Open Access Article
Open Access ArticleMassive quantum regions for simulations on bio-nanomaterials: synthetic ferritin nanocages†
Juan
Torras
 * and
Carlos
Alemán
* and
Carlos
Alemán

Department of Chemical Engineering (EEBE) and Barcelona Research Center for Multiscale Science and Engineering, Universitat Politècnica de Catalunya, C/Eduard Maristany 10-14, Ed I2, 08019, Barcelona, Spain. E-mail: Joan.torras@upc.edu
First published on 1st February 2018
Abstract
QM/MM molecular dynamics simulations on the 4His-ΔC* protein cage have been performed using multiple active zones (up to 86 quantum regions). The regulation and nanocage stability exerted by the divalent transition metal ions in the monomer-to-cage conversion have been understood by comparing high level quantum trajectories obtained using Cu2+ and Ni2+ coordination ions.
Biomacromolecules with highly ordered architectures, which play an important role in living organisms, have inspired the creation of new biomaterials with diverse functionalities. Among them, synthetic protein cages (PCs) have received increasing attention since the multi-functionality that each cage can hold is compatible with a wide range of applications.1 PCs can transport important drugs in the interior voids, while the exterior surface can protect the interior load or can be modified with functional ligands for targeting goals. These distinctive features have made modified PCs potentially interesting vehicles for drug delivery.2
Ferritin PCs are particularly attractive because their subunits self-assemble into spherical nanoparticles with outer and inner diameters of 12 and 8 nm, respectively.3 Ferritin nanocages, which remain intact under non-native conditions (e.g. high pH and temperature), are broken down into individual subunits under acidic conditions, recomposing at neutral pH.3 Due to this phenomenon, ferritin has been proposed as a carrier to transport encapsulated drugs through the human body.4
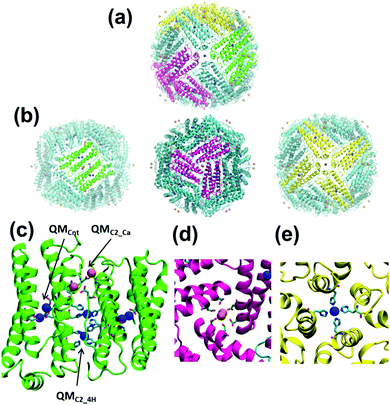
Modified ferritin PCs are highly cooperative symmetrical structures made from a small number of protein building blocks.5 Huard et al.6 recently engineered a PC, named 4His-ΔC* (Fig. 1a), by self-assembling a recombinant ferritin variant. Although 4His-ΔC* is indistinguishable from the wild-type protein in terms of assembly, stability and function, its monomer-to-cage conversion is selectively regulated by Cu2+ at pH 7 (i.e. other divalent ions, such as Zn2+ and Ni2+, produce a much lower yield of 24-mer formation). Accordingly, 4His-ΔC* opens the possibility of hiding therapeutic agents inside modified cages under physiological conditions for targeting delivery.7,8 However, the roles played by metal–ligands in the assembly and disassembly of cages, which are relevant for future developments, remain unclear. Explanatory simulations of this complex system require accurate description of the metal ligand associated with each subunit and consideration of the synergies among the 24-subunits. This can be successfully achieved using a multiscale strategy based on the maz-QM/MM-MD scheme, whose QM region is defined as the sum of several disjoint QM subregions (or active zones, AZs) but following the general QM/MM approach to obtain the energy and its gradients (forces). Thus, any particle within an AZ is subject to QM forces from the electronic wave function. However, the interactions with the other AZs are treated through electrostatic interactions (forces from point charges), similar to the rest of the MM region (see the ESI†).9,10
In this work we have used the maz-QM/MM-MD method to examine the stability of both the C2 dimer interface and the complete cage of 4His-ΔC*. The former consists of two four-helix bundle monomers linked by four metallic coordination sites involving residues of both monomers, while the latter is made of 24 four-helix bundle monomers arranged in octahedral (432) symmetry. The influence of the coordination ion on the stability of the dimer interface has been analysed considering Cu2+ and Ni2+ coordination ions. Each system, hereafter denoted as Cu- and Ni-dimers, involved six coordination environments (quantum regions) with a total of 188 quantum atoms (Fig. 1b and Fig. S2, ESI†): two coordination sites involving Cu2+ or Ni2+ transition metal ions (QMC2_4H), two sites coordinating Ca2+ ions (QMC2_Ca), and two coordinated metal regions in the center of each monomer (QMcnt). Besides, the study on the stability of the whole 4His-ΔC* sphere was conducted considering 86 quantum regions (i.e., all coordination spheres of either Cu2+ or Ni2+, and Ca2+ ions in the PC) with a total amount of 2830 quantum atoms. The resulting model (Fig. 1c), denoted as the Cu-cage, comprised 12 C2 dimer interfaces, 8 C3 pore interfaces, and 6 C4 pore interfaces. The average time used for each MD step was about 270 s using 256 processors for the dimer and 1900 s using 928 processors for the whole protein. The present work is the most ambitious and mature application of the PUPIL11 software so far. To the best of our knowledge, this is the first attempt to face a simulation with such a massive number of quantum regions within the QM/MM framework and, therefore, results have provided relevant information on both the 4His-ΔC* PC and the viability of the maz-QM/MM-MD approach when applied to huge biological systems.
Analysis of the maz-QM/MM-MD trajectory of the Cu-dimer in explicit water indicates that the distances between the metal ions and the atoms from the coordination groups are in good agreement with X-ray crystallographic values for the Cu–ferritin cage, though the former are more homogeneous than the latter (Table S1, ESI†). However, metal⋯metal distances between closer AZs experience a significant increase with respect to experimental data. For example, Cu2+⋯Cu2+ from QMC2_4H regions and Ca2+⋯Ca2+ from QMC2_Ca are overestimated by 0.26 Å (3%) and 1.06 Å (13%) with respect to experimental data. The largest difference corresponds to the Cu2+⋯Cu2+ distances of atoms located at the center of each monomer in QMCnt regions, which are 0.80 and 0.94 Å (23.5% and 27.6%, respectively) larger than the equivalent X-ray values. These differences are due to the highly solvated environment of the simulated system when compared with experimental data derived from hydrated protein crystals. However, we want to note the shorter metal–ligand distances of QMCnt AZ obtained from maz-QM/MM-MD calculations considering the solvated environment that accentuates the separation between the two metals.
On the other hand, metal–ligand distances from simulations on the Ni-dimer are on average larger than those obtained for the Cu-dimer in a range of about 0.5–2% depending of the AZ (Table S1, ESI†). Differences are particularly acute in one of the two QMC2_Ca regions in which Ca2+ experiences a change in the coordination pattern (i.e. from three protein ligands in the Cu-dimer to two protein ligands in the Ni-dimer). Besides, metal–metal distances in the QMC2_4H and QMC2_Ca regions are larger for the Ni-dimer than for the Cu-dimer by 5.3% and 8.9%, respectively, whereas the AZs of QMCnt shrink by 1% and 3.5%. These geometric differences reflect the different metal–ligand behavior between Cu2+ and Ni2+ and support the hypothesis that Ni2+ introduces changes in the stability of the C2 interface in 4His-ΔC*.
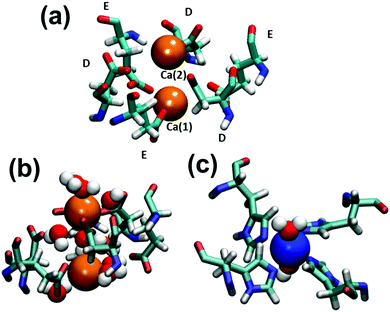
Table 1 describes the coordination of water molecules to metal ions in Cu- and Ni-dimers, which were derived from the radial distribution functions (RDFs) calculated for metal–O(water) atom pairs. Interestingly, no significant difference is detected between Cu2+ and Ni2+ hydration patterns. The square planar organization of the QMC2_4H AZ presents one coordinated water molecule that remains stable during the simulation at a distance of ∼1.9 Å. This Cu2+–water coordination site was also observed in the crystal structure, even though the Cu2+–O(water) distance was of 2.96 Å. The QMCnt AZ shows a fractional coordination number since one of the three existing water molecules is shared between the two metal ions. The three water molecules located in the latter AZ were already found in the crystallographic structure with metal–O(water) distances of 1.88, 1.99 and 2.26 Å. Finally, although Ca2+ ions in QMC2_Ca AZ are not explicitly coordinated, they interact with five water molecules that form the first solvation shell. The average metal–O(water) distance is 2.50 Å, while in the crystal structure the closer water is at 3.96 Å.
| Interface | Cu-dimer | Ni-dimer | |||
|---|---|---|---|---|---|
| d max | CN | d max | CN | ||
| QMC2_4H | 1.85 | 1.0 | 1.87 | 1.0 | |
| QMC2_Ca | 2.50 | 4.8 | 2.50 | 4.5 | |
| QMCnt | M1 | 1.86 | 1.3 | 1.84 | 1.5 |
| M2 | 1.84 | 2.5 | 1.86 | 2.5 | |
Hydrogen bonds involving the Tyr39 and Asn74 residues of Cu-4His-ΔC* were reported to be away from the C2 interface, even though such residues are located near the inner cage surface.6 This hydrogen bonding network, which is involved in the stability of the cage structure and the C2 interface, is sensitive to the divalent metal. In fact, monomer-to-cage conversion was observed for Cu-4His-ΔC* in the presence of divalent ions (e.g. Cu2+, Zn2+ and Ni2+), whereas the ferritin variant MIC1 (4His-ΔC* with Y39E, N74E and P88A mutations) exhibited a drastic dependence on the metal ion for the formation of cages (i.e. the cage was not formed in the presence of Ni2+ while the highest extent of cage formation was observed with Cu2+). In the MIC1, hydrogen bonds involving Tyr39 and Asn74 were suppressed from the C2 interface while Glu39, Glu74 and Asp42 added some additional electrostatic interactions.6
Simulations on Cu- and Ni-dimers of the 4His-ΔC* protein have clarified the influence of the metal ion on the interface, which can be difficult to model using a simple classical MD approach due to the complex metal–ligand bond coordination. In addition, maz-M/MM-MD trajectories take into account the influence of environmental effects on the stability of the C2 interface.
Table S2 (ESI†) sorts by occupation the main hydrogen bonds in both Cu- and Ni-dimers for values over 10% (i.e. percentage of the stored snapshots in which the hydrogen bond is formed). Although the Ni2+ instability on the ferritin variant MIC1 seems to occur due to the Y39E and N74E mutations, analysis of hydrogen bonds reflects differences between the metal ions before mutation from 4His-ΔC* to MIC1. Thus, the importance of the Tyr39⋯Asn74 hydrogen bond in the C2 interface stability is much higher for the Ni-dimer (83% and 77% for Y39⋯N74′ and Y39′⋯N74, respectively) than the Cu-dimer (38% and 25% for Y39⋯N74′ and Y39′⋯N74, respectively). This is consistent with the fact that, after mutation, the new Glu39 and Glu74 residues induce major repulsive charge interactions within the C2 interface of the Ni-dimer compared to that of the Cu-dimer. Besides, Asn74⋯Asp42, Tyr39⋯His67 and Tyr39⋯Lys71 hydrogen bonds contribute to the instability of the cage in the presence of Ni.2
The whole Cu-cage metalloprotein was modelled to determine the main interactions on the rest of the ferritin interfaces: C4 and C3 (Fig. 1). The former is responsible for the union of four monomers by means of a Cu2+ ion, and the latter for the union of three monomers by means of two Ca2+ ions. This simulation, which represented a very challenging task due to the huge amount of AZs involved in the system, should also be considered as a proof of principle of the maz-QM/MM MD methodology for large and complex biological systems. Thus, the 86 AZs include the Cu2+ and Ca2+ ions contained in all the C2, C3 and C4 interfaces of the cage.
Simulation of the Cu-cage involved a huge computational effort since 63% of AZs (i.e. the 54 zones with coordinated Cu2+ ions) have a difficult convergence. Furthermore, the communications between different parts of the program were under stress due not only to the vast size of the simulated system but also due to the differences between AZs (i.e. the computational time varies with the number of atoms and metallic ions). As the algorithm responsible for sharing computational resources for minimizing the execution time plays an important role, the software11,12 was re-engineered to parallelize the construction and simulation mechanism of each AZ independently using the distributed client–server architecture. A client (QMWorker) was created to build, simulate, and manage the information of each AZ, which is exchanged and updated at each iteration cycle with the main program. QMWorkers were distributed equitably between the different calculation nodes of the system in order to avoid memory problems and accelerate the process. On the other hand, the best performance was achieved when the number of calculation nodes is equal to the number of AZs (QMWorker) for an equivalent load distribution among them.
Table 2 lists the metal–ligand distances and the water coordination number on the C3 and C4 pore interfaces, derived from the RDFs calculated for the Cu-cage trajectory. Although the square planar organization of the C4 pore interface is very similar to the QMC2_4H AZs of the C2 interface, two coordinated water molecules were observed in the former while the latter presents a single water molecule only due to steric effects. Moreover, the coordination of Cu2+ at the C4 interface is similar to that observed by crystallography (Fig. 2c). However, Ca2+ ions at the C3 interface present some difference in water coordination: one is coordinated by 3 Glu residues and 4 water molecules meanwhile the other is surrounded by 3 Asp residues and 5 water molecules (Fig. 2a). The ligands are arranged in an alternated order with maximum probability distances of 2.37–2.38 Å to the closest carboxylic oxygens of Glu and Asp residues. Indeed, the C3 interface exhibits three additional water molecules compared with X-ray crystallography (Fig. 2b). These are located among Ca2+ ions acting as a bridge between the metal ions and the more distant carboxylic oxygens of the ligands, which increases the stability of the C3 pore interface.
| Interface | M–X | d max | CN | |
|---|---|---|---|---|
| QMC4_4H | Cu2+–H@N | 2.18 | 4.0 | |
| Cu2+–WAT@O | 1.83 | 2.0 | ||
| QMC3_Ca | M1 | Ca2+–E@O1 | 2.38 | 3.0 |
| Ca2+–WAT@O | 2.40 | 4.2 | ||
| M2 | Ca2+–D@O1 | 2.37 | 3.0 | |
| Ca2+–WAT@O | 2.41 | 5.0 | ||
| Ca2+ (1)–Ca2+ (2) | 4.01 | 1.0 |
In summary, multi-scale simulations presented in this work prove that the C2 interface of 4His-ΔC* is responsible for the stability of the system in the presence of divalent metallic ions. Ni2+ ions affect the hydrogen bonding network at the C2 interface, inducing the loss of interactions promoted by Cu2+ ions. This explains that 4His-ΔC* mutants do not form cage structures when Cu2+ is replaced by Ni2+.6 The multiscale relaxation of the entire Cu-cage protein, in which all Cu2+ and Ca2+ ions as well as their coordination spheres have been represented at ab-initio level, corroborates the great potential of the maz-QM/MM-MD methodology to model the chemical synergies induced by the different subsystems in very large biological systems. Results have evidenced the key role of C3 and C4 interfaces in the stability of the whole nanocage.
This work was supported by MINECO (MAT2015-69367-R), PRACE for access to Curie TN and HN (France at GENCI@CEA and Mare Nostrum at Spain), RES-BSC (Mare Nostrum at Spain), and “ICREA Academia” (C. A.).
Conflicts of interest
There are no conflicts to declare.Notes and references
- M. Uchida, M. T. Klem, M. Allen, P. Suci, M. Flenniken, E. Gillitzer, Z. Varpness, L. O. Liepold, M. Young and T. Douglas, Adv. Mater., 2007, 19, 1025–1042 CrossRef CAS.
- E. J. Lee, N. K. Lee and I.-S. Kim, Adv. Drug Delivery Rev., 2016, 106(Part A), 157–171 CrossRef CAS PubMed.
- M. Kim, Y. Rho, K. S. Jin, B. Ahn, S. Jung, H. Kim and M. Ree, Biomacromolecules, 2011, 12, 1629–1640 CrossRef CAS PubMed.
- Z. Zhen, W. Tang, H. Chen, X. Lin, T. Todd, G. Wang, T. Cowger, X. Chen and J. Xie, ACS Nano, 2013, 7, 4830–4837 CrossRef CAS PubMed.
- X. Liu and E. C. Theil, Acc. Chem. Res., 2005, 38, 167–175 CrossRef CAS PubMed.
- D. J. E. Huard, K. M. Kane and F. A. Tezcan, Nat. Chem. Biol., 2013, 9, 169–176 CrossRef CAS PubMed.
- Z. Zhen, W. Tang, C. Guo, H. Chen, X. Lin, G. Liu, B. Fei, X. Chen, B. Xu and J. Xie, ACS Nano, 2013, 7, 6988–6996 CrossRef CAS PubMed.
- S. Kim, G. S. Kim, J. Seo, G. Gowri Rangaswamy, I.-S. So, R.-W. Park, B.-H. Lee and I.-S. Kim, Biomacromolecules, 2016, 17, 12–19 CrossRef CAS PubMed.
- J. Torras, Phys. Chem. Chem. Phys., 2015, 17, 9959–9972 RSC.
- J. Torras, B. P. Roberts, G. M. Seabra and S. B. Trickey, in Adv. Protein Chem. Struct. Biol., ed. K.-C. Tatyana, Academic Press, 2015, vol. 100, pp. 1–31 Search PubMed.
- J. Torras, E. Deumens and S. B. Trickey, J. Comput. – Aided Mater. Des., 2006, 13, 201–212 CrossRef CAS.
- J. Torras, G. d. M. Seabra, E. Deumens, S. B. Trickey and A. E. Roitberg, J. Comput. Chem., 2008, 29, 1564–1573 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Methodology description of maz-QM/MM-MD, the molecular system, the simulation protocol, the averaged distances of metal–residue coordination, the hydrogen bond occupancy and partition scheme for the modelling of a large system. See DOI: 10.1039/c7cc09512k |
| This journal is © The Royal Society of Chemistry 2018 |