Microfluidic methods for aptamer selection and characterization
Sean K.
Dembowski
and
Michael T.
Bowser
 *
*
Department of Chemistry, University of Minnesota, Minneapolis, MN 55455, USA. E-mail: bowser@umn.edu; Tel: +(612) 624-0873
First published on 20th October 2017
Abstract
Nucleic acid aptamers have tremendous potential as molecular recognition elements in biomedical targeting, analytical arrays, and self-signaling sensors. However, practical limitations and inefficiencies in the process of selecting novel aptamers (SELEX) have hampered widespread adoption of aptamer technologies. Many factors have recently contributed to more effective aptamer screening, but no influence has done more to increase the efficiency, scale, and automation of aptamer selection than that of new microfluidic SELEX techniques. This review introduces aptamers as a powerful chemical and biological tool, briefly highlights traditional SELEX techniques and their limitations, covers in detail the recent advancements in microfluidic methods of aptamer selection and characterization, and suggests possible future directions of the field.
Introduction
Aptamers
Aptamers are synthetic sequences of RNA or single-stranded DNA that fold into a particular three-dimensional conformation that allows them to bind a molecular target with high affinity and specificity. Because nucleic acids possess an innate flexibility and adopt a wide variety of structural motifs, aptamers have been developed for virtually every type of target imaginable: metal ions,1–3 small molecules,4–7 lipids,8,9 proteins,10–14 viruses,15 and whole cells.16,17 Though often likened to antibodies, aptamers have a number of distinct advantages over their protein counterparts. Most notably, specific aptamers are generated by in vitro solid-phase synthesis, an inexpensive, scalable, and easily automated technique with excellent batch-to-batch reproducibility. This avoids the use of model animals required for antibody production and the associated time and expertise required. Additionally, aptamers are highly stable over wide ranges of pH, temperature and ionic strength, and they refold spontaneously after denaturation. Finally, aptamers are easy to chemically link or modify during the synthesis process at virtually any position for customized applications.Aptamers are utilized in a variety of laboratory affinity assays and separations, especially those previously dominated by antibodies such as biomolecule purification,18,19 chiral separation,20 and enzyme-linked immunosorbent assays (ELISA).21–23 Because nucleic acid aptamers can undergo conformational changes upon binding, they are well suited to the development FRET- or quenching-based biosensors for their target, while antibodies are too inflexible for such behavior.2,24,25 Aptamers are also gaining favor in biomedical applications as both diagnostic and therapeutic tools. Aptamers have been used extensively to target imaging agents,26–29 siRNA,30,31 and drugs32–34 both in vitro and in vivo. While targeting is the most common medical use, modified aptamers have also had success as primary therapeutic agents, most notably Macugen for the treatment of age-related macular degeneration (AMD). While Macugen is thus far the only FDA approved aptamer-based drug, several aptamer therapeutics are in clinical trials and numerous others are in preclinical stages.35–37
Given the tremendous applicability and the advantages of aptamers, it is not surprising that the number of aptamer-related articles published annually has increased more than 10-fold since 2001.35 However, the selection of aptamers for novel targets is less well represented in the literature, primarily because of the time, resources, and expertise required to combine the disparate steps in aptamer development. In addition, successful selection of useful and applicable aptamers is never a guarantee, even in the most careful and well-intentioned cases.
One innovation that has dramatically lessened the time- and labor-intensiveness of aptamer technologies is the incorporation of novel microfluidic strategies; no single advancement has led to as much improvement in speed, throughput, and automation in aptamer selection and characterization. Microfluidics have also led to notable advances in aptamer-based sensing (aptasensors), especially involving electrochemical detection38–41 and low-cost printed or paper-based devices;42–44 however, microfluidic applications of aptamers will not be covered here. Aptasensors have been recently reviewed elsewhere.45–48
The current review provides a detailed overview of the different types of microfluidic aptamer selection schemes, highlighting key examples of these families and the major contributions of the work. Current work in aptamer microarrays for selection and characterization are also covered, and the authors offer possible future directions of the field.
The SELEX method
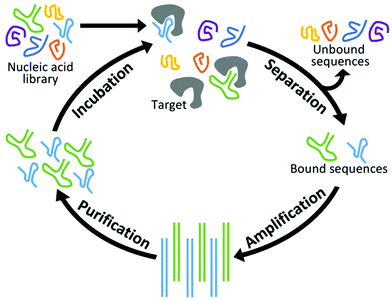
New aptamers are selected by an iterative combinatorial process called Systematic Evolution of Ligands by Exponential Enrichment (SELEX),49,50 which is outlined in Fig. 1. Briefly, a library of randomized RNA or DNA oligonucleotide sequences (oligos) is incubated with the target of interest. The large number of sequences creates sufficient structural diversity for numerous sequences to compete for binding with a limited amount of target. The in vitro nature of this incubation allows parameters such as temperature and ionic strength to be adjusted for optimized binding under the specified conditions. Next, unbound oligos are separated by one of a variety of partitioning techniques and discarded while those sequences complexed with the target are amplified by PCR. (In the case of RNA aptamers, sequences are reverse transcribed prior to PCR and transcribed back to RNA afterward.) Finally, the PCR products are purified and complementary strands are removed. This results in a new single-stranded pool that has been enriched with sequences that display affinity for the target. At the end of each cycle, the average affinity of the pool for the target of interest is measured, and the cycle is repeated until optimal binding affinity is reached. Finally, the nucleotide pool is sequenced and analyzed, and a subset of those sequences is synthesized and characterized in terms of their affinity, selectivity, and physical properties.Conventional SELEX
Nitrocellulose filtration was the first major partitioning technique employed because of its ability to pass nucleic acid strands while retaining proteins.49 While the technique is simple in that it requires no immobilization of the target and little adjustment from protein to protein, filtration is relatively inefficient and time-consuming, requiring a dozen or more rounds of SELEX and weeks to months to achieve suitable affinity. Nitrocellulose filtration is also only effective for proteins, making it unsuitable for many proposed targets. Affinity chromatography-based techniques, which rely on immobilizing the target of interest on a solid support, were introduced to address challenges presented by nitrocellulose SELEX and are now the technique of choice for many. The most frequently used supports are microbeads,51–53 which are easily retained by a microcolumn filter during selection, but other surfaces such as microplates are also used.54,55 In both cases, the target can be attached covalently with linker chemistry,56 adsorbed non-specifically,54 or linked with biotin to a streptavidin-coated surface.57 While this technique enhances flexibility and the number of allowable targets, macroscale affinity chromatography on the whole does not substantially increase the efficiency of the selection and often uses relatively large amounts of material. Additionally, the solid supports, and any linker or capping molecules used, present potential surfaces with which aptamers may develop unwanted, non-specific affinity, requiring negative selections to obtain target-specific aptamers.Microfluidic SELEX techniques
While different SELEX techniques offer certain pros and cons, there are a number of advantages that hold true of all microfluidic SELEX methods. First, because most separation techniques typically only operate at a surface interface, shrinking the system to yield a larger surface-to-volume ratio dramatically increases separation efficiency while reducing material consumption. In the case of affinity chromatography, this decrease in size also reduces the potential for non-specific and off-target binding, in many cases allowing negative selections to be removed altogether. Second, it has been shown that reducing the amount of target (thereby increasing the stringency of the selection) results in faster convergence to the aptamer pool to a small number of sequences.58 Microfluidic methods excel at handling small amounts of reagents, both increasing stringency and cutting material costs. Finally, a major drawback to SELEX in general is that each phase of the cycle can be labor-intensive, time-consuming, and require special expertise. Microfluidic techniques have a much higher potential for automation and multiplexing of this process, which will reduce the time and energy invested into each new aptamer and streamline the SELEX process. This has led to the development of many microfluidic SELEX separation techniques, including those based on capillary electrophoresis, microfluidic bead-based separations, and sol–gel encapsulation.Capillary electrophoresis SELEX
The first microfluidic technique introduced to enhance SELEX partitioning was capillary electrophoresis (CE). CE separates components of a mixture based on their electrophoretic mobility. Unbound DNA or RNA migrate together while oligo-target complexes, because of their altered size and charge, separate from the unbound sequences and are collected (Fig. 2). Because DNA and RNA have a higher negative charge density than virtually any target molecule of interest, complexation results in a significant and predictable decrease in mobility. This, together with the dramatic increase in the performance and prevalence of CE technology during the 1990s in response to the Human Genome Project, made CE the natural choice for SELEX's stringent nucleic acid separation requirements.CE-based aptamer selection (CE-SELEX) was first implemented by Mendonsa and Bowser seeking a DNA aptamer for human immunoglobulin E (IgE).59 Because of the high partitioning efficiency of CE, only four rounds of SELEX were required to obtain multiple aptamer sequences with dissociation constants (Kd) below 30 nM, drastically shortening the overall selection time. Additionally, the entire incubation and separation process can be performed in free solution, which allows the process to be done without tagging or immobilizing either molecule. Similar CE-SELEX techniques have been used to select aptamers for a variety of proteins, including human immunodeficiency virus-reverse transcriptase (HIV-RT),60 human vascular endothelial growth factor 165 (hVEGF165),61 α-fetoprotein,62 and human epididymis protein 4 (HE4).63 Although the separation between bound and unbound sequences is more substantial with larger targets, CE-SELEX has also been used successfully on smaller targets including the 4.3 kDa neuropeptide-Y64 and the 580 Da N-methyl mesoporphyrin (NMM),65 demonstrating the flexibility and robust nature of the technique.
An additional advantage of CE-SELEX is that CE provides an excellent technique for determining the binding affinity of both the mixed selection pool and individual aptamer sequences. However, this is typically done in an additional post-selection step. The Krylov group developed Non-Equilibrium Capillary Electrophoresis of Equilibrium Mixtures (NECEEM), which applied a novel analysis to extract kinetic and thermodynamic parameters of the nucleic acid pool.66 By supplying target-free buffer at the capillary inlet, the migrating complexes slowly unbound depending on the aptamer dissociation equilibrium and rate constants (Kd and koff, respectively), which were analyzed in the separation electropherogram. Krylov further reduced the required time by removing the intermediate amplification and purification steps altogether. This “non-SELEX” method facilitated three successive NECEEM separation steps in one hour and improved the affinity of the DNA pool for the h-RAS protein by over four orders of magnitude.67 While the selection stringency of this technique may be limited by the need to collect sufficient DNA for subsequent partitioning steps, the utility of non-SELEX has been further demonstrated in identifying aptamers for three signal transduction proteins, each with at least three orders of magnitude improvement in affinity.68
One major concern over CE-SELEX is the small sample volume that CE can handle. Injections are often less than 10 nL,61 so even when very high DNA concentrations are used, only about 1012–1013 oligos are sampled per run. One way around this has been utilizing bulk separation techniques like nitrocellulose filtration prior to CE to remove a large portion of non-binding sequences before CE-SELEX.69 This effectively samples more relevant sequences by removing a large fraction of non-binders before the CE step, although it does so at the cost of additional time and complexity.
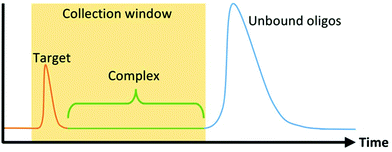
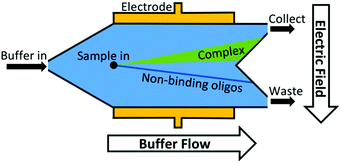
Another approach to increase the number of sequences assessed is to move from discrete separations to continuous partitioning techniques. One example of this is micro-free flow electrophoresis (μFFE). In this technique, buffer and sample are pressure-driven through a planar channel while an electric field is applied across the channel, perpendicular to the direction of flow (Fig. 3). Because species are deflected laterally based on their mobility, sample can be streamed continuously through the device rather than injected in discrete plugs. Binding sequences are collected through a separate exit port than non-binders. This continuous approach eliminates the complicated manual timing required in CE to obtain reproducible sample collections. A μFFE-SELEX technique was demonstrated that sampled nearly 2 × 1014 DNA sequences in a 30-minute separation, a 300-fold improvement over CE-SELEX.70 Aptamers for human IgE with a Kd of 20–30 nM were selected in only two rounds, indicating no loss in performance versus CE-based techniques.
Bead-based microfluidic SELEX
The Soh group was the first to implement magnetic microparticle SELEX on the microfluidic scale. Their first device, a continuous-flow magnetic activated chip separation device (CMACS), used a multi-stream laminar flow architecture to keep the bulk sample at the edges of the channel and a fresh buffer stream along the center (Fig. 4). This layout had less lateral diffusion across the interface than in a single-stream flow, which enhanced separation efficiency.73 Tiny ferromagnetic structures created strong, precise magnetic fields to guide the beads into the central buffer stream for collection. This microfluidic addition enhanced recovery of beads to over 99.5% under optimal conditions and achieved a bulk Kd of 33 nM in just one round.74 Despite the excellent results, CMACS suffered from a number of difficulties in practice. Ideal flow conditions were only reached with careful adjustment under microscopic monitoring, undermining the robustness of the technique. Microbubbles and blockages also occurred frequently, which disrupted the flow streams and had a detrimental impact on aptamer separation.
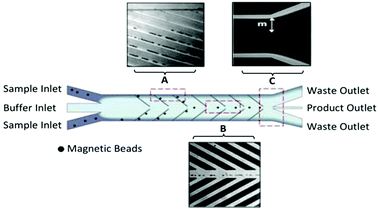 | ||
| Fig. 4 Schematic of the CMACS device. Laminar flow architecture keeps the sample stream near the channel walls and flows to waste. Angled magnetized Ni strips guide the magnetic beads containing bound oligos into the central buffer stream which is collected through a separate outlet. Optical micrographs show the device during operation at (A) the channel wall, (B) the buffer stream, and (C) the outlet. The large distance m prevents diffusion of unwanted sequences into the central stream. Adapted from Lou et al.73 Copyright 2009 National Academy of Sciences. | ||
To combat these issues, a new design was developed: the micromagnetic separation chip (MMS). The MMS relied on grids of patterned titanium and nickel, which were magnetized externally, to catch target-coated magnetic beads as they flowed through the channel. Once enough sample had passed through the chip, the channel was rinsed thoroughly to remove non-specifically adhered oligos. The external magnets were then removed, releasing the microbeads and target-binding sequences into solution for collection (Fig. 5).74 The MMS chip demonstrated recovery and partition efficiency as good as or better than those of CMACS (roughly 99.5% and 106, respectively) while offering significantly more robust and reproducible operation. MMS was first demonstrated using the protein streptavidin as the target, and later applied to platelet-derived growth factor-BB to obtain three aptamers with Kd's below 3 nM.75 The process has also been incorporated into special selections, such as obtaining orthogonal aptamer pairs for sandwich assays,76 aptamers with a long binding lifetime,77 and self-reporting aptamer biosensors.78
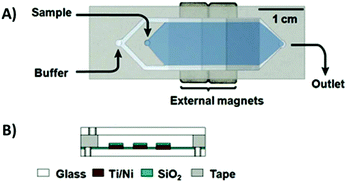 | ||
| Fig. 5 Schematic of the MMS device. (A) Top view and (B) side view. Sequential buffer and sample inlets flow sample towards the outlet. The patterned Ti/Ni grids become magnetized by external rare-earth magnets and traps magnetic beads containing bound oligos. Adapted from Qian et al.74 with permission from the American Chemical Society. | ||
Magnetic microbead SELEX continues to be perhaps the most popular microfluidic SELEX mechanism, with constant tweaking and adjustment pushing the boundaries of the technique. Recently, a device by Hong et al.79 incorporated both positive and negative selection chambers into the chip separated by a microvalve, reducing the time and hassle of performing a negative selection. A quantitative fluorescence binding analysis was performed on a separate microfluidic chip using magnetic target immobilization, demonstrating a trend towards miniaturization of additional steps of the aptamer selection process.
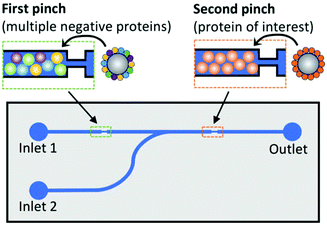 | ||
| Fig. 6 Schematic of dual positive–negative selection chip. Beads in the first pinch are coated with multiple non-target proteins for negative selection, while beads in the second pinch are coated with the target of interest. The oligos are flowed through inlet 1, and only sequences with affinity for the target but not for any of the negative proteins will bind in the second pinch. These oligos are then eluted by the addition of DDT to inlet 2. Adapted from Wang et al.80 with permission from the American Chemical Society. | ||
Since these beads do not move throughout the separation, similar strategies have been employed that forego the microbeads altogether, immobilizing the target directly on the device surface with similar effect. Liu et al. constructed a circuitous microfluidic path over a protein microarray so that oligos passed over four different negative selection proteins before reaching the region displaying the target, lactoferrin.81 Only those sequences bound to lactoferrin were eluted, achieving multiple sequences with Kd's below 10 nM.
In some cases though, the microbeads don't just provide a static surface but are actually a dynamic part of the device as with many magnetic systems. Recently, a continuous separation device was developed using migration by acoustic waves (acoustophoresis) for aptamer selection.82 Similarly to the CMACS device, the incubated sample of target-coated beads and DNA was introduced at the edges of the channel with a central buffer stream holding the sample stream against the channel walls. As sample passed down the channel, an acoustic standing wave field focused the beads to the node in the central buffer stream for collection while leaving smaller species such as unbound DNA in the waste-bound edge streams. An aptamer was obtained with sub-nanomolar affinity for prostate-specific antigen (PSA) after eight rounds of SELEX. As with other techniques not requiring a separate elution step (μFFE, CMACS), this device can overcome a common weakness of microfluidic SELEX, the small number of sequences sampled, by simply extending the time of continuous operation.
Sol–gel-based microfluidic SELEX
While SELEX techniques that rely on immobilizing the target have undoubtedly been successful in many cases, there has always been some concern over whether immobilization affects target conformation, occludes possible binding sites, or has other unintended consequences on the selection. One approach to circumvent covalent immobilization has been the use of sol–gel nanocomposites to trap proteins or other biomolecules.83,84 Briefly, monomers or nanoparticles of the gel precursor (often one or more silica derivatives) are dissolved to create an aqueous sol. Chemical additives, along with the target, are then mixed into the sol, causing the silica to coalesce and solidify into a porous framework, trapping the target in the growing gel. Gels with pores of two different sizes are favored; the smaller pores trap the macromolecule in place while the larger pores allow easy diffusion of aptamers or other ligands throughout the network. It has been shown that enzymes in the gel not only maintain their function but can be more stable against chemical and thermal denaturation than in free solution.85,86Sol–gel encapsulation was first demonstrated as a SELEX technique by Park et al.87 The microfluidic device consisted of a narrow channel connecting five identical chambers, each constructed over an aluminium microheater. A droplet of the protein-containing sol–gel was spotted into each chamber and allowed to gel overnight, then a polydimethylsiloxane (PDMS) cover sealed the chip. Incubated oligos were observed to bind with high efficiency to the trapped target but not significantly to the gel matrix. Applying current through the heater electrodes yielded enough heat to denature and elute the oligo sequences. A strength of the technique is high target density, with each 300 μm droplet holding about 0.6 ng of protein. The device isolated RNA aptamers with affinity for a yeast transcription factor87 and TATA binding protein88 with high affinity and in fewer rounds than with conventional SELEX. The high target density and heat-triggered release lend themselves well to a multiplexed selection, but the first attempts at simultaneous selection were fraught with issues of contamination. However, the design was improved by independent sample and elution lines as well as pneumatic valves between the reaction chambers (Fig. 7). The new device performed selections against five targets simultaneously in less time than the previous system without contamination.89 The capabilities and applicability of sol–gel-based SELEX have continued to expand, as the technique has selected aptamers against the small molecule xanthine, even when traditional SELEX could not,90 and against Bisphenol-A, which is too insoluble for other SELEX methods.91
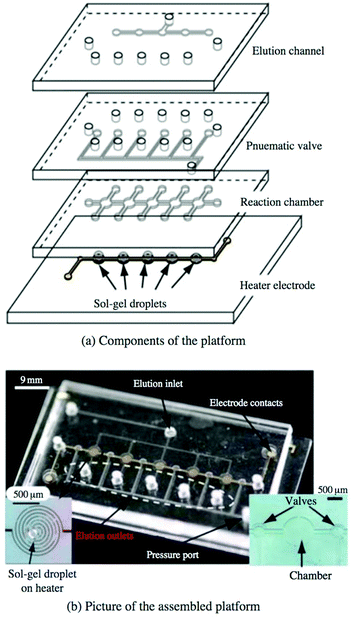 | ||
| Fig. 7 Multiplex sol–gel-based SELEX chip, showing (a) a schematic of the individual layers and (b) a picture of the assembled chip. Electrode heaters (b, left inset) are patterned onto the bottom glass layer and spotted with the sol–gel. The pneumatic layer controls the valves between reaction chambers to prevent cross-contamination during elution, while the remaining layers contain the main fluidic network. With valves open, sample is flowed through the reaction chambers to incubate with each sol–gel droplet, followed by washing to remove non-specific binders. Valves are closed and aptamers are eluted by running current through the electrode heaters and collecting at each elution outlet. Reproduced from Lee et al.89 with permission from Springer. | ||
Integrated microfluidic SELEX systems
As discussed previously, the SELEX cycle consists of four main steps: incubation of oligos with the target to compete for binding; partitioning of free oligos from those bound to the target; amplification of the oligo pool by PCR; and purification, whereby the complementary strand is removed to achieve a single-stranded pool again. (Additional transcription and reverse transcription steps are necessary when RNA aptamers are sought.) The techniques discussed thus far have focused on incorporating the partitioning and sometimes the incubation steps into a microfluidic environment. However, one of the main draws of a microfluidic SELEX technique, the enhanced potential for automation, is significantly limited until all parts of this cycle can be linked in a single integrated system. For this reason, many groups have pursued not only the microfluidic partitioning methods described above but also miniaturization of the amplification, purification, and intermediate steps as well.The first major step to this end was a prototype by Hybarger et al. that incorporated transcription, incubation, selection, reverse transcription, and amplification into a system of microlines, all controlled by valves actuated in LabView.92 The main achievement of the prototype was not just a seamless transition between the steps but also the use of pressurized reagent manifolds to fill, rinse, and clear sections of the system accordingly with no manual intervention. The full prototype still had a large footprint and its own share of practical difficulties, but it nonetheless set a high bar for future integrated SELEX systems.
The first integrated microfluidic SELEX chip, developed in the lab of Gwo-Bin Lee, was a three-layer, four-chamber PDMS and glass chip.93 The processes of incubation, separation, and amplification were performed in a central sample chamber connected to reagent and waste chambers by pneumatic microvalves (Fig. 8). The system used typical magnetic bead SELEX, first demonstrated with C-reactive protein (CRP) as the target conjugated to superparamagnetic polystyrene beads. Efficient mixing of the sample chamber was achieved within three seconds by deflection of the PDMS membrane layer with pressurized air. After incubation, the microbeads and any bound aptamers were held by an external magnetic field and rinsed thoroughly from the wash buffer chamber. PCR reagents were then introduced and the temperature was cycled in the sample chamber using microheaters and microsensors patterned on the glass substrate. The inherent automation coupled with the faster temperature cycling allowed by on-chip PCR achieved a round of incubation, selection and amplification in under 60 minutes.
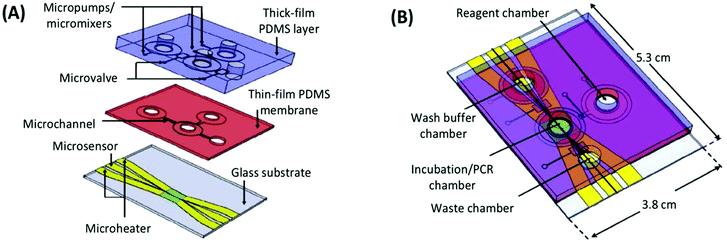 | ||
| Fig. 8 A schematic of the integrated SELEX chip by Huang et al. (A) The bottom glass layer is patterned with microheaters and sensors (platinum) with associated electrodes (gold) for temperature control during PCR. The thin-film PDMS membrane and thick PDMS block facilitate all liquid handling, including pumping and mixing by pneumatic membrane deflection. (B) The assembled device contains a main incubation/PCR chamber connected to chambers for wash buffer, PCR reagents, and waste by low-volume microchannels to prevent dead space and bubble formation. Adapted from Huang et al.93 with permission from Elsevier. | ||
The system was further enhanced to improve mixing and transport, and a competitive assay chip was designed to accompany the SELEX chip and further improve automation and speed of the overall process.94 Though the competitive assay chip was not a direct in-line addition and required intermediate manual processing, the increased efficiency of a chip-based assay provides a glimpse into the possible future of combined SELEX-aptamer characterization chips. The system has been continuously adapted and expanded to improve function and robustness. Example improvements include additional reagent and sample chambers, vacuum-driven pumps to accommodate live cells without lysis, and temperature-controlled regions of the device to speed cycling while avoiding reagent degradation.95–97
Purification of the PCR products to remove reagents and complementary nucleic acid strands has proven the most difficult piece to incorporate into microfluidic systems, either performed off-chip (reducing speed and automation) or absent altogether (reducing efficiency) in the previous devices. A new design by the Lin group utilized a bead-based PCR system to isolate the active aptamer from its complement generated during PCR.98 The device consisted of two chambers, one for incubation and selection, the other for amplification and purification (Fig. 9). The chambers were connected by a gel-filled channel to facilitate electrophoretic transfer of the DNA from one chamber to the other and back. Each chamber was surrounded by a weir to keep the beads in the appropriate chamber while allowing reagents and buffers to come and go freely. The key to purification was the use of reverse PCR primers immobilized onto microbeads in the amplification chamber. By performing PCR with immobilized reverse primers, only aptamer strands were transferred back to the selection chamber while the complementary strands remained bound to the PCR beads. The device performed three complete rounds of selection in under 10 hours with no off-chip activity, obtaining aptamers for both protein (IgE) and small molecule (bisboronic acid) targets. While this strategy provided an elegant solution to the purification problem, the electrophoretic transfer was not without practical issues, which led to a new design that incorporated both electrophoresis and pressure-driven flow into the system.99
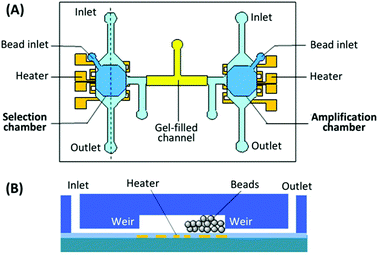 | ||
| Fig. 9 Schematic of the integrated SELEX chip by Kim et al. (A) A top view of the device, showing the selection and amplification chambers connected by a gel-filled channel. (B) A side view of each chamber, showing weirs to prevent bead movement and patterned microheater for elution or PCR. Adapted from Kim et al.98 | ||
Microfluidic aptamer characterization
Sequencing
The majority of microfluidic influence in SELEX has focused on optimizing the separation and partitioning methods used in the selection itself, with the goal of obtaining sequence pools with the highest average affinity and the most high-affinity binders in the least amount of time. However, even given highly efficient selection and enrichment, the ability to identify the best aptamers from the pool depends heavily on the nucleotide sequencing and binding characterization methods used.Traditionally, aptamer pools were cloned into bacterial vectors and sequenced by the Sanger method, which demanded labor-intensive handling of individual bacterial colonies and usually returned a few dozen to a hundred sequences at most, a tiny percent of the selected pool. A small number of those sequences were then synthesized and had their affinity tested individually. Though this technique is still commonplace in the field, finding the best aptamers by this method is largely governed by blind luck in picking the colonies. The introduction of high-throughput next-generation sequencing (NGS) techniques has allowed researchers to view a much larger fraction of the pool and to rationally choose sequences for further characterization.75,100 While sequence-based criteria (usually the copy number or enrichment-fold of a motif) are not always the best indicator of affinity,61,75 this strategy of rational sequence selection by NGS is arguably more effective than picking colonies randomly. As commercial NGS services become quicker and cheaper, choosing aptamer candidates via NGS analysis is rapidly becoming the method of choice.
Affinity measurement
Once individual sequences are chosen, some number of them, usually up to a few dozen, are synthesized in high purity, and the affinities of the individual sequences are determined for the target of interest as well as possibly for similar targets to determine selectivity. While NGS sequence analysis may hint at the best aptamers, it is no substitute for experimentally determining the affinity of a large number of sequences, as the true trends in affinity always deviate from those predicted by the informatics data. Even when using the microfluidic selection techniques above, affinity characterization has almost universally been done sequentially on large instrumentation. Microplate fluorescence assays, affinity capillary electrophoresis, and surface plasmon resonance are among the most common techniques for finding target affinity, in each case determining the amount of aptamer bound at many different concentrations of target to construct a binding curve and determine the aptamer dissociation constant. Because instrumental characterization techniques are robust and highly automated, it is unlikely that microfluidic characterization methods outside microarrays will see widespread adoption until they display significant advances in performance, automation, or multiplexing.For characterization of aptamers, arrays were first used to examine and improve existing sequences. For example, aptamers against IgE have been of particular interest because a long consensus region is highly conserved through many known aptamers. Microarrays with thousands of sequences systematically mutated from a few IgE aptamers have been used to interrogate the roles of specific bases.104,105 Mutations of certain bases or motifs completely eliminated binding behavior while other changes had no effect, allowing the original sequence to be truncated further.104 Similar work has been done using microarrays with over 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mutants to analyze and improve existing aptamer sequences binding to green fluorescent protein (GFP) and Drosophila negative elongation factor E (NELF-E).106 Ever improving technology has continued to increase both the number of sequences that can be tested and the amount of sample space that can be interrogated by more complex rational algorithms to control mutation. Work by Kinghorn et al.107 tested nearly 200
000 mutants to analyze and improve existing aptamer sequences binding to green fluorescent protein (GFP) and Drosophila negative elongation factor E (NELF-E).106 Ever improving technology has continued to increase both the number of sequences that can be tested and the amount of sample space that can be interrogated by more complex rational algorithms to control mutation. Work by Kinghorn et al.107 tested nearly 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mutants of previously selected aptamers against a malarial antigen. The mutants were determined using the public tool Resample, which not only recognized and generated preliminary mutants around motif families but also filtered out candidates based on predicted disadvantageous pairing and structural motifs. The process produced a truncated version of an existing sequence, as well as three novel sequences, all with Kd's below 10 nM, substantially better than the starting aptamers.
000 mutants of previously selected aptamers against a malarial antigen. The mutants were determined using the public tool Resample, which not only recognized and generated preliminary mutants around motif families but also filtered out candidates based on predicted disadvantageous pairing and structural motifs. The process produced a truncated version of an existing sequence, as well as three novel sequences, all with Kd's below 10 nM, substantially better than the starting aptamers.
Arrays are most commonly applied after SELEX in this manner, but their use as a part of the SELEX process is an interesting shortcut to condense the workflow and reduce backtracking. This was demonstrated by Cho et al. by combining the MMS device, NGS, and commercial microarrays into the quantitative parallel aptamer selection system (QPASS).108 Briefly, four rounds of SELEX were performed on the MMS against the protein angiopoietin-2 (Ang2). A small aliquot from each pool underwent NGS and returned ∼107 sequences, and the sequences were ranked by copy number. Eight identical arrays were then generated, each containing the 235 most common sequences from each pool, including different types of linkers and control sequences, with each spot in triplicate per array. By incubating each array with a different concentration of fluorescent target, an 8-point binding isotherm (and the associated Kd) was determined simultaneously for each of the over 200 sequences. Characterizing that many sequences sequentially by standard methods would be totally unthinkable, demonstrating the power of array-based SELEX.
The researchers have since used microarrays for rapid discovery of aptamer pairs that bind to different epitopes of the same target.109 Such pairs are necessary for some highly selective analytical assays or for development of bidentate aptamer binding schemes. Isolation of these aptamer pairs was difficult and relied largely on luck before this technique. By first generating an array of the Ang2 aptamers found previously, then incubating the target protein with the array, Ang2 was now presented for potential binding but with the binding epitope of the first aptamer occluded. A new library was then flowed over this protein-bound array to identify new sequences that bound to an independent epitope. Screening of this huge number of sequences allowed identification of multiple pairs with the strongest original aptamer sequence. By connecting these two aptamer sequences with a poly-T linker, the resulting aptamer exhibited an astounding 62 pM Kd, 300-fold stronger affinity than either sequence individually. There is still tremendous room for growth, as these examples can be scaled up to current commercially-available arrays of 1–2 million features with almost no increase in time or labor. As the scale, complexity, and economics of microfluidic DNA arrays continue to improve, it seems inevitable that microarrays will become inextricably linked with the SELEX process itself.
Conclusions
Despite many advantages of aptamers over other affinity reagents such as antibodies, conventional SELEX methods for isolating new aptamers are labor-intensive and fickle at best, which has bottlenecked progress in the field. However, incorporating new microfluidic SELEX techniques, either by scaling down existing procedures or by inventing novel strategies only feasible at the microscale, has produced tremendous growth in all parts of the selection and characterization process. While it is likely that no single SELEX technique will emerge as the optimal method in all cases (given the variety of targets, selection requirements, and intended aptamer uses), these abundant new microfluidic SELEX methods all have improved efficiency and speed over the older bulk techniques they have begun to replace.Based on the current state of the field, we predict two significant future directions. First, extensive work has already been done on specific selection techniques, and we anticipate a continued shift toward integrated microfluidic methods combining selection with purification, characterization, negative selection, and/or multiplexing to take full advantage of the superb automation that microfluidics provides. Second, the trend has thus far been towards increased complexity in prototype design to maximize speed and elegance, creating a divide between method-focused groups designing new selection techniques and those groups selecting aptamers for use in their research. We predict a future movement toward simpler designs that are more cost-effective and accessible to these application-focused groups with less microfluidics expertise, such as 3D printed devices or commercially available disposable devices. In both cases, we eagerly await exciting future microfluidic developments to meet the growing aptamer screening needs in bioanalytical and medical fields.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors gratefully acknowledge funding from the National Institutes of Health (R01 GM063533).References
- H. Qu, A. T. Csordas, J. P. Wang, S. S. Oh, M. S. Eisenstein and H. T. Soh, ACS Nano, 2016, 10, 7558–7565 CrossRef CAS PubMed.
- M. Rajendran and A. D. Ellington, Anal. Bioanal. Chem., 2008, 390, 1067–1075 CrossRef CAS PubMed.
- Y. S. Cho, E. J. Lee, G. H. Lee and S. S. Hah, Bioorg. Med. Chem. Lett., 2015, 25, 5536–5539 CrossRef CAS PubMed.
- D. Mann, C. Reinemann, R. Stoltenburg and B. Strehlitz, Biochem. Biophys. Res. Commun., 2005, 338, 1928–1934 CrossRef CAS PubMed.
- X. Y. Ma, W. F. Wang, X. J. Chen, Y. Xia, N. Duan, S. J. Wu and Z. P. Wang, Food Control, 2015, 47, 545–551 CrossRef CAS.
- X. J. Chen, Y. K. Huang, N. Duan, S. J. Wu, X. Y. Ma, Y. Xia, C. Q. Zhu, Y. Jiang and Z. P. Wang, Anal. Bioanal. Chem., 2013, 405, 6573–6581 CrossRef CAS PubMed.
- R. Stoltenburg, N. Nikolaus and B. Strehlitz, J. Anal. Methods Chem., 2012, 14 Search PubMed.
- A. Khvorova, Y. G. Kwak, M. Tamkun, I. Majerfeld and M. Yarus, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 10649–10654 CrossRef CAS.
- A. Vlassov, A. Khvorova and M. Yarus, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 7706–7711 CrossRef CAS PubMed.
- J. D. Toscano-Garibay, M. L. Benitez-Hess and L. M. Alvarez-Salas, in Cervical Cancer: Methods and Protocols, ed. D. Keppler and A. W. Lin, Humana Press Inc., Totowa, 2015, vol. 1249, pp. 221–239 Search PubMed.
- L. C. Bock, L. C. Griffin, J. A. Latham, E. H. Vermaas and J. J. Toole, Nature, 1992, 355, 564–566 CrossRef CAS PubMed.
- Y. Nonaka, K. Sode and K. Ikebukuro, Molecules, 2010, 15, 215–225 CrossRef CAS PubMed.
- S. E. Lupold, B. J. Hicke, Y. Lin and D. S. Coffey, Cancer Res., 2002, 62, 4029–4033 CAS.
- S. Shigdar, J. Lin, Y. Yu, M. Pastuovic, M. Wei and W. Duan, Cancer Sci., 2011, 102, 991–998 CrossRef CAS PubMed.
- H. M. Kwon, K. H. Lee, B. W. Han, M. R. Han, D. H. Kim and D. E. Kim, PLoS One, 2014, 9, 9 Search PubMed.
- M. Lu, L. Zhou, X. H. Zheng, Y. Quan, X. L. Wang, X. N. Zhou and J. Ren, Cancer Biomarkers, 2015, 15, 163–170 CrossRef CAS PubMed.
- K. L. Ji, W. S. Lim, S. F. Y. Li and K. Bhakoo, Anal. Bioanal. Chem., 2013, 405, 6853–6861 CrossRef CAS PubMed.
- S. Javaherian, M. U. Musheev, M. Kanoatov, M. V. Berezovski and S. N. Krylov, Nucleic Acids Res., 2009, 37, e62 CrossRef PubMed.
- Q. Zhao, X. F. Li, Y. H. Shao and X. C. Le, Anal. Chem., 2008, 80, 7586–7593 CrossRef CAS PubMed.
- J. Ruta, C. Ravelet, J. Desire, J.-L. Decout and E. Peyrin, Anal. Bioanal. Chem., 2008, 390, 1051–1057 CrossRef CAS PubMed.
- S. Y. Toh, M. Citartan, S. C. B. Gopinath and T. H. Tang, Biosens. Bioelectron., 2015, 64, 392–403 CrossRef CAS PubMed.
- D. W. Drolet, L. MoonMcDermott and T. S. Romig, Nat. Biotechnol., 1996, 14, 1021–1025 CrossRef CAS PubMed.
- J. Nie, Y. Deng, Q. P. Deng, D. W. Zhang, Y. L. Zhou and X. X. Zhang, Talanta, 2013, 106, 309–314 CrossRef CAS PubMed.
- R. Nutiu and Y. F. Li, J. Am. Chem. Soc., 2003, 125, 4771–4778 CrossRef CAS PubMed.
- D. Li, S. Song and C. Fan, Acc. Chem. Res., 2010, 43, 631–641 CrossRef CAS PubMed.
- C. A. Dougherty, W. B. Cai and H. Hong, Curr. Top. Med. Chem., 2015, 15, 1138–1152 CrossRef CAS PubMed.
- H. Hong, S. Goel, Y. Zhang and W. Cai, Curr. Med. Chem., 2011, 18, 4195–4205 CrossRef CAS PubMed.
- T. C. Chu, F. Shieh, L. A. Lavery, M. Levy, R. Richards-Kortum, B. A. Korgel and A. D. Ellington, Biosens. Bioelectron., 2006, 21, 1859–1866 CrossRef CAS PubMed.
- Z. Q. Cui, Q. Ren, H. P. Wei, Z. Chen, J. Y. Deng, Z. P. Zhang and X. E. Zhang, Nanoscale, 2011, 3, 2454–2457 RSC.
- J. O. McNamara, E. R. Andrechek, Y. Wang, K. D. Viles, R. E. Rempel, E. Gilboa, B. A. Sullenger and P. H. Giangrande, Nat. Biotechnol., 2006, 24, 1005–1015 CrossRef CAS PubMed.
- Y. Tan and X. M. Zhang, Prog. Biochem. Biophys., 2012, 39, 410–415 CrossRef CAS.
- Z. J. Chen, Z. G. Tai, F. F. Gu, C. L. Hu, Q. G. Zhu and S. Gao, Eur. J. Pharm. Biopharm., 2016, 107, 130–141 CrossRef CAS PubMed.
- L. S. Liu, X. L. Lu and Y. X. Zhao, J. Nanosci. Nanotechnol., 2016, 16, 6611–6621 CrossRef CAS.
- Z. H. Wang, J. F. Xia, F. Cai, F. F. Zhang, M. Yang, S. Bi, R. J. Gui, Y. H. Li and Y. Z. Xia, Colloids Surf., B, 2015, 134, 40–46 CrossRef CAS PubMed.
- C. M. C. Mattice and M. C. DeRosa, Biodrugs, 2015, 29, 151–165 CrossRef CAS PubMed.
- M. Hanout, D. Ferraz, M. Ansari, N. Maqsood, S. Kherani, Y. J. Sepah, N. Rajagopalan, M. Ibrahim, D. V. Do and N. Quan Dong, BioMed Res. Int., 2013, 830837 Search PubMed.
- A. Parashar, J. Clin. Diagn. Res., 2016, 10, BE1–BE6 CrossRef.
- T. Limi, S. Y. Lee, J. Yang, S. Y. Hwang and Y. Ahn, BioChip J., 2017, 11, 109–115 CrossRef.
- J. M. Moon, D. M. Kim, M. H. Kim, J. Y. Han, D. K. Jung and Y. B. Shim, Biosens. Bioelectron., 2017, 91, 128–135 CrossRef CAS PubMed.
- S. R. Jin, Z. Z. Ye, Y. X. Wang and Y. B. Ying, Sci. Rep., 2017, 7, 8 CrossRef PubMed.
- Z. Matharu, D. Patel, Y. D. Gao, A. Hague, Q. Zhou and A. Revzin, Anal. Chem., 2014, 86, 8865–8872 CrossRef CAS PubMed.
- H. Y. Fu, J. H. Yang, L. Guo, J. F. Nie, Q. B. Yin, L. Zhang and Y. Zhang, Biosens. Bioelectron., 2017, 96, 194–200 CrossRef CAS PubMed.
- P. Jiang, M. Y. He, L. Shen, A. N. Shi and Z. H. Liu, Sens. Actuators, B, 2017, 239, 319–324 CrossRef CAS.
- S. Sathish, S. G. Ricoult, K. Toda-Peters and A. Q. Shen, Analyst, 2017, 142, 1772–1781 RSC.
- A. Dhiman, P. Kaira, V. Bansal, J. G. Bruno and T. K. Sharma, Sens. Actuators, B, 2017, 246, 535–553 CrossRef CAS.
- N. Duan, S. J. Wu, S. L. Dai, H. J. Gu, L. L. Hao, H. Ye and Z. P. Wang, Analyst, 2016, 141, 3942–3961 RSC.
- M. Hasanzadeh, N. Shadjou and M. de la Guardia, Trac, Trends Anal. Chem., 2017, 89, 119–132 CrossRef CAS.
- Y. S. Kim, N. H. A. Raston and M. B. Gu, Biosens. Bioelectron., 2016, 76, 2–19 CrossRef PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CAS.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- J. G. Bruno, M. P. Carrillo, T. Phillips and B. King, In Vitro Cell. Dev. Biol.: Anim., 2008, 44, 63–72 CrossRef CAS PubMed.
- E. Kowalska, F. Bartnicki, K. Pels and W. Strzalka, Anal. Bioanal. Chem., 2014, 406, 5495–5499 CrossRef CAS PubMed.
- D. R. Latulippe, K. Szeto, A. Ozer, F. M. Duarte, C. V. Kelly, J. M. Pagano, B. S. White, D. Shalloway, J. T. Lis and H. G. Craighead, Anal. Chem., 2013, 85, 3417–3424 CrossRef CAS PubMed.
- C. H. Kim, L. P. Lee, J. R. Min, M. W. Lim and S. H. Jeong, Biosens. Bioelectron., 2014, 51, 426–430 CrossRef CAS PubMed.
- V. Mudili, S. S. Makam, N. Sundararaj, C. Siddaiah, V. K. Gupta and P. V. L. Rao, Sci. Rep., 2015, 5, 12 Search PubMed.
- D. K. Yang, L. C. Chen, M. Y. Lee, C. H. Hsu and C. S. Chen, Biosens. Bioelectron., 2014, 62, 106–112 CrossRef CAS PubMed.
- A. Grozio, V. M. Gonzalez, E. Millo, L. Sturla, T. Vigliarolo, L. Bagnasco, L. Guida, C. D'Arrigo, A. De Flora, A. Salis, E. M. Martin, M. Bellotti and E. Zocchi, Nucleic Acid Ther., 2013, 23, 322–331 CrossRef CAS PubMed.
- H. A. Levine and M. Nilsen-Hamilton, Comput. Biol. Chem., 2007, 31, 11–35 CrossRef CAS PubMed.
- S. D. Mendonsa and M. T. Bowser, J. Am. Chem. Soc., 2004, 126, 20–21 CrossRef CAS PubMed.
- R. K. Mosing, S. D. Mendonsa and M. T. Bowser, Anal. Chem., 2005, 77, 6107–6112 CrossRef CAS PubMed.
- M. Jing and M. T. Bowser, Anal. Chem., 2013, 85, 10761–10770 CrossRef CAS PubMed.
- L. L. Dong, Q. W. Tan, W. Ye, D. L. Liu, H. F. Chen, H. W. Hu, D. Wen, Y. Liu, Y. Cao, J. W. Kang, J. Fan, W. Guo and W. Z. Wu, Sci. Rep., 2015, 5, 10 Search PubMed.
- R. M. Eaton, J. A. Shallcross, L. E. Mael, K. S. Mears, L. Minkoff, D. J. Scoville and R. J. Whelan, Anal. Bioanal. Chem., 2015, 407, 6965–6973 CrossRef CAS PubMed.
- S. D. Mendonsa and M. T. Bowser, J. Am. Chem. Soc., 2005, 127, 9382–9383 CrossRef CAS PubMed.
- J. Yang and M. T. Bowser, Anal. Chem., 2013, 85, 1525–1530 CrossRef CAS PubMed.
- M. Berezovski, A. Drabovich, S. M. Krylova, M. Musheev, V. Okhonin, A. Petrov and S. N. Krylov, J. Am. Chem. Soc., 2005, 127, 3165–3171 CrossRef CAS PubMed.
- M. Berezovski, M. Musheev, A. Drabovich and S. N. Krylov, J. Am. Chem. Soc., 2006, 128, 1410–1411 CrossRef CAS PubMed.
- J. Tok, J. Lai, T. Leung and S. F. Y. Li, Electrophoresis, 2010, 31, 2055–2062 CrossRef CAS PubMed.
- J. Ashley, K. L. Ji and S. F. Y. Li, Electrophoresis, 2015, 36, 2616–2621 CrossRef CAS PubMed.
- M. Jing and M. T. Bowser, Lab Chip, 2011, 11, 3703–3709 RSC.
- J. G. Bruno, Biochem. Biophys. Res. Commun., 1997, 234, 117–120 CrossRef CAS PubMed.
- Z. J. Xi, R. R. Huang, Z. Y. Li, N. Y. He, T. Wang, E. B. Su and Y. Deng, ACS Appl. Mater. Interfaces, 2015, 7, 11215–11223 CAS.
- X. H. Lou, J. R. Qian, Y. Xiao, L. Viel, A. E. Gerdon, E. T. Lagally, P. Atzberger, T. M. Tarasow, A. J. Heeger and H. T. Soh, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 2989–2994 CrossRef CAS PubMed.
- J. R. Qian, X. H. Lou, Y. T. Zhang, Y. Xiao and H. T. Soh, Anal. Chem., 2009, 81, 5490–5495 CrossRef PubMed.
- M. Cho, Y. Xiao, J. Nie, R. Stewart, A. T. Csordas, S. S. Oh, J. A. Thomson and H. T. Soh, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 15373–15378 CrossRef CAS PubMed.
- A. T. Csordas, A. Jorgensen, J. Y. Wang, E. Gruber, Q. Gong, E. R. Bagley, M. A. Nakamoto, M. Eisenstein and H. T. Soh, Anal. Chem., 2016, 88, 10842–10847 CrossRef CAS PubMed.
- S. S. Oh, K. M. Ahmad, M. Cho, S. Kim, Y. Xiao and H. T. Soh, Anal. Chem., 2011, 83, 6883–6889 CrossRef CAS PubMed.
- S. S. Oh, K. Plakos, X. H. Lou, Y. Xiao and H. T. Soh, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14053–14058 CrossRef CAS PubMed.
- S. L. Hong, Y. T. Wan, M. Tang, D. W. Pang and Z. L. Zhang, Anal. Chem., 2017, 89, 6535–6542 CrossRef CAS PubMed.
- Q. Wang, W. Liu, Y. Q. Xing, X. H. Yang, K. M. Wang, R. Jiang, P. Wang and Q. Zhao, Anal. Chem., 2014, 86, 6572–6579 CrossRef CAS PubMed.
- X. H. Liu, H. Li, W. C. Jia, Z. Chen and D. K. Xu, Lab Chip, 2017, 17, 178–185 RSC.
- J. W. Park, S. J. Lee, S. Ren, S. Lee, S. Kim and T. Laurell, Sci. Rep., 2016, 6, 9 CrossRef PubMed.
- D. Avnir, T. Coradin, O. Lev and J. Livage, J. Mater. Chem., 2006, 16, 1013–1030 RSC.
- R. B. Bhatia, C. J. Brinker, A. K. Gupta and A. K. Singh, Chem. Mater., 2000, 12, 2434–2441 CrossRef CAS.
- D. T. Nguyen, M. Smit, B. Dunn and J. I. Zink, Chem. Mater., 2002, 14, 4300–4306 CrossRef CAS.
- W. Jin and J. D. Brennan, Anal. Chim. Acta, 2002, 461, 1–36 CrossRef CAS.
- S. M. Park, J. Y. Ahn, M. Jo, D. K. Lee, J. T. Lis, H. G. Craighead and S. Kim, Lab Chip, 2009, 9, 1206–1212 RSC.
- J. Y. Ahn, M. Jo, P. Dua, D. K. Lee and S. Kim, Oligonucleotides, 2011, 21, 93–100 CrossRef CAS PubMed.
- S. Lee, J. Kang, S. Ren, T. Laurell, S. Kim and O. C. Jeong, BioChip J., 2013, 7, 38–45 CrossRef CAS.
- H. Bae, S. Ren, J. Kang, M. Kim, Y. Jiang, M. M. Jin, I. M. Min and S. Kim, Nucleic Acid Ther., 2013, 23, 443–449 CrossRef PubMed.
- J. Y. Ahn, S. Lee, M. Jo, J. Kang, E. Kim, O. C. Jeong, T. Laurell and S. Kim, Anal. Chem., 2012, 84, 2647–2653 CrossRef CAS PubMed.
- G. Hybarger, J. Bynum, R. F. Williams, J. J. Valdes and J. P. Chambers, Anal. Bioanal. Chem., 2006, 384, 191–198 CrossRef CAS PubMed.
- C. J. Huang, H. I. Lin, S. C. Shiesh and G. B. Lee, Biosens. Bioelectron., 2010, 25, 1761–1766 CrossRef CAS PubMed.
- C. J. Huang, H. I. Lin, S. C. Shiesh and G. B. Lee, Biosens. Bioelectron., 2012, 35, 50–55 CrossRef CAS PubMed.
- H. C. Lai, C. H. Wang, T. M. Liou and G. B. Lee, Lab Chip, 2014, 14, 2002–2013 RSC.
- L. Y. Hung, C. H. Wang, K. F. Hsu, C. Y. Chou and G. B. Lee, Lab Chip, 2014, 14, 4017–4028 RSC.
- C. H. Weng, I. S. Hsieh, L. Y. Hung, H. I. Lin, S. C. Shiesh, Y. L. Chen and G. B. Lee, Microfluid. Nanofluid., 2013, 14, 753–765 CrossRef CAS.
- J. Kim, T. R. Olsen, J. Zhu, J. P. Hilton, K. A. Yang, R. Pei, M. N. Stojanovic and Q. Lin, Sci. Rep., 2016, 6, 10 Search PubMed.
- T. Olsen, J. Zhu, J. Kim, R. J. Pei, M. N. Stojanovic and Q. Lin, SLAS Technol., 2017, 22, 63–72 Search PubMed.
- A. Berezhnoy, C. A. Stewart, J. O. McNamara, W. Thiel, P. Giangrande, G. Trinchieri and E. Gilboa, Mol. Ther., 2012, 20, 1242–1250 CrossRef CAS PubMed.
- Y. L. Song, Y. Z. Shi, X. R. Li, Y. L. Ma, M. X. Gao, D. Liu, Y. Mao, Z. Zhu, H. Lin and C. Y. Yang, Anal. Chem., 2016, 88, 8294–8301 CrossRef CAS PubMed.
- R. Bumgarner, Curr. Protoc. Mol. Biol., 2013, 101, 22.1.1–22.1.11 Search PubMed.
- L. Wang and P. C. H. Li, Anal. Chim. Acta, 2011, 687, 12–27 CrossRef CAS PubMed.
- E. Katilius, C. Flores and N. W. Woodbury, Nucleic Acids Res., 2007, 35, 7626–7635 CrossRef CAS PubMed.
- N. O. Fischer, J. B. H. Tok and T. M. Tarasow, PLoS One, 2008, 3, 9 Search PubMed.
- J. M. Tome, A. Ozer, J. M. Pagano, D. Gheba, G. P. Schroth and J. T. Lis, Nat. Methods, 2014, 11, 683–688 CrossRef CAS PubMed.
- A. B. Kinghorn, R. M. Dirkzwager, S. L. Liang, Y. W. Cheung, L. A. Fraser, S. C. C. Shiu, M. S. L. Tang and J. A. Tanner, Anal. Chem., 2016, 88, 6981–6985 CrossRef CAS PubMed.
- M. Cho, S. S. Oh, J. Nie, R. Stewart, M. Eisenstein, J. Chambers, J. D. Marth, F. Walker, J. A. Thomson and H. T. Soh, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 18460–18465 CrossRef CAS PubMed.
- M. Cho, S. S. Oh, J. Nie, R. Stewart, M. J. Radeke, M. Eisenstein, P. J. Coffey, J. A. Thomson and H. T. Soh, Anal. Chem., 2015, 87, 821–828 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |