Minimalistic peptide supramolecular co-assembly: expanding the conformational space for nanotechnology
Abstract
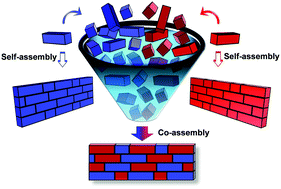
Molecular self-assembly is a ubiquitous process in nature and central to bottom-up nanotechnology. In particular, the organization of peptide building blocks into ordered supramolecular structures has gained much interest due to the unique properties of the products, including biocompatibility, chemical and structural diversity, robustness and ease of large-scale synthesis. In addition, peptides, as short as dipeptides, contain all the molecular information needed to spontaneously form well-ordered structures at both the nano- and the micro-scale. Therefore, peptide supramolecular assembly has been effectively utilized to produce novel materials with tailored properties for various applications in the fields of material science, engineering, medicine, and biology. To further expand the conformational space of peptide assemblies in terms of structural and functional complexity, multicomponent (two or more) peptide supramolecular co-assembly has recently evolved as a promising extended approach, similar to the structural diversity of natural sequence-defined biopolymers (proteins) as well as of synthetic covalent co-polymers. The use of this methodology was recently demonstrated in various applications, such as nanostructure physical dimension control, the creation of non-canonical complex topologies, mechanical strength modulation, the design of light harvesting soft materials, fabrication of electrically conducting devices, induced fluorescence, enzymatic catalysis and tissue engineering. In light of these significant advancements in the field of peptide supramolecular co-assembly in the last few years, in this tutorial review, we provide an updated overview and future prospects of this emerging subject.
- This article is part of the themed collection: Peptide and protein nanotechnology


 Please wait while we load your content...
Please wait while we load your content...