly, the activity of the chlorine-substituted propargyl-ASS derivatives at COX-1 cannot be increased upon coordination. At COX-2, however, all chlorinated Co-ASS derivatives showed the same effects as the parent compound Co-ASS. This finding clearly documents that chlorine substituents reduced the unwanted COX-1 inhibition and shifted therefore the selectivity to COX-2.
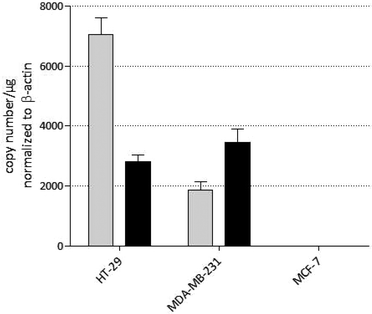
The MDA-MB-231 (breast cancer) and the HT-29 (colon carcinoma) cell lines express both COX-1 and COX-2, though at different levels. The highest COX-1/2 expression was found in HT-29 cells. In MDA-MB-231 cells, total copy numbers per μg are only the half compared to HT-29 cells. This is the consequence of a distinctly reduced COX-1 level. The expression of COX-2 is comparable in both cell lines. Interestingly, the MCF-7 cell line used in this study is negative for both, COX-1 and COX-2, and allows therefore the monitoring of possible COX independent cytotoxic effects.
The precursors (ASS and propargyl-ASS derivatives), the chlorine-substituted complexes, and Co-ASS were screened against the HT-29, MDA-MB-231, and MCF-7 cell lines. Cisplatin was used as positive control. The IC50 values presented in Table 2 were determined after an incubation period of 72 h. Concentration–activity curves are submitted as ESI.† As expected, ASS and propargyl-ASS were inactive up to a concentration of 100 μM. All precursors were excluded from the extended studies on metabolic activity and apoptosis because chlorination of the ASS and the propargyl-ASS moiety did not increase the antiproliferative effects.
| Compound | IC50 [μM] | ||
|---|---|---|---|
| HT-29 | MDA-MB-231 | MCF-7 | |
IC50 values are calculated as the concentration where 50% of the compounds maximum antiproliferative activity compared to the vehicle treated control were achieved and are presented as mean ± SD of ≥![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 independent experiments. 4 independent experiments. |
|||
| ASS | ≥![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
≥![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
≥![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
| Propargyl-ASS | ≥![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
≥![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
≥![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
| 3-Cl-Co-ASS | 2.67 ± 0.47 | 8.04 ± 0.30 | 22.9 ± 1.9 |
| 4-Cl-Co-ASS | 1.51 ± 0.12 | 5.24 ± 0.33 | 15.2 ± 1.3 |
| 5-Cl-Co-ASS | 2.04 ± 0.11 | 7.85 ± 0.93 | 20.7 ± 2.9 |
| 6-Cl-Co-ASS | 21.1 ± 4.0 | 22.4 ± 1.7 | 41.2 ± 3.2 |
| Co-ASS | 1.15 ± 0.10 | 10.1 ± 0.6 | 16.4 ± 1.4 |
| Cisplatin | 3.52 ± 0.48 | 3.57 ± 1.06 | 4.02 ± 1.11 |
Cisplatin showed comparable activity at all cell lines with IC50 values of about 3.5–4.0 μM.
The activity of the dicobaltcarbonyl complexes correlates with the total COX level in the cells: the higher the COX level the higher the sensitivity of cell against the compounds. For instance, the lead compound Co-ASS was highly active against HT-29 cells (IC50 = 1.15 μM) but less active against MDA-MB-231 (IC50 = 10.1 μM) and especially MCF-7 (IC50 = 16.4 μM) cells.
After chlorination of the ASS moiety in position 3, 4 or 5, the effects on the HT-29 cell line remained nearly unchanged (IC50 = 1.5–2.7 μM) and slightly decreased against MDA-MB-231 cells in the order Co-ASS (IC50 = 10.1 μM) < 3-Cl-Co-ASS (IC50 = 8.04 μM) ≈ 5-Cl-Co-ASS (IC50 = 7.85 μM) < 4-Cl-Co-ASS (IC50 = 5.24 μM). In contrast, the 6-chloro derivative was considerably less potent with IC50 ≈![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 μM at both cell lines.
22 μM at both cell lines.
The 3-, 4-, and 5-Cl-substituted complexes and also Co-ASS showed distinctly weaker antiproliferative effects in MCF-7 cells (highest concentration tested: 40 μM) with IC50 values ranging from 15.2 to 22.9 μM. This finding is an indication but not the proof that COX-1 and COX-2 inhibition are involved in the mode of action. However, the 6-Cl-substituted compound (IC50 = 41.2 μM) displayed a far smaller effect in comparison to the other derivatives.
The determined effects against COX-negative MCF-7 cells demonstrate the interference in further pathways in the cells. Therefore, the growth inhibitory potency cannot be discussed in relation to the COX-1/2 inhibition only. Further parameters must be taken into consideration. This is especially true for 6-Cl-Co-ASS, which was the complex with the lowest antiproliferative effect, while being equally potent as the other complexes in the COX-1/2 assay at isolated enzymes.
In order to study the influence of the chlorine substituents on the stability in aqueous solution, the ASS derivatives and the ASS-propargyl esters were dissolved in 30% DMSO/0.1 M phosphate buffer (pH 7.4) to a concentration of 0.5 mg mL−1 and analyzed time-dependently for degradation at 37 °C by HPLC analysis. This mixture was used to guarantee that educts and all degradation products remain in solution. Unfortunately, the solubility of the respective cobalt complexes was not high enough to study them under in vitro conditions.
The position of the chlorine substituent at the aromatic moiety strongly influenced the deacetylation. If the substituent is located at position 4 or 5, the acetylsalicylic ester degraded into the salicylic acid (SS) with t1/2 = 8.5 h and 12.5 h, respectively.
The 3-chloro and 6-chloro derivatives were much more stable (t1/2 (3-Cl-ASS) = 32.5 h; t1/2 (6-Cl-ASS) = 41.7 h).
If the propargyl esters were comparably incubated, ASS, SS, and the SS-propargyl ester were identified as degradation products. The stability increased about 1.5-fold upon esterification to the propargyl ester in the case of the 3-, 4-, and 5-Cl-substituted compounds (t1/2 (propargyl-3-Cl-ASS) = 53.1 h; t1/2 (propargyl-4-Cl-ASS) = 13.5 h; t1/2 (propargyl-5-Cl-ASS) = 19.0 h). A very contrary result was obtained after condensation of 6-Cl-ASS with propargyl alcohol. The half-life decreased from 41.7 h to 7.9 h. After 48 h the 6-ASS propargyl ester was completely hydrolyzed. In contrast to the other derivatives, the hydrolysis of the propargyl ester was preferred and the cleavage of the acetyl group played a subordinate role.
Although 6-Cl-Co-ASS could not be investigated under comparable conditions, it can be assumed that it is hydrolyzed into 6-Cl-ASS and propargyl-Co2(CO)6 to a higher extent prior to uptake into cells compared to all other compounds.
The high-performance liquid chromatography (HPLC) chromatograms are submitted as ESI.†
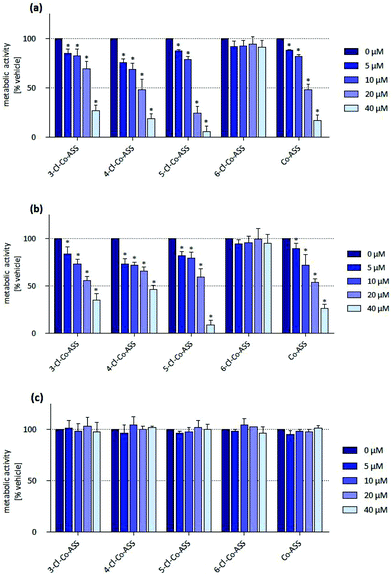
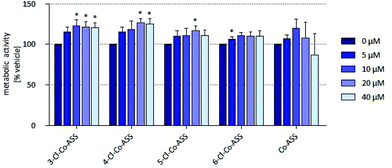
In HT-29 cells, 4-Cl-Co-ASS, and Co-ASS reduced the metabolic activity to approximately 50% at 20 μM. 5-Cl-Co-ASS was more active and reduced the metabolic activity at 20 μM to 24% and at 40 μM to 6%, while the 3-chloro derivative decreased the activity at these concentrations only to 69% (20 μM) and 27% (40 μM), respectively. 6-Cl-Co-ASS was completely inactive (Fig. 2a).
In MDA-MB-231 cells nearly the same tendency was observed: 5-Cl-Co-ASS > Co-ASS ≈ 3-Cl-Co-ASS > 4-Cl-Co-ASS > 6-Cl-Co-ASS (Fig. 2b). As against HT-29 cells, 6-Cl-Co-ASS was inactive. 5-Cl-Co-ASS as most active complex reduced the metabolic activity to 60% (20 μM) and 10% (40 μM) documenting that MDA-MB-231 cells are slightly less sensitive to the chlorine-substituted compounds.
Interestingly, the metabolic activity of the MCF-7 cells was not significantly affected by any cobalt complex (Fig. 2c), not even at the highest concentration (40 μM) used.
In summary, cells expressing COX-1/2 (HT-29, MDA-MB-231) were more sensitive against the cobalt complexes than the cell line lacking COX-1/2 (MCF-7). It should be noted, that the metabolic activity is reduced in all cell lines only at relatively high concentrations, which were far above the IC50 value determined in the proliferation assay. These findings suggest that the cobalt complexes have a cytostatic effect on the cells already at low concentrations without interfering with their mitochondrial metabolic turnover.
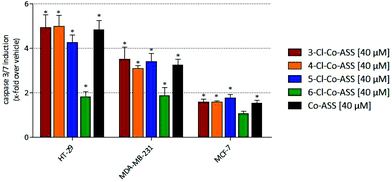
A significant induction of caspase-3/7 was only observed at a concentration of 40 μM, why only these data are presented in Fig. 3. Further data are available as ESI.†
In accordance with the results of the MTT assay, in MCF-7 cells the cobalt complexes only marginally induced caspase-3/7-related apoptosis. In MDA-MB-231 and HT-29 cells, 3-, 4-, and 5-Cl-Co-ASS as well as Co-ASS increased the caspase-3/7 content 3- to 5-fold independent on the position of the chlorine substituent.
In general, the caspase-3/7-inductive effect was more pronounced in HT-29 cells compared to the MDA-MB-231 cell line. These data correlate with the results of the MTT assay.
It is obvious that Co-ASS and its chlorine derivatives show cytostatic activity at concentrations <![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 μM as the cells neither lose their metabolic capacity nor migrate to an apoptotic state.
20 μM as the cells neither lose their metabolic capacity nor migrate to an apoptotic state.
COX-1/2 catalyze the double oxygenation of added arachidonic acid to PGG2 and the subsequent reduction to PGH2 by its peroxidase (POX) active site. PGH2 is then converted to PGE2 by tissue- and cell-specific isomerases. PGE2 is relatively stable upon release of the cell to the medium and is therefore a good analyte to quantify the arachidonic acid turnover by COX-1/2. Results of this assay are presented in Fig. 4.
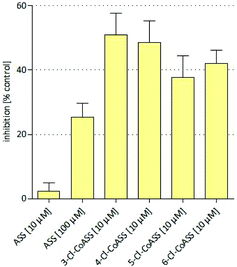 | ||
| Fig. 4 Inhibition of intracellular PGE2 synthesis assessed in HT-29 colon carcinoma cells treated with ASS and cobalt complexes at 10 μM. Values represent the mean ± SD of 3 independent assays. | ||
Treatment of HT-29 cells with ASS reduced the intracellular PGE2 synthesis (25.4%) only at 100 μM. The respective propargyl ester (ASS-propargyl) caused marginal PGE2 inhibition (18%) at 10 μM. Coordination to Co2(CO)6 led to the lead structure Co-ASS, which is the most active compound in this study (57.5% at 10 μM). Chlorine substituents at the aromatic moiety slightly decreased the influence on the PGE2 release if they are in position 3 (37.7%), 4 (48.5%), or 5 (41.9%). 6-Cl-Co-ASS reduced the PGE2 level only by 18%. These findings correlate well with the results obtained in the other tests against this cell line and clearly point to a participation of COX inhibition in the mode of action. It is worthy to mention that PGE2 inhibition distinctly higher than 20% is necessary to influence the cell growth because ASS as well ASS-propargyl were inactive and 6-Cl-Co-ASS was only marginally active at the used concentrations.
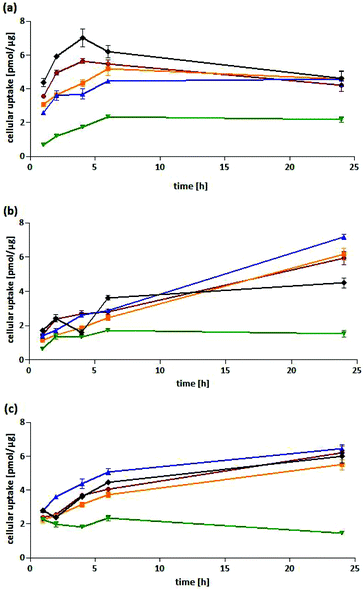
Cellular uptake into HT-29 cells. All complexes showed a fast cellular uptake with a maximum after 4–6 h. While the level remained nearly constant for 5-Cl-Co-ASS and 6-Cl-Co-ASS, it slowly decreased for Co-ASS, 3-Cl-Co-ASS as well as 4-Cl-Co-ASS after 6 h of incubation. After 24 h, with the exception of 6-Cl-Co-ASS all complexes showed nearly the same cobalt content of about 4.5 pmol μg−1 protein. For 6-Cl-Co-ASS only the half amount was determined (2.2 pmol μg−1) and could be the explanation of the lesser PGE2 level measured in the above mentioned PGE2 assay.
Cellular uptake into MDA-MB-231 cells. In MDA-MB-231 cells the complexes were taken up to a lesser extent compared to HT-29 cells during the first 6 h of incubation. In case of Co-ASS and 6-Cl-Co-ASS the maximum was reached and the cobalt content did not change during the following 18 h of incubation. After 24 h, 4.5 pmol μg−1 (Co-ASS) and 1.5 pmol μg−1 (6-Cl-Co-ASS) were determined. Interestingly, the cobalt amount further increased for 3-Cl-Co-ASS, 4-Cl-Co-ASS, and 5-Cl-Co-ASS to 6.0 pmol μg−1, 6.2 pmol μg−1, and 7.2 pmol μg−1, respectively. Saturation could not be observed. After a fast initial uptake within the first 2 h, 3-/4-/5-Cl-Co-ASS displayed nearly linear uptake kinetics expressed by their coefficients of determinations (r2). The values of r2 are 0.9696, 0.9982, and 0.9934, respectively.
Cellular uptake into MCF-7 cells. Co-ASS and 3/4/5-Cl-Co-ASS revealed in MCF-7 cells comparable uptake profiles. They reached after 6 h cobalt levels of 3.7–5.1 pmol μg−1, which further increased until the end of the experiment (24 h). After 24 h, for 3-Cl-Co-ASS an intracellular amount of 6.2 pmol μg−1 and 4-Cl-Co-ASS of 5.5 pmol μg−1 were measured. The highest uptake showed 5-Cl-Co-ASS with 6.4 pmol μg−1 after 24 h, slightly higher than Co-ASS (6.0 pmol μg−1). Again, the lowest level was determined for 6-Cl-Co-ASS. Interestingly, it reached the highest level of 2.0 pmol μg−1 after 2 h that decreased during the following incubation period to 1.5 pmol μg−1.
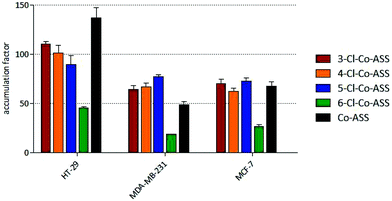
If the protein contents are correlated with the cell volumes, the accumulation grades can be calculated.29,30 Compared to the extracellular concentration of 10 μM, the complexes were strongly accumulated in the tumor cells (Fig. 6). The highest accumulation grades were measured within 24 h for 3-/4-/5-Cl-Co-ASS in HT-29 cells (90- to 110-fold), while MDA-MB-231 and MCF-7 cells accumulated the complexes to a lower extent (64- to 77-fold and 62- to 73-fold, respectively). The values of 6-Cl-Co-ASS are less than 50% of its derivatives (45-fold, 18-fold, and 27-fold, respectively).
Conclusions
In continuation of our previously published study about the unselective COX-1/2 inhibitor Co-ASS, we synthesized its 3/4/5/6-chlorine-substituted derivatives. The aim of this study was to increase the selectivity for the COX-2 isoenzyme. The chlorine substituents at the Co-ASS should utilize on the one hand the larger binding cave of COX-2 to promote activity at this isoenzyme and the other hand they should reduce the binding to COX-1 due to steric repulsion. Indeed, all complexes had high inhibitory potency against isolated COX-2 (60–80% inhibition at 10 μM) and low effects at COX-1 (about 20% at 10 μM). Biological effects (inhibition of proliferation and metabolic activity, induction of apoptosis) were only determined in cells with COX expression. Especially the inactivity against COX-negative MCF-7 cell points to an interference in the arachidonic acid oxidation cascade as part of the mode of action. The missing effects of 6-Cl-Co-ASS in cellular systems can be explained by its low stability in aqueous solution and/or by a distinctly reduced cellular uptake. Co-ASS and its Cl-substituted derivatives demonstrated high selectivity against tumor cells, because they were completely inactive at the healthy, non-tumorigenic human bone marrow stromal cell line HS-5.Experimental section
General material and methods
All educts, solvents, and other chemicals were purchased from Sigma-Aldrich, TCI Chemicals, Fluka, or Alfa Aesar. Solvents were purchased in appropriate purity or distilled prior to utilization. Milli-Q water was obtained from a Milli-Q Gradient A10 water purification system (Merck Millipore, Billerica, USA). Column chromatography was performed using silica gel 60 (particle size 40–63 μm). Thin layer chromatography (TLC) was carried out using Polygram® SIL G/UV254 polyester foils with a 0.2 mm layer of silica gel with fluorescence indicator (Macherey-Nagel, Düren, Germany). Detection was performed by observation under UV-light at 254 and/or 365 nm. Melting points were measured with a Kofler heatable microscope. 1H-Nuclear magnetic resonance spectra (1H-NMR-spectra) were recorded using a Gemini 2000 NMR Spectrometer (Varian, Palo Alto, USA, now part of Agilent Technologies, Santa Clara, USA) at 200 MHz in DMSO-d6 with tetramethylsilane as internal standard. Chemical shifts are given in parts per million (ppm). Coupling constants are given in Hertz (Hz). High resolution mass spectrometry (HR-MS) was performed using an Orbitrap Elite (Thermo Fisher Scientific, Waltham, USA). Infrared (IR) spectra were recorded using an Alpha FT-IR Spektrometer (Bruker, Billerica, USA). HPLC measurements were conducted with an instrument from Shimadzu (Duisburg, Germany) equipped with an autosampler SIL-20A HT, a pump LC20 AD, a column oven CT0-10AS VP and a diode-array detector SPD-M20A with a deuterium and tungsten lamp. Separation was performed over an Eurospher 100-5 C18 (250 × 4 mm) column (Knauer, Berlin, Germany). Purity of all final products was above 95%. Absorbance and luminescence in biological assays were measured with an Enspire multimodal plate reader (PerkinElmer Life Sciences, Waltham, USA).Chemistry
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) as eluent. The coloured fractions were collected. The first fraction contained the alkinyl cobalt complex of the salicylic acid derivative and the second one contained the chloroacetylsalicylic acid ester cobalt complex.
1) as eluent. The coloured fractions were collected. The first fraction contained the alkinyl cobalt complex of the salicylic acid derivative and the second one contained the chloroacetylsalicylic acid ester cobalt complex.
[2-Acetoxy-3-chloropropargylbenzoate]dicobalthexacarbonyl (3-Cl-Co-ASS). Red crystals; yield: 0.052 g (0.096 mmol, 48.8%); mp >
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 °C. 1H-NMR (200 MHz, CDCl3): δ 7.98 (d, 1H, ArH-6), 7.64 (d, 1H, ArH-4), 7.22 (dd, 1H, ArH-5), 6.12 (s, 1H, CH), 5.48 (s, 1H, CH2), 2.41 (s, 3H, CH3); IR (ν cm−1): 3091 w (Aryl-C–H), 2917 w, 2852 w (C–H), 2094 m, 2012 s, 2001 s (C
150 °C. 1H-NMR (200 MHz, CDCl3): δ 7.98 (d, 1H, ArH-6), 7.64 (d, 1H, ArH-4), 7.22 (dd, 1H, ArH-5), 6.12 (s, 1H, CH), 5.48 (s, 1H, CH2), 2.41 (s, 3H, CH3); IR (ν cm−1): 3091 w (Aryl-C–H), 2917 w, 2852 w (C–H), 2094 m, 2012 s, 2001 s (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C), 1775 s, 1719 s (C
C), 1775 s, 1719 s (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1595 m, 1572 w (C
O), 1595 m, 1572 w (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1364 m (δ –CH3), 1251 s, 1183 s, 1105 s (C–O); HR-MS: m/z calculated for C18H9ClCo2O10 [2M + Na]+: 1098.6892, found: 1098.6979; HPLC [MeOH/water (70
C), 1364 m (δ –CH3), 1251 s, 1183 s, 1105 s (C–O); HR-MS: m/z calculated for C18H9ClCo2O10 [2M + Na]+: 1098.6892, found: 1098.6979; HPLC [MeOH/water (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30)]: purity 99.7%.
30)]: purity 99.7%.
[2-Acetoxy-4-chloropropargylbenzoate]dicobalthexacarbonyl (4-Cl-Co-ASS). Red crystals; yield: 0.050 g (0.093 mmol, 50%); mp >
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 °C. 1H-NMR (200 MHz, CDCl3): δ 8.04 (s, 1H, ArH-6), 7.28 (s, 2H, ArH-3, ArH-5), 6.12 (s, 1H, CH), 5.47 (s, 1H, CH2), 2.36 (s, 3H, CH3); IR (ν cm−1): 3092 w (Aryl-C–H), 2925 w, 2852 w (C–H), 2097 m, 2053 s, 2006 s (C
150 °C. 1H-NMR (200 MHz, CDCl3): δ 8.04 (s, 1H, ArH-6), 7.28 (s, 2H, ArH-3, ArH-5), 6.12 (s, 1H, CH), 5.47 (s, 1H, CH2), 2.36 (s, 3H, CH3); IR (ν cm−1): 3092 w (Aryl-C–H), 2925 w, 2852 w (C–H), 2097 m, 2053 s, 2006 s (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C), 1775 s, 1725 s (C
C), 1775 s, 1725 s (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1597 s, 1568 (C
O), 1597 s, 1568 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1369 m (δ –CH3), 1184 s, 1092 s (C–O); HR-MS: m/z calculated for C18H9ClCo2O10 [2M + Na]+: 1098.6892, found: 1098.6977; HPLC [MeOH/water (70
C), 1369 m (δ –CH3), 1184 s, 1092 s (C–O); HR-MS: m/z calculated for C18H9ClCo2O10 [2M + Na]+: 1098.6892, found: 1098.6977; HPLC [MeOH/water (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30)]: purity 95.6%.
30)]: purity 95.6%.
[2-Acetoxy-5-chloropropargylbenzoate]dicobalthexacarbonyl (5-Cl-Co-ASS). Red crystals; yield: 0.065 g (0.121 mmol, 61%); mp >
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 °C. 1H-NMR (200 MHz, CDCl3): δ 8.08 (s, 1H, ArH-6), 7.54 (d, 1H, 3J = 7.8 Hz, ArH-4), 7.08 (d, 3J = 8.0 Hz, 1H, ArH-3), 6.12 (s, 1H, CH), 5.47 (s, 1H, CH2), 2.35 (s, 3H, CH3); IR (ν cm−1): 3128 w (Aryl-C–H), 2962 w, 2928 w, 2853 w (C–H), 2094 m, 2026 s, 2003 s (C
150 °C. 1H-NMR (200 MHz, CDCl3): δ 8.08 (s, 1H, ArH-6), 7.54 (d, 1H, 3J = 7.8 Hz, ArH-4), 7.08 (d, 3J = 8.0 Hz, 1H, ArH-3), 6.12 (s, 1H, CH), 5.47 (s, 1H, CH2), 2.35 (s, 3H, CH3); IR (ν cm−1): 3128 w (Aryl-C–H), 2962 w, 2928 w, 2853 w (C–H), 2094 m, 2026 s, 2003 s (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C), 1762 s, 1730 s (C
C), 1762 s, 1730 s (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1603 w, 1579 w (C
O), 1603 w, 1579 w (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1369 m (δ –CH3), 1188 s, 1069 s (C–O); HR-MS: m/z calculated for C18H9ClCo2O10 [2M + Na]+: 1098.6892, found: 1098.6980; HPLC [MeOH/water (70
C), 1369 m (δ –CH3), 1188 s, 1069 s (C–O); HR-MS: m/z calculated for C18H9ClCo2O10 [2M + Na]+: 1098.6892, found: 1098.6980; HPLC [MeOH/water (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30)]: purity 99.6%.
30)]: purity 99.6%.
[2-Acetoxy-6-chloropropargylbenzoate]dicobalthexacarbonyl (6-Cl-Co-ASS). Red crystals; yield: 0.053 g (0.098 mmol, 50%); mp >
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 °C. 1H-NMR (200 MHz, CDCl3): δ 7.39 (dd, 3J = 8.4 Hz, 1H, ArH-4), 7.33 (d, 1H, ArH-5), 7.12 (d, 3J = 8.0 Hz, 1H, ArH-3), 6.11 (s, 1H, CH), 5.23 (s, 1H, CH2), 2.28 (s, 3H, CH3); IR (ν cm−1): 3094 w (Aryl-C–H), 2959 w, 2923 w, 2851 w (C–H), 2098 m, 2054 s, 2004 s (C
150 °C. 1H-NMR (200 MHz, CDCl3): δ 7.39 (dd, 3J = 8.4 Hz, 1H, ArH-4), 7.33 (d, 1H, ArH-5), 7.12 (d, 3J = 8.0 Hz, 1H, ArH-3), 6.11 (s, 1H, CH), 5.23 (s, 1H, CH2), 2.28 (s, 3H, CH3); IR (ν cm−1): 3094 w (Aryl-C–H), 2959 w, 2923 w, 2851 w (C–H), 2098 m, 2054 s, 2004 s (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C), 1774 s, 1736 s (C
C), 1774 s, 1736 s (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1596 m (C
O), 1596 m (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1369 m (δ –CH3), 1259 s, 1183 m, 1100 s (C–O); HR-MS: m/z calculated for C18H9ClCo2O10 [2M + Na]+: 1098.6892, found: 1098.6980; HPLC [MeOH/water (70
C), 1369 m (δ –CH3), 1259 s, 1183 m, 1100 s (C–O); HR-MS: m/z calculated for C18H9ClCo2O10 [2M + Na]+: 1098.6892, found: 1098.6980; HPLC [MeOH/water (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30)]: purity 100%.
30)]: purity 100%.
Stability studies
The 0.1 M phosphate buffer was prepared by diluting 2.26 mL of a NaH2PO4·H2O solution (13.8 g NaH2PO4·H2O in 100 mL of water) and 15.48 mL of a Na2HPO4 solution (8.9 g Na2HPO4 in 100 mL of water) with water to a final volume of 100 mL. 5 mg of the test substance were weighed in a 10 mL volumetric flask, dissolved in 3 mL of DMSO and filled up with 0.1 M phosphate buffer (pH 7.4) to the mark. A solution of 4.01 mg 5-chlorosalicylic acid dissolved in 3 mL of DMSO was filled up to 10 mL with phosphate buffer and was used as reference solution.The mobile phase was a two-component solvent (acetonitrile and acetic acid 1%), which was run in a gradient. The gradient specifications can be found in Table S3 of the ESI.†
Biology
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 independent experiments were calculated and the effects of the compounds were expressed as percentage of the cell mass of a vehicle-treated control, which was set at 100%. The IC50 values were calculated with Prism 7.0 (GraphPad, San Diego, CA) using nonlinear regression and the decadal logarithm of the inhibitor versus variable slope response equation. The top constraint was set to 100%.
4 independent experiments were calculated and the effects of the compounds were expressed as percentage of the cell mass of a vehicle-treated control, which was set at 100%. The IC50 values were calculated with Prism 7.0 (GraphPad, San Diego, CA) using nonlinear regression and the decadal logarithm of the inhibitor versus variable slope response equation. The top constraint was set to 100%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 independent experiments. Untreated control was set to 0% inhibition.
3 independent experiments. Untreated control was set to 0% inhibition.
Cellular uptake studies. Exponentially growing HT-29, MCF-7, and MDA-MB-231 cells, respectively, were seeded in 6-well tissue culture plates in 2 mL of completed DMEM and incubated at 37 °C in a 5% CO2/95% air atmosphere. The medium was aspirated after 24 h and completed DMEM containing the respective drug (10 μM) or the vehicle DMSO was added. After the appropriate incubation periods (1 h, 2 h, 4 h, 6 h, 24 h), the medium was removed and the cells were detached by washing once with PBS followed addition of accutase (GE Healthcare BioSciences, Pasching, Austria) and harvested by collecting them with PBS. The cell suspension was centrifuged (8000 rcf, 3 min, RT) and the supernatant was discarded. The isolated cell pellet was washed with PBS and stored at −20 °C until further analysis. Immediately after thawing, the cell pellet was resuspended in twice distilled water and lysed by sonification (20 s, nine cycles, 80–85% power). The cell lysate was appropriately diluted with Triton-X-100 (final concentration: 0.1%) and stabilized with HNO3 13% (final concentration: 10%) to be analyzed for cobalt content by HR CS AAS. The cellular concentration of the compounds is expressed as the amount of compound (pmol) related to cellular protein mass (μg). Values were calculated as the mean of 3 independent experiments.
HR CS AAS. For the cobalt quantification, a contrAA 700 high-resolution continuum-source atomic absorption spectrometer (Analytik Jena AG) was employed. Pure samples of the respective complexes were utilized as standards and calibration was conducted in a matrix matched manner. The mean integrated absorbances of triplicate injections were used throughout the studies. Samples were directly injected into coated standard graphite tubes (Analytik Jena AG). The time-temperature furnace program applied for the determination of cobalt was optimized regarding pyrolysis and atomization temperatures. It can be found as ESI (Table S2†). Cobalt was quantified at a wavelength of 240.7254 nm.
Quantification of protein content. The protein content of cell lysates was quantified by the method of Bradford.31 In brief, the lysates were appropriately diluted with twice distilled water. To each 20 μL thereof 200 μL of Bradford reagent were added respectively in 96-well plates. After incubation for 5 min at RT, the absorbances were read at 595 nm in a microplate reader (Thermo Scientific Multiskan GO). Solutions of albumin bovine fraction V diluted with twice distilled water (31.25–1500 μg mL−1) were used as calibration standards. Samples were measured in duplicate.
Abbreviations
| ASS | Acetylsalicylic acid |
| COX | Cyclooxygenase |
| DMEM | Dulbecco's modified eagle medium |
| DMSO | Dimethyl sulfoxide |
| EIA | Enzyme immunoassay |
| FCS | Fetal calf serum |
| HPLC | High-performance liquid chromatography |
| HR CS AAS | High-resolution continuum-source atomic absorption spectrometry |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NSAID | Nonsteroidal anti-inflammatory drug |
| PG | Prostaglandin |
| qPCR | Quantitative real time polymerase chain reaction |
| SD | Standard deviation |
| SE | Standard error |
| THF | Tetrahydrofurane. |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Philipp Schuster and Sina Katharina Götzfried (University of Innsbruck, Department of Pharmaceutical Chemistry) for resynthesizing the compounds and for performing the HPLC analysis, Peter Enoh (University of Innsbruck, Department of Pharmaceutical Chemistry) for carrying out the high-resolution mass spectrometry and Caroline Gallati (University of Innsbruck, Department of Pharmaceutical Chemistry) for measuring the 1H-NMR spectra.References
- D. L. Simmons, R. M. Botting and T. Hla, Pharmacol. Rev., 2004, 56, 387–437 CrossRef CAS PubMed.
- A. L. Blobaum and L. J. Marnett, J. Med. Chem., 2007, 50, 1425–1441 CrossRef CAS PubMed.
- K. Kawaguchi, R. W. Hickey, M. E. Rose, L. Zhu, J. Chen and S. H. Graham, Brain Res., 2005, 1050, 130–137 CrossRef CAS PubMed.
- T. Yang, Acta Physiol. Scand., 2003, 177, 417–421 CrossRef CAS PubMed.
- M. Murakami and I. Kudo, Prog. Lipid Res., 2004, 43, 3–35 CrossRef CAS PubMed.
- M. N. A. Khan and Y. S. Lee, Med. Res. Rev., 2011, 31, 161–201 CrossRef CAS PubMed.
- M. Regulski, K. Regulska, W. Prukała, H. Piotrowska, B. Stanisz and M. Murias, Drug Discovery Today, 2016, 21, 598–615 CrossRef CAS PubMed.
- F. J. Giles, H. M. Kantarjian, B. N. Bekele, J. E. Cortes, S. Faderl, D. A. Thomas, T. Manshouri, A. Rogers, M. J. Keating, M. Talpaz, S. O'Brien and M. Albitar, Br. J. Haematol., 2002, 119, 38–45 CrossRef CAS PubMed.
- S. Gupta, M. Srivastava, N. Ahmad, D. G. Bostwick and H. Mukhtar, Prostate, 2000, 42, 73–78 CrossRef CAS PubMed.
- K. Uefuji, T. Ichikura and H. Mochizuki, Clin. Cancer Res., 2000, 6, 135–138 CAS.
- F. L. Queiroga, I. Pires, M. Parente, H. Gregório and C. S. Lopes, Vet. J., 2011, 189, 77–82 CrossRef CAS PubMed.
- H. Sheng, J. Shao, J. D. Morrow, R. D. Beauchamp and R. N. DuBois, Cancer Res., 1998, 58, 362–366 CAS.
- D. F. Legler, M. Bruckner, E. Uetz-von Allmen and P. Krause, Int. J. Biochem. Cell Biol., 2010, 42, 198–201 CrossRef CAS PubMed.
- M. Nakanishi and D. W. Rosenberg, Semin. Immunopathol., 2013, 35, 123–137 CrossRef CAS PubMed.
- T. M. Brasky, M. R. Bonner, K. B. Moysich, H. M. Ochs-Balcom, C. Marian, C. B. Ambrosone, J. Nie, M. H. Tao, S. B. Edge, M. Trevisan, P. G. Shields and J. L. Freudenheim, Breast Cancer Res. Treat., 2011, 126, 157–165 CrossRef CAS PubMed.
- F. M. Robertson, M. Bondy, W. Yang, H. Yamauchi, S. Wiggins, S. Kamrudin, S. Krishnamurthy, H. Le-Petross, L. Bidaut, A. N. Player, S. H. Barsky, W. A. Woodward, T. Buchholz, A. Lucci, N. Uenoand and M. Cristofanilli, CA-Cancer J. Clin., 2010, 60, 351–375 CrossRef PubMed.
- K. Ng, J. A. Meyerhardt, A. T. Chan, K. Sato, J. A. Chan, D. Niedzwiecki, L. B. Saltz, R. J. Mayer, A. B. Benson, P. L. Schaefer, R. Whittom, A. Hantel, R. M. Goldberg, A. P. Venook, S. Ogino, E. L. Giovannucci and C. S. Fuchs, J. Natl. Cancer Inst., 2015, 107, dju345 Search PubMed.
- P. Li, H. Wu, H. Zhang, Y. Shi, J. Xu, Y. Ye, D. Xia, J. Yang, J. Cai and Y. Wu, Gut, 2015, 64, 1419–1425 CrossRef CAS PubMed.
- K. Schmidt, M. Jung, R. Keilitz, B. Schnurr and R. Gust, Inorg. Chim. Acta, 2000, 306, 6–16 CrossRef CAS.
- I. Ott, B. Kircher, C. P. Bagowski, D. H. W. Vlecken, E. B. Ott, J. Will, K. Bensdorf, W. S. Sheldrick and R. Gust, Angew. Chem., Int. Ed., 2009, 48, 1160–1163 CrossRef CAS PubMed.
- B. Hinz and K. Brune, J. Pharmacol. Exp. Ther., 2002, 300, 367–375 CrossRef CAS PubMed.
- C. A. Rouzer and L. J. Marnett, J. Lipid Res., 2009, 50, S29–S34 CrossRef PubMed.
- N. Chandrasekharan and D. L. Simmons, Genome Biol., 2004, 5, 241 CrossRef CAS PubMed.
- L. J. Marnett and A. S. Kalgutkar, Trends Pharmacol. Sci., 1999, 20, 465–469 CrossRef CAS PubMed.
- K. Umezu, F. Tabuchi and Y. Kimura, J. Fluorine Chem., 2003, 121, 97–99 CrossRef CAS.
- B. Neises and W. Steglich, Angew. Chem., 1978, 90, 556–557 CrossRef CAS.
- I. Ott, K. Schmidt, B. Kircher, P. Schumacher, T. Wiglenda and R. Gust, J. Med. Chem., 2005, 48, 622–629 CrossRef CAS PubMed.
- V. Obermoser, M. E. Urban, M. S. Murgueitio, G. Wolber, U. Kintscher and R. Gust, Eur. J. Med. Chem., 2016, 124, 138–152 CrossRef CAS PubMed.
- R. Gust, B. Schnurr, R. Krauser, G. Bernhardt, M. Koch, B. Schmid, E. Hummel and H. Schönenberger, J. Cancer Res. Clin. Oncol., 1998, 124, 585–597 CrossRef CAS PubMed.
- I. Ott, H. Scheffler and R. Gust, ChemMedChem, 2007, 2, 702–707 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
Footnotes |
† Electronic supplementary information (ESI) available: Synthesis and characterization of intermediate products; graphic presentation of crystal violet proliferation assay (HT-29, MDA-MB-231, MCF-7); graphic presentation of caspase-3/7 induction at concentrations ≤![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 μM (HT-29, MDA-MB-231, MCF-7); tabular data of COX-1/2 inhibition; HPLC chromatograms of stability tests including specifications of solvent gradient; HPLC chromatograms of purity tests; 1H-NMR spectra, mass spectra and HPLC chromatograms of final products; graphite furnace time–temperature program. See DOI: 10.1039/c7dt04790h 40 μM (HT-29, MDA-MB-231, MCF-7); tabular data of COX-1/2 inhibition; HPLC chromatograms of stability tests including specifications of solvent gradient; HPLC chromatograms of purity tests; 1H-NMR spectra, mass spectra and HPLC chromatograms of final products; graphite furnace time–temperature program. See DOI: 10.1039/c7dt04790h |
| ‡ These authors contributed equally to the publication. |
| This journal is © The Royal Society of Chemistry 2018 |