Open Access Article
Open Access ArticleHighly efficient base editing in Staphylococcus aureus using an engineered CRISPR RNA-guided cytidine deaminase†
Tongnian
Gu‡
a,
Siqi
Zhao‡
bc,
Yishuang
Pi
a,
Weizhong
Chen
a,
Chuanyuan
Chen
bc,
Qian
Liu
d,
Min
Li
d,
Dali
Han
*bc and
Quanjiang
Ji
 *a
*a
aSchool of Physical Science and Technology, ShanghaiTech University, Shanghai 201210, China. E-mail: quanjiangji@shanghaitech.edu.cn
bKey Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China. E-mail: handl@big.ac.cn
cCollege of Future Technology, Sino-Danish College, University of Chinese Academy of Science, Beijing 100049, China
dDepartment of Laboratory Medicine, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai 200127, China
First published on 22nd February 2018
Abstract
Novel therapeutic means against Staphylococcus aureus infections are urgently needed due to the emergence of drug-resistant S. aureus. We report the development of a CRISPR RNA-guided cytidine deaminase (pnCasSA–BEC), enabling highly efficient gene inactivation and point mutations in S. aureus. We engineered a fusion of a Cas9 nickase (Cas9D10A) and a cytidine deaminase (APOBEC1) that can be guided to a target genomic locus for gene inactivation via generating a premature stop codon. The pnCasSA–BEC system nicks the non-edited strand of the genomic DNA, directly catalyzes the conversion of cytidine (C) to uridine (U), and relies on DNA replication to achieve C → T (G → A) conversion without using donor repair templates. The development of the base-editing system will dramatically accelerate drug-target exploration in S. aureus and provides critical insights into the development of base-editing tools in other microbes.
Introduction
As a major human pathogen, Staphylococcus aureus is the leading cause of hospital- and community-acquired infections. This pathogen can cause a wide variety of infections, including minor skin infections and life-threatening diseases, such as endocarditis, necrotizing pneumonia, and toxic shock syndrome.1,2 The emergence of drug-resistant S. aureus, such as methicillin-resistant and vancomycin-intermediate S. aureus (MRSA and VISA), has posed a severe public crisis worldwide.3,4 Hence it is urgently needed to develop novel therapeutic means against drug-resistant S. aureus infections.We have developed a CRISPR/Cas9-mediated genome editing system (pCasSA).5 This method allows for rapid and efficient genetic manipulation in S. aureus, accelerating bacterial physiology studies, such as pathogenesis and drug resistance, and boosting novel drug-target exploration and new therapeutic method development.
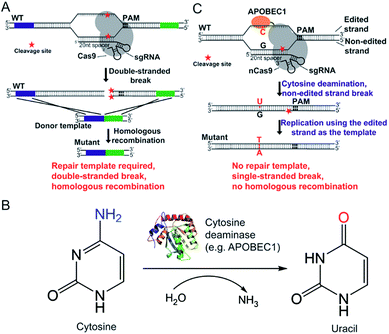
The CRISPR/Cas9 system was originally discovered in bacterial immune systems and has been engineered for genome editing in a variety of organisms.5–15 The system is composed of a Cas9 nuclease and a synthetic guide RNA (sgRNA) (Fig. 1A). The Cas9 nuclease forms a complex with sgRNA and can be directed to any genomic locus via base pairing of the 20-nucleotide sequence at the 5 prime of sgRNA with the genomic DNA when an adjacent protospacer-adjacent motif (PAM) is present in the target locus (Fig. 1A). After targeting, the Cas9 nuclease cleaves the genomic DNA by its enzymatic active residues (Asp10 and His840 in the case of Streptococcus pyogenes Cas9), generating a double-stranded break in the target genome (Fig. 1A). Given that the double-stranded DNA break is lethal to bacteria because of the lack of the non-homologous end-joining (NHEJ) DNA repair mechanism, only the cells that undergo the homologous recombination repair process can survive after editing. Hence the pCasSA system is capable of selecting separate double-crossover events that occur in the traditional S. aureus genome editing methods (e.g. pKOR1, pMAD, and pIMAY)16–18 in one step, achieving convenient genome editing in S. aureus (Fig. 1A). However, the weak intrinsic homologous recombination capacity of S. aureus prevents the high-rate recovery of survival cells after the double-stranded break of the genome, thus hampering the applications of the pCasSA system in strains with low transformation efficiencies, such as many MRSA strains directly isolated from patients.
Fortunately, the recent development of “base editors” allows for direct mutagenesis of bases inside the genome, opening a new avenue for genome editing in biology.19–24 Until now, two kinds of base editors have been developed (the cytidine editor and the adenosine editor) and each one is composed of a dead Cas9 (Cas9D10A or H840A) protein or a Cas9 nickase (Cas9D10A) and a deaminase fused to the Cas9 protein (Fig. 1B and C). The deaminase can be directed to any genomic locus by the Cas9/sgRNA complex to achieve base editing in the single-stranded DNA generated upon Cas9/sgRNA binding (Fig. 1B and C). The CRISPR/Cas9 genome editing method creates a double-stranded break, relies on the homologous recombination repair mechanism (in bacteria), and requires a donor repair template. In contrast, base editors directly catalyze the conversion of cytidine to uridine through a deamination reaction (adenosine to guanosine conversion in the case of recently developed adenine editors), nick the non-edited strand, and use the DNA replication mechanism to achieve editing (Fig. 1B and C). Hence we envision that “base editors” will allow for efficient genetic manipulation in S. aureus without sacrificing transformation CFUs.
In this study, we have developed a highly efficient and convenient base-editing system (pnCasSA–BEC) in S. aureus via engineering a fusion of a Cas9 nickase and a cytidine deaminase. The tool allows for gene inactivation and point mutations in the genomes of S. aureus, thereby dramatically accelerating bacterial physiology studies and drug-target exploration. The development of the pnCasSA–BEC system will provide critical insights into base-editing system development in other microbes.
Results and discussion
To harness “base editors” for base editing in S. aureus, we designed and constructed the plasmid pnCasSA–BEC (Fig. 2A). Inspired by the success of using cap 1A and rpsL promoters to drive the expressions of the sgRNA and the Cas9 protein, respectively, in the pCasSA5 plasmid, we used the same promoters to drive the expressions of the sgRNA and the fusion protein of a Cas9 nickase (Streptococcus pyogenes Cas9D10A) and a deaminase (rat APOBEC1). The deaminase was linked to the N-terminus of the Cas9 protein via an XTEN linker19 (Fig. 2A). We chose the Cas9 nickase instead of the dead Cas9 for base editing because bacterial cells lack nick-directed mismatch repair machinery.25 The nick generated by the Cas9 nickase in the non-edited strand of the genome will promote the cells to use the deaminated strand for DNA replication (Fig. 1C), thereby enhancing the base-editing efficiency. We introduced two BsaI sites in the plasmid for convenient and seamless cloning of the 20 bp spacer by Golden Gate assembly.26 In addition, we introduced an S. aureus temperature-sensitive replication origin repF in the plasmid to facilitate plasmid curing after editing.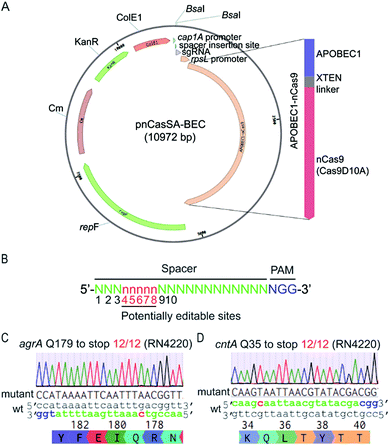 | ||
| Fig. 2 The engineered base-editing system enables highly efficient C → T conversion in the S. aureus RN4220 strain. (A) Map of the base-editing plasmid pnCasSA–BEC. APOBEC1-nCas9, a fusion protein composed of a deaminase APOBEC1 at the N terminus and a Cas9 nickase Cas9D10A at the C terminus; rpsL promoter, an S. aureus strong promoter used to drive the expression of APOBEC1-nCas9; BsaI sites, Golden Gate assembly of spacers; cap 1A promoter, the sgRNA expression promoter; ColE1, an E. coli replication origin. KanR, the antibiotic marker kanamycin used in E. coli; Cm, the chloramphenicol-resistance marker used in S. aureus; repF, an S. aureus temperature-sensitive origin for plasmid curing after editing. (B) The potential editable sites in the genome. The prediction of the potential editable sites is based on the mammalian “base editor” BE3.19 (C and D) The pnCasSA–BEC system enables highly efficient base editing in the RN4220 strain. Both the mutation efficiencies of agrA Q179 to stop codon (C) and cntA Q35 to stop codon (D) were 100%. The PAM sites are colored blue. The spacers are colored green. The mutation sites are colored red. A representative sequencing chromatogram for each mutation is shown. | ||
The mammalian C → T conversion “base editor” has an editable window from positions 4 to 8 in the spacer sequence (Fig. 2B) with the favorable editing site at position 7.19 To assess the capacity of the pnCasSA–BEC system in the conversion of C → T in S. aureus, we assembled different spacers of agrA, cntA, and esaD genes that contain potentially editable C(s). Next, we transformed the assembled plasmids into the laboratory S. aureus strain RN4220. As shown in Fig. 2C, D and S1,† the C at position 7 of agrA, the C at position 5 of cntA, and the C at position 6 of esaD were mutated to T successfully with 100% efficiency, resulting in the generation of premature stop codons. However, the Cs at positions 3 and 4 of agrA (Fig. 2C) and the C at position 3 of esaD (Fig. S1†) could not be mutated to T in the experiment.
Because of the different capacities of the pnCasSA–BEC system in mutating C(s) at different positions, we systematically investigated the editing window of the pnCasSA–BEC system in S. aureus. We assembled 11 different spacers containing C(s) at different positions (from positions 2 to 12) into the pnCasSA–BEC plasmid and transformed them individually into the RN4220 strain to examine the editing efficiencies. As shown in Fig. S2,† the Cs at positions 4 to 8 were mutated to Ts with 100% efficiency whereas the Cs at positions 2, 3, and 9–12 could not be mutated to T by the pnCasSA–BEC system, indicating that the editable window of the pnCasSA–BEC system is from positions 4 to 8. The discrepancy in the editing efficiencies at position 4 of the agrA spacer (Fig. 2C) and spacer 3 (Fig. S2†) indicates that other factors, such as adjacent bases or the sequence content of the spacers, can affect the editing efficiency of the pnCasSA–BEC system at position 4. In fact, the in vitro activity of the base editors follows the order TC ≥ CC ≥ AC > GC.19
Achieving a desired amino acid mutation may require the mutations of multiple Cs. We thus assessed the capacity of the pnCasSA–BEC system in mutating two successive Cs in S. aureus. Four different spacers containing two successive Cs at positions 4–5, 5–6, 6–7, and 7–8 were assembled into the pnCasSA–BEC plasmid, respectively. They were transformed into the RN4220 strain to test the editing efficiencies. As shown in Fig. S3,† both the successive Cs except the C at position 4 of spacer 1 could be mutated to Ts by the pnCasSA–BEC system with high efficiencies from 4/6 to 6/6 (or 18/18), demonstrating the great capacity of the pnCasSA–BEC system in mutating two successive Cs in the editable window. The C at position 4 of spacer 1 could not be mutated to T by the pnCasSA–BEC system (Fig. S3†).
The pnCasSA–BEC plasmid contains the same temperature-sensitive replication origin repF as the pCasSA plasmid. We used the same method5 to cure the pnCasSA–BEC plasmid after editing as that of the pCasSA plasmid. As shown in Fig. S4,† after culturing the cells at a non-permissive temperature (42 °C), all four randomly picked colonies could grow normally in the absence of chloramphenicol whereas none of them could grow in the presence of the antibiotic, confirming the successful removal of the pnCasSA–BEC plasmid from the cells (Fig. S4A and B†).
In agree with our expectation, the transformation of the spacer-introduced pnCasSA–BEC plasmid did not reduce the CFU (Fig. S5A and B†). However, the transformation of the spacer- or both the spacer and repair arm-introduced pCasSA plasmid drastically reduced the CFU (Fig S5A and B†).5 Thereby the pnCasSA–BEC system may possess a greater capacity than the pCasSA system in genome editing in strains with low transformation efficiencies, such as some MRSA strains directly isolated from patients.
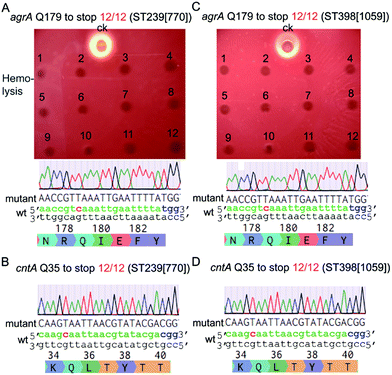
Next, we tested the base-editing efficiency of the pnCasSA–BEC system in four clinically isolated S. aureus strains, including ST239 (770), ST398 (1059), Newman, and USA300. ST239 (770) and ST398 (1059) are two MRSA strains isolated from patients in Ren Ji Hospital, China.27,28 The agrA-spacer and the cntA-spacer introduced pnCasSA–BEC plasmids isolated from the RN4220 strain were transformed into the four aforementioned strains by electroporation.29 Several S. aureus restriction-system-modified E. coli strains, such as IM08B and ALC7885, probably can also be used for the restriction modification of the pnCasSA–BEC plasmid.30,31 As shown in Fig. 3A–D and S6A–D,† the efficiencies of C → T conversions at position 7 of the agrA spacer and position 5 of the cntA spacer in all four strains were 100%, demonstrating the great capacity of the pnCasSA–BEC system in base editing in the clinically isolated S. aureus strains. Because the disruption of the agrA gene in S. aureus abolishes the pathogen's hemolysis activity,32 the C → T mutation in the agrA gene (generation of a premature stop codon) in all four strains was also confirmed phenotypically by the hemolysis assay (Fig. 3A, C and S6A, C†). In agree with the results obtained in the RN4220 strain, the spacer-introduced pnCasSA–BEC plasmid exhibited similar transformation efficiencies to the empty pnCasSA–BEC plasmid in all four strains (Fig S7A and B†).
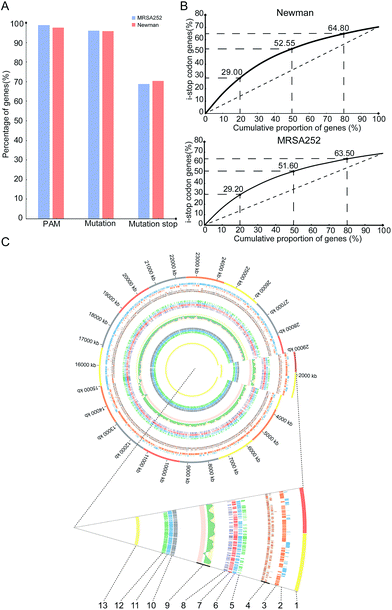
To gain insights into overall editable sites of the pnCasSA–BEC system in S. aureus, we performed bioinformatics scanning against the genomes of the MRSA252 and Newman strains. The results showed that almost all the genes (98.81% of MRSA252 and 97.6% of Newman) contain at least one PAM site (Fig. 4A). 96.12% of the genes of the MRSA252 strain and 95.7% of the genes of the Newman strain contain possibly editable C(s) (Fig. 4A). The potential codons that can be changed and the potential changed amino acids are listed in Fig. S8A and B.† In addition, 68.8% of the genes of the MRSA252 strain and 70.36% of the genes of the Newman strain possess possibly editable stop sites (CAA [Gln], CAG [Gln], CGA [Arg], and TGG [Trp]) (Fig. 4A). All the possibly editable stop sites are listed in Tables S1 and S2.† Because the closer the editable stop sites are to start codons, the higher possibility the translated product would be inactive, we analyzed the distribution of the possibly editable stop sites inside the genes. As shown in Fig. 4B, in the Newman strain, 29%, 52.55%, and 64.8% of the possibly editable stop sites locate at the top 20%, 50%, and 80% of the gene bodies, respectively, while in the MRSA252 strain, 29.2%, 51.6%, and 63.5% of the possibly editable stop sites locate at the top 20%, 50%, and 80% of the gene bodies, respectively. Further bioinformatics analysis revealed that the editable sites of the pnCasSA–BEC system are almost evenly distributed across the genome without chain preference (Fig. 4C). Together, these analyses unveiled that numerous editable sites of the pnCasSA–BEC system are present in S. aureus genomes and many genes can be inactivated using this system.
Conclusions
We have developed a highly efficient and convenient base-editing system in S. aureus by engineering a fusion of a Cas9 nickase and a cytidine deaminase. Unlike our previously developed pCasSA system, the pnCasSA–BEC system can achieve efficient genome editing without using repair templates or sacrificing transformation CFUs. Given its ease of use and high efficiency, this system can probably be further engineered to be a multiplexed editing system that cannot be achieved by the pCasSA system. Besides gene inactivation, the pnCasSA–BEC system is also capable of editing other sites in the genomes, such as the promoter regions and the regions coding for small RNAs. Future applications of the pnCasSA–BEC system can dramatically accelerate both the basic-science and applied investigations in S. aureus, in particular in strains with low-transformation efficiencies. The development of the pnCasSA–BEC system will shed light on base-editing system development in other microbes.Experimental
Strains, plasmids, primers, and growth conditions
Plasmids and strains used in this study are listed in Table S3.† Primers used in this study are listed in Table S4.†E. coli strains and S. aureus strains were grown in Luria–Bertani broth (LB) and tryptic soy broth (TSB), respectively. Antibiotics were used at the following concentrations: kanamycin 50 μg mL−1 for E. coli, chloramphenicol 5 μg mL−1 for the S. aureus RN4220 strain, and 10 μg mL−1 for other S. aureus strains.Construction of the pnCasSA–BEC plasmid
To achieve the construction of the pnCasSA–BEC plasmid, we first constructed the pnCasSA plasmid, which contained a Cas9 nickase (Cas9D10A) gene. The D10A mutation in the Cas9 protein was achieved by Gibson assembly.33 The pCasSA plasmid was separated into two parts. Each part was PCR amplified using the primers containing the desired mutation. The two parts were assembled by Gibson assembly, resulting in the pnCasSA plasmid. The successful construction of the pnCasSA was verified by sequencing.To construct the pnCasSA–BEC plasmid, we separated the plasmid into four parts. The first fragment contained the Cas9 nickase gene. The second fragment contained the repF origin, the chloramphenicol and kanamycin resistance markers, and the ColE1 origin. The third fragment contained the sgRNA expression cassette. The aforementioned three fragments were PCR amplified from the pnCasSA plasmid, respectively, using the primers listed in Table S4.† The fourth fragment of the E. coli codon optimized rat APOBEC1 gene was synthesized by GENEWIZ (Suzhou, China) and PCR amplified using the primers listed in Table S4.† The first and the second fragments were assembled first by Gibson assembly, resulting in the pnCas plasmid. The pnCas plasmid was linearized by BsaI. The fourth fragment containing the APOBEC1 gene was inserted into the pnCas plasmid by Gibson assembly, resulting in the pnCas–BEC plasmid. The pnCas–BEC plasmid was linearized by BsaI again. The third fragment containing the sgRNA expression cassette was inserted into the pnCas–BEC plasmid by Gibson assembly, resulting in the final plasmid pnCasSA–BEC. The successful construction of the pnCasSA–BEC plasmid was confirmed by PCR, enzyme digestion, and sequencing.
Electroporation
Electrocompetent cells of the S. aureus strains were prepared using the following protocol. A single colony of each S. aureus strain was inoculated into 2 mL TSB and incubated at 30 °C for 12 h. The cells were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 diluted into 100 mL fresh TSB medium. The cells were grown to OD600 of ∼0.3–0.4 and chilled on ice for 20 min. The cells were harvested by centrifugation (5000 rpm) at 4 °C for 6 min, and washed twice with 20 mL sterile ice-cold 0.5 M sucrose. Finally, the cells were resuspended into 1 mL 0.5 M sucrose. 50 μL aliquots were frozen in liquid nitrogen and stored at −80 °C.
100 diluted into 100 mL fresh TSB medium. The cells were grown to OD600 of ∼0.3–0.4 and chilled on ice for 20 min. The cells were harvested by centrifugation (5000 rpm) at 4 °C for 6 min, and washed twice with 20 mL sterile ice-cold 0.5 M sucrose. Finally, the cells were resuspended into 1 mL 0.5 M sucrose. 50 μL aliquots were frozen in liquid nitrogen and stored at −80 °C.
Before electroporation, 50 μL competent cells and the pnCasSA–BEC plasmid were thawed on ice for 5 min. The cells were mixed with 1 μg of the pnCasSA–BEC plasmid, and transferred into a 1 mm electroporation cuvette (Bio-Rad) at room temperature. After being pulsed at 2.1 kV mm−1, 100 Ω, and 25 μF, the cells were immediately supplied with 950 μL TSB and recovered at 30 °C for 1–2 h. The cells were plated onto a TSB plate containing chloramphenicol and incubated at 30 °C for 24 h.
Plasmid curing
The cells containing the desired mutation were cultured in TSB containing 10 μg mL−1 chloramphenicol. They were incubated at 30 °C overnight. The next day, the cells were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 diluted and incubated at 42 °C for 12 h without adding chloramphenicol. The bacteria were plated onto a TSB plate in the absence of chloramphenicol. The plate was incubated at 37 °C overnight. The next day, several colonies were randomly picked and resuspended individually into 10 μL ddH2O. The cells were streaked onto two different TSB plates in the presence and absence of 5 μg mL−1 chloramphenicol. The successful curing of the plasmid was confirmed when the cells could only grow on the plate without chloramphenicol.
1000 diluted and incubated at 42 °C for 12 h without adding chloramphenicol. The bacteria were plated onto a TSB plate in the absence of chloramphenicol. The plate was incubated at 37 °C overnight. The next day, several colonies were randomly picked and resuspended individually into 10 μL ddH2O. The cells were streaked onto two different TSB plates in the presence and absence of 5 μg mL−1 chloramphenicol. The successful curing of the plasmid was confirmed when the cells could only grow on the plate without chloramphenicol.
Hemolysis assay
Various S. aureus strains were grown overnight in TSB at 30 °C. The next day, 2 μL cells were loaded onto a sheep blood plate. The plate was incubated at 30 °C for 24 h and further stored at 4 °C for 24 h before being photographed.Editing efficiency evaluation
The genomic DNA of each colony was purified individually. The PCR reaction was performed individually using the genomic DNA as the template. The PCR products covering the editable site(s) were sent out for sequencing. The editing efficiency was calculated using the following method: the number of PCR products that contained the desired C → T conversion divided by the number of all the PCR products sent out for sequencing.Bioinformatics analysis
The genome sequences of S. aureus MRSA252 and Newman strains were obtained from NCBI (accession numbers: 299279 and 299277). The locations of all PAM (NGG) sites were scanned across the entire genome for the first time. Editable sites with the presence of C(s) at the 13–17 bp upstream of NGG were selected for further analysis. The sites that could introduce stop codons (UAG, UAA, and UGA) after the C → T conversion were treated as editable stop sites. Integrative Genomics Viewer (IGV)34 was used for the visualization of the genes with editable stop sites. Circos (http://circos.ca)35 was used for visualizing editable genes of the pnCasSA–BEC system in the genome of the S. aureus MRSA252 strain.Conflicts of interest
A patent has been submitted for the S. aureus base-editing method.Acknowledgements
We thank Professor Taeok Bae for reading the manuscript and giving critical comments. Q. J. acknowledges the financial support from the National Key R&D Program of China (2017YFA0506800), the National Natural Science Foundation of China (91753127 and 31700123), the Shanghai Committee of Science and Technology, China (17ZR1449200), the ShanghaiTech Startup Funding, and the “Young 1000 Talents” Program. D. H. acknowledges the financial support from the CAS Strategic Priority Research Program (XDA16010108), the CAS Hundred Talent Program, and the National Natural Science Foundation of China (31741074). W. C. acknowledges the financial support from the China Postdoctoral Science Foundation (2017M620178).References
- F. D. Lowy, N. Engl. J. Med., 1998, 339, 520–532 CrossRef CAS PubMed.
- G. L. Archer, Clin. Infect. Dis., 1998, 26, 1179–1181 CAS.
- L. B. Rice, Am. J. Infect. Control, 2006, 34, S11–S19 CrossRef PubMed ; discussion S64–S73.
- R. M. Klevens, M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey and S. K. Fridkin, M. I. Active Bacterial Core surveillance, JAMA, 2007, 298, 1763–1771 CrossRef CAS PubMed.
- W. Chen, Y. Zhang, W. S. Yeo, T. Bae and Q. Ji, J. Am. Chem. Soc., 2017, 139, 3790–3795 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS PubMed.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini and F. Zhang, Science, 2013, 339, 819–823 CrossRef CAS PubMed.
- J. E. DiCarlo, J. E. Norville, P. Mali, X. Rios, J. Aach and G. M. Church, Nucleic Acids Res., 2013, 41, 4336–4343 CrossRef CAS PubMed.
- W. Jiang, D. Bikard, D. Cox, F. Zhang and L. A. Marraffini, Nat. Biotechnol., 2013, 31, 233–239 CrossRef CAS PubMed.
- P. Mali, L. Yang, K. M. Esvelt, J. Aach, M. Guell, J. E. DiCarlo, J. E. Norville and G. M. Church, Science, 2013, 339, 823–826 CrossRef CAS PubMed.
- R. E. Cobb, Y. Wang and H. Zhao, ACS Synth. Biol., 2015, 4, 723–728 CrossRef CAS PubMed.
- H. Wang, M. La Russa and L. S. Qi, Annu. Rev. Biochem., 2016, 85, 227–264 CrossRef CAS PubMed.
- Y. Mao, H. Zhang, N. Xu, B. Zhang, F. Gou and J. K. Zhu, Mol. Plant, 2013, 6, 2008–2011 CrossRef CAS PubMed.
- C. Zhao, X. Shu and B. Sun, Appl. Environ. Microbiol., 2017, 83, e00291 Search PubMed.
- Q. Liu, Y. Jiang, L. Shao, P. Yang, B. Sun, S. Yang and D. Chen, Acta Biochim. Biophys. Sin., 2017, 49, 764–770 CrossRef PubMed.
- T. J. Foster, Methods Microbiol., 1998, 27, 433–454 CrossRef CAS.
- T. Bae and O. Schneewind, Plasmid, 2006, 55, 58–63 CrossRef CAS PubMed.
- I. R. Monk, I. M. Shah, M. Xu, M. W. Tan and T. J. Foster, mBio, 2012, 3, e00277 CrossRef CAS PubMed.
- A. C. Komor, Y. B. Kim, M. S. Packer, J. A. Zuris and D. R. Liu, Nature, 2016, 533, 420–424 CrossRef CAS PubMed.
- K. Nishida, T. Arazoe, N. Yachie, S. Banno, M. Kakimoto, M. Tabata, M. Mochizuki, A. Miyabe, M. Araki, K. Y. Hara, Z. Shimatani and A. Kondo, Science, 2016, 353, aaf8729 CrossRef PubMed.
- A. C. Komor, K. T. Zhao, M. S. Packer, N. M. Gaudelli, A. L. Waterbury, L. W. Koblan, Y. B. Kim, A. H. Badran and D. R. Liu, Sci. Adv., 2017, 3, eaao4774 CrossRef PubMed.
- Y. Zong, Y. Wang, C. Li, R. Zhang, K. Chen, Y. Ran, J. L. Qiu, D. Wang and C. Gao, Nat. Biotechnol., 2017, 35, 438–440 CrossRef CAS PubMed.
- N. M. Gaudelli, A. C. Komor, H. A. Rees, M. S. Packer, A. H. Badran, D. I. Bryson and D. R. Liu, Nature, 2017, 551, 464–471 CrossRef CAS PubMed.
- P. Billon, E. E. Bryant, S. A. Joseph, T. S. Nambiar, S. B. Hayward, R. Rothstein and A. Ciccia, Mol. Cell, 2017, 67, 1068–1079 CrossRef CAS PubMed.
- K. Fukui, J. Nucleic Acids, 2010, 2010, 260512 Search PubMed.
- C. Engler, R. Gruetzner, R. Kandzia and S. Marillonnet, PLoS One, 2009, 4, e5553 Search PubMed.
- M. Li, X. Du, A. E. Villaruz, B. A. Diep, D. Wang, Y. Song, Y. Tian, J. Hu, F. Yu, Y. Lu and M. Otto, Nat. Med., 2012, 18, 816–819 CrossRef CAS PubMed.
- X. Hong, J. Qin, T. Li, Y. Dai, Y. Wang, Q. Liu, L. He, H. Lu, Q. Gao, Y. Lin and M. Li, Front. Microbiol., 2016, 7, 951 Search PubMed.
- G. R. Kraemer and J. J. Iandolo, Curr. Microbiol., 1990, 21, 373–376 CrossRef CAS.
- I. R. Monk, J. J. Tree, B. P. Howden, T. P. Stinear and T. J. Foster, mBio, 2015, 6, e00308 CrossRef CAS PubMed.
- M. J. Jones, N. P. Donegan, I. V. Mikheyeva and A. L. Cheung, PLoS One, 2015, 10, e0119487 Search PubMed.
- R. P. Novick, Mol. Microbiol., 2003, 48, 1429–1449 CrossRef CAS PubMed.
- D. G. Gibson, L. Young, R. Y. Chuang, J. C. Venter, C. A. Hutchison III and H. O. Smith, Nat. Methods, 2009, 6, 343–345 CrossRef CAS PubMed.
- J. T. Robinson, H. Thorvaldsdóttir, W. Winckler, M. Guttman, E. S. Lander, G. Getz and J. P. Mesirov, Nat. Biotechnol., 2011, 29, 24–26 CrossRef CAS PubMed.
- M. Krzywinski, J. Schein, I. Birol, J. Connors, R. Gascoyne, D. Horsman, S. J. Jones and M. A. Marra, Genome Res., 2009, 19, 1639–1645 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc00637g |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2018 |