Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
2-Oxoglutarate regulates binding of hydroxylated hypoxia-inducible factor to prolyl hydroxylase domain 2†
Martine I.
Abboud
 a,
Tom E.
McAllister
a,
Tom E.
McAllister
 a,
Ivanhoe K. H.
Leung
a,
Ivanhoe K. H.
Leung
 ab,
Rasheduzzaman
Chowdhury
ab,
Rasheduzzaman
Chowdhury
 a,
Christian
Jorgensen
a,
Christian
Jorgensen
 c,
Carmen
Domene
ad,
Jasmin
Mecinović
c,
Carmen
Domene
ad,
Jasmin
Mecinović
 ae,
Kerstin
Lippl
a,
Rebecca L.
Hancock
a,
Richard J.
Hopkinson
ae,
Kerstin
Lippl
a,
Rebecca L.
Hancock
a,
Richard J.
Hopkinson
 af,
Akane
Kawamura
af,
Akane
Kawamura
 a,
Timothy D. W.
Claridge
a,
Timothy D. W.
Claridge
 *a and
Christopher J.
Schofield
*a and
Christopher J.
Schofield
 *a
*a
aChemistry Research Laboratory, University of Oxford, 12 Mansfield Road, Oxford, OX1 3TA, UK. E-mail: tim.claridge@chem.ox.ac.uk; christopher.schofield@chem.ox.ac.uk
bSchool of Chemical Sciences, The University of Auckland, New Zealand
cDepartment of Chemistry, Britannia House, Kings College London, UK
dDepartment of Chemistry, University of Bath, Claverton Down, Bath, BA2 7AY, UK
eInstitute for Molecules and Materials, Radboud University, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
fLeicester Institute of Structural and Chemical Biology and Department of Chemistry, University of Leicester, Leicester, LE1 7RH, UK
First published on 9th March 2018
Abstract
Prolyl hydroxylation of hypoxia inducible factor (HIF)-α, as catalysed by the Fe(II)/2-oxoglutarate (2OG)-dependent prolyl hydroxylase domain (PHD) enzymes, has a hypoxia sensing role in animals. We report that binding of prolyl-hydroxylated HIF-α to PHD2 is ∼50 fold hindered by prior 2OG binding; thus, when 2OG is limiting, HIF-α degradation might be inhibited by PHD binding.
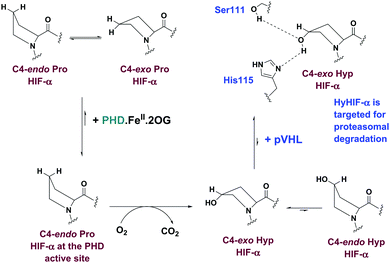
The chronic response to limiting O2 in animals involves upregulation of multiple genes as enabled by the α,β-heterodimeric hypoxia inducible factors (HIFs). Under normoxia, HIF-α subunits are efficiently degraded by proteasomes; in hypoxia, HIF-α subunits accumulate, so enabling the α,β-HIF complex to promote expression of HIF target genes, including genes for biomedicinally important proteins, such as the vascular endothelial growth factor and erythropoietin.1 There is thus a considerable interest in the therapeutic manipulation of the HIF system (Fig. S1, ESI†).2,3trans-4-Prolyl hydroxylation of HIF-α substantially (∼103 fold) increases the strength of its binding to the von Hippel–Lindau protein (pVHL), the targeting component of a ubiquitin E3 ligase.4–7 HIF-α prolyl hydroxylation occurs at P402 and P564 (HIF-1α) in the N- and C-terminal oxygen dependent degradation domains (NODD/CODD, respectively), and is catalysed by human Fe(II)- and 2-oxoglutarate (2OG)-dependent oxygenases (PHD1-3),8,9 the most important of which is likely PHD2 (Fig. S2, ESI†).10 Evidence from studies with proteins, cells, and animals supports the proposal that PHD activity is limited by O2 availability. PHD catalysis involves binding of 2OG, HIF-α, then O2, to the active site, with CO2 and succinate being produced as coproducts (Fig. S3, ESI†).11–13 Evidence has been presented that the PHDs act as hypoxia sensors, including by the manifestation of an unusually slow reaction with O2.11,12 PHD catalysis has the potential to be regulated by Fe(II) and 2OG availability, as well as by succinate and other TCA cycle intermediates.14,15 C4 trans prolyl hydroxylation increases the fraction of the C4-exo prolyl conformation, as observed when HIF-α is complexed with pVHL,4–7,16 likely in part due to the ‘gauche’ stereoelectronic effect.17 The interactions of HIF-α with pVHL and the PHDs are of interest from a chemical perspective because of the profound effect of hydroxylation on biological function. Here, we report evidence that the presence of the 2OG cosubstrate at the PHD2 active site can regulate binding of unhydroxylated versus prolyl hydroxylated HIF-α (Fig. 1).
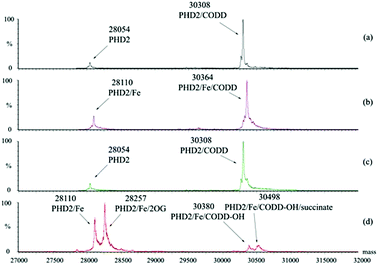
We initially investigated the binding of 19-mer HIF-1α CODD and hydroxylated CODD (hyCODD) peptides to recombinantly expressed PHD2181–426 (tPHD2) using non-denaturing mass spectrometry (ESI-MS).18,19 The results indicated that CODD binds strongly not only to tPHD2·Fe, but also to apo-tPHD2 (Fig. 2). Addition of 2OG to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of apo-tPHD2·CODD did not lead to observation of an apo-tPHD2·2OG complex, consistent with 2OG binding to the Fe(II) in catalysis.20 Analysis of an equimolar mixture of apo-tPHD2, Fe(II), 2OG, and CODD manifested masses corresponding to complexes of tPHD2·Fe·CODD-OH and tPHD2·Fe·CODD-OH·succinate, but not any observable quaternary complex with 2OG (Fig. 2). These results indicate that CODD and hyCODD bind to apo-tPHD2/tPHD2·Fe (Fig. 2).
1 mixture of apo-tPHD2·CODD did not lead to observation of an apo-tPHD2·2OG complex, consistent with 2OG binding to the Fe(II) in catalysis.20 Analysis of an equimolar mixture of apo-tPHD2, Fe(II), 2OG, and CODD manifested masses corresponding to complexes of tPHD2·Fe·CODD-OH and tPHD2·Fe·CODD-OH·succinate, but not any observable quaternary complex with 2OG (Fig. 2). These results indicate that CODD and hyCODD bind to apo-tPHD2/tPHD2·Fe (Fig. 2).
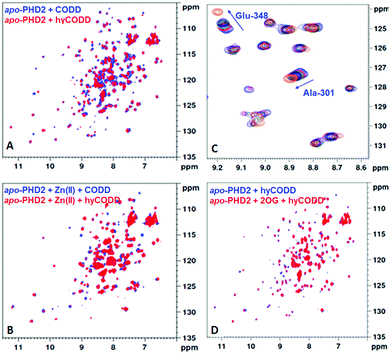
Having shown that the interaction between tPHD2 and CODD is preserved in the gas phase, we then carried out solution studies. NMR studies21,22 using 15N-labelled PHD2181–402 showed that CODD and hyCODD bind with approximately equal affinity to apo-PHD2181–402 and PHD2181–402·Zn (i.e. both saturated apo-PHD2181–402 and PHD2181–402·Zn at 2.7-fold excess) (Fig. 3A and B). The spectrum of the PHD2181–402·Zn·2OG·CODD complex was observed as reported.22 By contrast, hyCODD only showed weak binding to PHD2181–402·Zn·2OG (Fig. 3C). The 1H–15N HSQC spectra of apo-PHD2181–402·hyCODD and apo-PHD2181–402·2OG·hyCODD are similar in the presence of excess hyCODD (Fig. 3D). The overall results imply hyCODD binding is blocked by the presence of 2OG; as anticipated, binding of CODD to PHD2·Zn is not hindered by 2OG.22,23
To investigate the relative affinities of hyCODD and CODD to tPHD2, a fluorescence polarisation assay with N-terminal fluorescein-tagged CODD (CODD*) was developed (Fig. S4, ESI†). Consistent with the NMR and MS data, CODD and hyCODD bind to apo- and metallated-tPHD2 equally strongly (within a 2-fold difference). With tPHD2·Zn·2OG, hyCODD was observed to bind ∼50-fold less strongly than CODD (Fig. S5, ESI†). Isothermal calorimetry results showed that both CODD and hyCODD bind with similar affinity to apo-tPHD2 (9.4 ± 4.5 and 4.6 ± 1.4 μM, respectively); by contrast, CODD, but not hyCODD, was observed to bind to tPHD2·Zn·2OG (KD = 1.8 ± 0.4 μM) (Fig. S6, ESI†). Addition of hyCODD displaced 2OG from tPHD2·Zn·2OG as observed by NMR, implying hyCODD and 2OG compete for binding to the metal complexed tPHD2 (Fig. S7, ESI†). CODD does not displace 2OG, consistent with the FP observations (Fig. S5, ESI†) and the binding requirements of the catalytic cycle (Fig S2, ESI†).
Mutation of genes encoding for TCA cycle enzymes are observed in tumours with consequent effects on metabolite levels, resulting in proposed inhibition of human 2OG oxygenases, including the PHDs, so promoting tumorigenesis.14,15 The effects of TCA cycle intermediates (Fig. S8, ESI†) on CODD/hyCODD binding to tPHD2·Zn were investigated by NMR; of the tested compounds, only fumarate and succinate were observed to bind to tPHD2·Zn, consistent with previous reports.15 By contrast with 2OG, the binding of fumarate or succinate was only weakly disrupted by hyCODD (Fig. S9, ESI†), implying the ‘additional’ carbonyl of 2OG has a role in hindering hyCODD, but not CODD, binding to the tPHD2·metal complex. The observation of simultaneous binding of succinate and hyCODD to tPHD2 is consistent with the MS results (Fig. 2d).
Analysis of crystallographic data of tPHD2·HIF-α complexes indicates the metal chelated 2OG C1 carboxylate may hinder hyCODD, but not CODD, binding (Fig. 4). By contrast, modelling of succinate binding (using tPHD2 structures and those of succinate complexed with other 2OG oxygenases)15,22 indicates that it will not manifest a steric clash with hyCODD (Fig. S10, ESI†). Crystallographic analysis implies that in the tPHD2·Mn·2OG·ODD complexes, the substrate prolyl ring adopts the C4-endo conformation which changes to the C4-exo on hydroxylation.4,5,17 This proposal is supported by the reported observation that cis, but not trans, 4-fluoro prolyl CODD is a tPHD2 substrate17 and by the 19F NMR studies which show a lack of efficient binding of trans-4-fluoro prolyl CODD to tPHD2·Fe·2OG (Fig. S11, ESI†). Modelling studies imply the C4-exo, rather than the C4-endo, conformation of trans-4-prolyl hydroxylated CODD will lead to a clash between the hyCODD alcohol and the 2OG C1 carboxylate (Fig. S12–S18, ESI†).
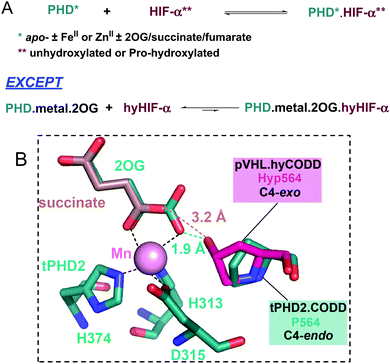 | ||
| Fig. 4 Summary of PHD-HIF/hyHIF-α binding results, highlighting the proposed role of 2OG in blocking hyHIF-α binding. (A) HIF and hydroxylated HIF (hyHIF) bind equally strongly to apo-tPHD2 and metallated tPHD2; 2OG differentiates between the binding of HIF and hyHIF. (B) View from a structure of tPHD2·Mn·2OG·CODD complex (PDB ID: 5L9B).22 The view was overlaid with the hyCODD conformation (fuchsia) as bound to pVHL (PDB ID: 1LQB)5 showing the potential steric clash between the 2OG C1 carboxylate and the C4-exo hydroxyl group in hyCODD (1.9 Å). No steric clash is predicted between the carboxylate of succinate and the C4-exo hydroxyl group in hyCODD (3.2 Å). The succinate (light brown) binding mode was based on 2OG binding as in PDB ID: 5L9B.22 | ||
Whether hyCODD competes with 2OG and/or NODD was investigated using 1D CLIP HSQC (with selective 13C-inversion) NMR using 13C-2OG and 13C-NODD.24,25 The addition of unlabelled CODD to tPHD2·Zn·2OG·NODD leads to NODD, but not 2OG, displacement, suggesting that the affinity of CODD to tPHD2 is higher than NODD, consistent with previous work.12,22 Addition of hyCODD manifests displacement of 2OG and NODD (Fig. S19, ESI†), implying competition with both. Inhibition of the tPHD2-catalysed 2OG turnover to succinate was observed by 1H NMR in the presence of hyCODD with CODD as a substrate and, to a greater extent, with NODD as a substrate (Fig. S20, ESI†).
The results reveal 2OG binding hinders the binding of hyCODD, but not CODD/NODD, to the tPHD2·metal·2OG complex. Thus, competition with 2OG can promote release of prolyl hydroxylated HIF-1α from tPHD2 (and by implication other PHD/HIF-α isoform combinations), so enabling HIF-α degradation.4,5 Once the trans-4-hydroxyproline is formed, the ‘gauche’ stereoelectronic effect biases the conformation towards the C4-exo form, as observed in the hyCODD·pVHL complex.4,5,17 Formation of the C4-exo conformation at the tPHD active site may thus promote a clash between the hyCODD alcohol and the 2OG C1 carboxylate, which is involved in Fe(II) chelation (Fig. S10, ESI†).
Hydroxylated prolyl HIF (hyHIF)-α is upregulated in many tumour cells.26 Under cellular circumstances when there is accumulation of hyHIF-α (e.g. due to mutations to the gene encoding for pVHL as occurs in the VHL disease),27,28 or if iron (e.g. in anaemia) or 2OG availability is limiting (e.g. potentially due to mutations in the genes encoding for isocitrate dehydrogenase),29,30 cellular formation of the PHD·Fe·hyHIF-α or PHD·hyHIF-α complexes may become substantial, with consequent potential limitation of the HIF-mediated hypoxic response. The results thus suggest a negative feedback mechanism for PHD activity, which is linked to TCA cycle metabolism.
We thank the Biochemical Society (Krebs Memorial Award), the Wellcome Trust (106244/Z/14/Z), Cancer Research UK (C8717/A18245), the British Heart Foundation Centre of Research Excellence, Oxford (RE/13/130181), the John Fell Fund (143/075), a Junior Research Fellowship from Kellogg College, Oxford, and the Biotechnology and Biological Sciences Research Council for funding our work. We thank EPSRC and PRACE for providing computational resources through access to Archer and other European HPC facilities.
Conflicts of interest
The authors declare no conflicts of interest.Notes and references
- W. G. Kaelin, Annu. Rev. Biochem., 2005, 74, 115–128 CrossRef CAS PubMed.
- M. C. Chan, J. P. Holt-Martyn, C. J. Schofield and P. J. Ratcliffe, Mol. Aspects Med., 2016, 47–48, 54–75 CrossRef CAS PubMed.
- S. E. Wilkins, M. I. Abboud, R. L. Hancock and C. J. Schofield, ChemMedChem, 2016, 11, 773–786 CrossRef CAS PubMed.
- W. C. Hon, M. I. Wilson, K. Harlos, T. D. W. Claridge, C. J. Schofield, C. W. Pugh, P. H. Maxwell, P. J. Ratcliffe, D. I. Stuart and E. Y. Jones, Nature, 2002, 417, 975–978 CrossRef CAS PubMed.
- P. Jaakkola, D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. von Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh and P. J. Ratcliffe, Science, 2001, 292, 468–472 CrossRef CAS PubMed.
- C. J. Illingworth, C. Loenarz, C. J. Schofield and C. Domene, Biochemistry, 2010, 49, 6936–6944 CrossRef CAS PubMed.
- C. E. Stebbins, W. G. Kaelin, Jr. and N. P. Pavletich, Science, 1999, 284, 455–461 CrossRef CAS PubMed.
- C. J. Schofield and P. J. Ratcliffe, Nat. Rev. Mol. Cell Biol., 2004, 5, 343–354 CrossRef CAS PubMed.
- C. J. Schofield and P. J. Ratcliffe, Biochem. Biophys. Res. Commun., 2005, 338, 617–626 CrossRef CAS PubMed.
- R. J. Appelhoff, Y. M. Tian, R. R. Raval, H. Turley, A. L. Harris, C. W. Pugh, P. J. Ratcliffe and J. M. Gleadle, J. Biol. Chem., 2004, 279, 38458–38465 CrossRef CAS PubMed.
- H. Tarhonskaya, R. Chowdhury, I. K. Leung, N. D. Loik, J. S. McCullagh, T. D. Claridge, C. J. Schofield and E. Flashman, Biochem. J., 2014, 463, 363–372 CrossRef CAS PubMed.
- E. Flashman, E. A. L. Bagg, R. Chowdhury, J. Mecinovic, C. Loenarz, M. A. McDonough, K. S. Hewitson and C. J. Schofield, J. Biol. Chem., 2008, 283, 3808–3815 CrossRef CAS PubMed.
- L. A. McNeill, E. Flashman, M. R. Buck, K. S. Hewitson, I. J. Clifton, G. Jeschke, T. D. Claridge, D. Ehrismann, N. J. Oldham and C. J. Schofield, Mol. BioSyst., 2005, 1, 321–324 RSC.
- M. A. Selak, S. M. Armour, E. D. MacKenzie, H. Boulahbel, D. G. Watson, K. D. Mansfield, Y. Pan, M. C. Simon, C. B. Thompson and E. Gottlieb, Cancer Cell, 2005, 7, 77–85 CrossRef CAS PubMed.
- K. S. Hewitson, B. M. Lienard, M. A. McDonough, I. J. Clifton, D. Butler, A. S. Soares, N. J. Oldham, L. A. McNeill and C. J. Schofield, J. Biol. Chem., 2007, 282, 3293–3301 CrossRef CAS PubMed.
- C. Domene, C. Jorgensen, K. Vanommeslaeghe, C. J. Schofield and A. MacKerell, Jr., J. Chem. Theory Comput., 2015, 11, 3946–3954 CrossRef CAS PubMed.
- C. Loenarz, M. L. Coleman, A. Boleininger, B. Schierwater, P. W. Holland, P. J. Ratcliffe and C. J. Schofield, EMBO Rep., 2011, 12, 63–70 CrossRef CAS PubMed.
- J. Mecinovic, R. Chowdhury, B. M. Lienard, E. Flashman, M. R. Buck, N. J. Oldham and C. J. Schofield, ChemMedChem, 2008, 3, 569–572 CrossRef CAS PubMed.
- J. Mecinovic, R. Chowdhury, E. Flashman and C. J. Schofield, Anal. Biochem., 2009, 393, 215–221 CrossRef CAS PubMed.
- M. A. McDonough, V. Li, E. Flashman, R. Chowdhury, C. Mohr, B. M. Lienard, J. Zondlo, N. J. Oldham, I. J. Clifton, J. Lewis, L. A. McNeill, R. J. Kurzeja, K. S. Hewitson, E. Yang, S. Jordan, R. S. Syed and C. J. Schofield, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 9814–9819 CrossRef CAS PubMed.
- N. M. Mbenza, P. G. Vadakkedath, D. J. McGillivray and I. K. H. Leung, J. Inorg. Biochem., 2017, 177, 384–394 CrossRef CAS PubMed.
- R. Chowdhury, I. K. Leung, Y. M. Tian, M. I. Abboud, W. Ge, C. Domene, F. X. Cantrelle, I. Landrieu, A. P. Hardy, C. W. Pugh, P. J. Ratcliffe, T. D. Claridge and C. J. Schofield, Nat. Commun., 2016, 7, 12673 CrossRef CAS PubMed.
- R. Chowdhury, M. A. McDonough, J. Mecinovic, C. Loenarz, E. Flashman, K. S. Hewitson, C. Domene and C. J. Schofield, Structure, 2009, 17, 981–989 CrossRef CAS PubMed.
- T.-L. Yeh, T. Leissing, M. I. Abboud, C. C. Thinnes, O. Atasoylu, J. P. Holt-Martyn, D. Zhang, A. Tumber, K. Lippl, C. T. Lohans, I. K. H. Leung, H. Morcrette, I. J. Clifton, T. D. W. Claridge, A. Kawamura, E. Flashman, X. Lu, P. J. Ratcliffe, R. Chowdhury, C. W. Pugh and C. J. Schofield, Chem. Sci., 2017, 8, 7651–7668 RSC.
- M. C. Chan, O. Atasoylu, E. Hodson, A. Tumber, I. K. Leung, R. Chowdhury, V. Gomez-Perez, M. Demetriades, A. M. Rydzik, J. Holt-Martyn, Y. M. Tian, T. Bishop, T. D. Claridge, A. Kawamura, C. W. Pugh, P. J. Ratcliffe and C. J. Schofield, PLoS One, 2015, 10, e0132004 Search PubMed.
- C. E. Snell, H. Turley, A. McIntyre, D. Li, M. Masiero, C. J. Schofield, K. C. Gatter, A. L. Harris and F. Pezzella, PLoS One, 2014, 9, e88955 Search PubMed.
- H. Cario, K. Schwarz, N. Jorch, U. Kyank, P. E. Petrides, D. T. Schneider, R. Uhle, K. M. Debatin and E. Kohne, Haematologica, 2005, 90, 19–24 CAS.
- V. R. Gordeuk, A. I. Sergueeva, G. Y. Miasnikova, D. Okhotin, Y. Voloshin, P. L. Choyke, J. A. Butman, K. Jedlickova, J. T. Prchal and L. A. Polyakova, Blood, 2004, 103, 3924–3932 CrossRef CAS PubMed.
- O. C. Andronesi, G. S. Kim, E. Gerstner, T. Batchelor, A. A. Tzika, V. R. Fantin, M. G. Vander Heiden and A. G. Sorensen, Sci. Transl. Med., 2012, 4, 116ra114 Search PubMed.
- L. Dang, D. W. White, S. Gross, B. D. Bennett, M. A. Bittinger, E. M. Driggers, V. R. Fantin, H. G. Jang, S. Jin, M. C. Keenan, K. M. Marks, R. M. Prins, P. S. Ward, K. E. Yen, L. M. Liau, J. D. Rabinowitz, L. C. Cantley, C. B. Thompson, M. G. Vander Heiden and S. M. Su, Nature, 2010, 465, 966 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of experimental procedures and additional experiments. See DOI: 10.1039/c8cc00387d |
| This journal is © The Royal Society of Chemistry 2018 |