Open Access Article
Open Access ArticleChemical proteomic profiling of protein N-homocysteinylation with a thioester probe†
Nan
Chen
 a,
Jinmin
Liu
a,
Zeyu
Qiao
a,
Yuan
Liu
a,
Yue
Yang
b,
Changtao
Jiang
b,
Xian
Wang
b and
Chu
Wang
a,
Jinmin
Liu
a,
Zeyu
Qiao
a,
Yuan
Liu
a,
Yue
Yang
b,
Changtao
Jiang
b,
Xian
Wang
b and
Chu
Wang
 *ac
*ac
aSynthetic and Functional Biomolecules Center, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, College of Chemistry and Molecular Engineering, Peking University, Beijing, 100871, China. E-mail: chuwang@pku.edu.cn
bDepartment of Physiology and Pathophysiology, School of Basic Medical Sciences, Peking University, Beijing, 100191, China
cPeking-Tsinghua Center for Life Sciences, Peking University, Beijing, 100871, China
First published on 16th February 2018
Abstract
Hyperhomocysteinemia (HHcy) refers to a medical condition of abnormally high level of homocysteine (Hcy) in blood (>15 μmol L−1) and has been clinically implicated with cardiovascular diseases and neurodegenerative disorders. Excessive Hcy can be converted to a reactive thioester intermediate, Hcy thiolactone (HTL), which selectively reacts with protein lysine residues (“N-homocysteinylation”) and this non-enzymatic modification largely contributes to manifestations of HHcy. However, the proteome-wide detection of protein N-homocysteinylation remains a challenge to date. In this work, we report a chemoselective reaction to label and enrich N-homocysteinylation from complex proteome samples as inspired by native chemical ligation for protein synthesis. Alkynyl thioester probes are synthesized and the reaction is validated with small molecule and purified protein models successfully. We performed quantitative chemical proteomics to identify more than 800 N-homocysteinylated proteins as well as 304 N-homocysteinylated sites directly from HTL-treated HeLa cells. The chemical proteomics strategies will facilitate functional study of protein N-homocysteinylations in the HHcy-implicated diseases.
Introduction
Homocysteine (Hcy) is an intermediary amino acid involved in the metabolism of methionine and cysteine.1 The normal concentration of Hcy in plasma is low (5–15 μmol L−1) and increased accumulation (up to 500 μmol L−1), namely, hyperhomocysteinemia (HHcy), is a contributing risk factor for cardiovascular diseases and neurodegenerative disorders.2–4 Among various hypotheses, protein N-homocysteinylation represents a major mechanism of Hcy toxicity.2,5–9 Protein N-homocysteinylation is a non-enzymatic post-translational modification of lysine residues mediated by homocysteine thiolactone (HTL), which is a reactive thioester intermediate generated from an error-editing reaction of Hcy with methionyl-tRNA synthetase (MetRS).10 Several studies have shown that N-homocysteinylation on select proteins will cause protein damage,11–13 aggregation9 and auto-immune responses.14,15Traditional methods for detecting N-homocysteinylation include high-performance liquid chromatography (HPLC)-based16 and antibody-based assays.17 They suffer from the limitations of poor sensitivity and lack of identity of modified proteins. Recently, mass spectrometry (MS)-based methods coupled with direct trypsin digestion18–20 or aldehyde chemical labeling21 have been developed to identify N-homocysteinylated proteins and sites. However, they were mostly applied on individual cases and about ten human proteins with N-homocysteinylated sites have been reported to date (Table S1†).
Chemical proteomics22 is an emerging platform to understand the reactivity of amino acids,23–26 targets of bioactive compounds,27–30 protein–protein interactions31,32 and post-translational modifications33–36 in biological systems. The key component is specific conjugation with targets by chemical probes, followed by enrichment and MS-based identification. Considering low abundance and sub-stoichiometry of protein N-homocysteinylation, a specific chemical probe will be necessary for its large-scale profiling from complex proteomes of cells or tissues.
Herein, we report the development of a chemoselective reaction between thioester and N-homocysteinylation as inspired by native chemical ligation (NCL).37 NCL is widely used for synthesizing native backbone proteins by ligating two peptides with an N-terminal cysteine and a C-terminal thioester, respectively.38 To overcome the limitation of requiring a cysteine in the ligated sequence, many variant methodologies have been developed, including methionine ligation,39 in which an N-terminal homocysteine is used for ligation, followed by S-methylation to form methionine (Scheme S1†). Given that the chemical structure of protein N-homocysteinylation is identical to that of an N-terminal homocysteine, we hypothesize that thioester could serve as a specific probe to react with protein N-homocysteinylation and enable its global profiling in complex proteomes. As shown in Scheme 1, alkynyl thioester probes react with N-homocysteinylated proteins through a reversible transthioesterification to form a six-member intermediate, followed by a rapid intramolecular S-to-N acyl shift. Consequently, an N-homocysteinylated lysine can be selectively functionalized with an alkyne handle through an amide bond. A fluorescent or biotin reporter tag will be conjugated for either visualization by SDS-PAGE or enrichment for MS-based proteomic analysis. To test this hypothesis, we first synthesize a series of thioester probes and validate the reactions with small molecule model. Next, we chose the best probe to demonstrate its chemoselective labeling of N-homocysteinylation on purified proteins. Finally, we apply this probe in a chemoproteomic strategy to globally profile protein N-homocysteinylation in cellular proteomes. These studies highlight the novel application of NCL in the profiling of protein post-translational modifications and to our best knowledge, for the first time, report quantification of N-homocysteinylated proteins and sites in complex proteomes of mammalian cells.
Results and discussion
Validation of reaction between N-homocysteinylation and thioester probes with a small molecule model
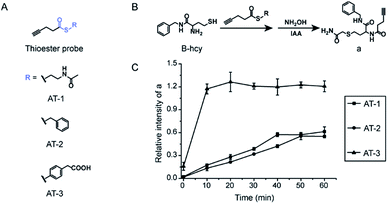
To test our hypothesis, three thioester probes were designed and synthesized (Fig. 1A and Scheme S2A†). AT-1 and AT-2 employ alkyl thiol and benzyl thioalcohol as leaving groups, both of which were commonly used as C-terminal thioester reactive groups in NCL.37,40 AT-3 was derived from mercaptophenylacetic acid, MPAA, which is a non-malodorous and water-soluble thiol used as an excellent thiol additive to speed up peptide ligation.41 In theory, AT-3 should have better solubility in aqueous buffer and better reactivity towards N-homocysteinylation. We first evaluated the reactivity of the three probes in a small molecule model. 2-Amino-N-benzyl-4-mercaptobutanamide (B-hcy) was synthesized as a mimic of N-homocysteinylation (Fig. 1B and Scheme S2B†) and reactions were carried out in a denaturing buffer condition commonly used for NCL. As shown in Fig. 1B and S1,† hydroxylamine (NH2OH) was added to quench excessive thioester probes and reaction intermediates (b), followed by alkylation of thiol groups with iodoacetamide (IAA). The amount of desired product (a) was monitored by LC-MS and plotted over time in Fig. 1C. As expected, AT-3 (1 mM) demonstrated the fastest reaction kinetic and B-hcy (100 μM) was consumed completely within 30 min at pH 7. Thus, AT-3 was chosen for labeling proteins next.Chemoselective labeling of N-homocysteinylation on purified proteins
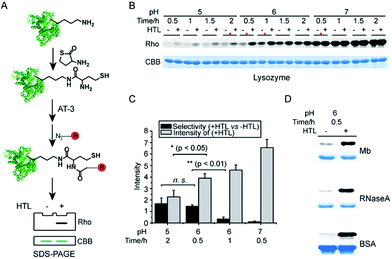
Unlike the small molecule model, proteins are composed of various amino acids such as nucleophilic lysine residues that have been reported to react with thioester under physiological conditions.42 Notably, thioester probes have been implemented to profile non-enzymatic acylation in proteomes.43 We therefore tried to optimize the condition in order to achieve chemoselective labeling of N-homocysteinylation on purified proteins. As depicted in Fig. 2A, N-homocysteinylated proteins were firstly generated by incubating purified proteins with HTL at room temperature and the modified proteins were further verified by ESI-MS analysis (Fig. S2†). N-homocysteinylated proteins were dissolved in denaturing buffer and labeled with AT-3 (1 mM). After removal of excessive probes, the labeled proteins were conjugated with rhodamine-azide via copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC)44 and separated by SDS-PAGE for direct visualization of in-gel fluorescence. As shown in Fig. 2B, lysozyme was labeled at various pH conditions for different amounts of time. The overall labeling intensity rose with increasing pH and time regardless of HTL modifications, indicating potential off-target reaction with lysine residues. We quantified the labeling selectivity (+HTL versus −HTL) for four selected conditions (marked with red asterisks, Fig. 2B) and the results showed that the selectivity (+HTL versus −HTL) dropped sharply at higher pH or with increased time of labeling. We then subjected the non-HTL-induced samples to trypsin digestion and analysis by liquid chromatography-tandem MS (LC-MS/MS), which did reveal nonspecific labeling of lysine residues (Table S2†). Therefore, to balance between the labeling selectivity and overall intensity, we chose pH 6 and 0.5 h for selective labeling of N-homocysteinylated proteins (Fig. 2C). Under this condition, other HTL-induced proteins (Mb, RNaseA and BSA) were also labeled with good selectivity (Fig. 2D). We further identified the AT-3 labeled adducts on sites of N-homocysteinylation in these proteins by LC-MS/MS (Fig. S3†).Profiling of N-homocysteinylated proteins and sites with a quantitative, MS-based chemical proteomic platform
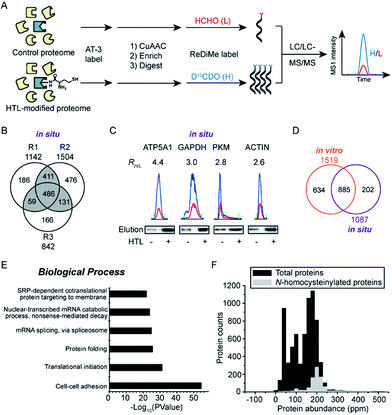
We next set out to apply the AT-3 probe to characterize N-homocysteinylated proteins and sites in complex cellular proteomes using a quantitative, MS-based chemical proteomic platform (Fig. 3A). Soluble proteomes of HeLa cells treated with H2O or HTL (0.2 mM) were labeled by AT-3 (1 mM), conjugated with biotin-azide via CuAAC, enriched by streptavidin beads and proteolytically digested by trypsin. The tryptic peptides from H2O- and HTL-treated samples were then isotopically differentiated by reductive dimethylation (ReDiMe),45 combined pairwise and fractionated by high-pH HPLC for LC-MS/MS analysis. The selective labeling of HTL-modified proteomes were confirmed by western blotting (Fig. S4A†) and the proteomic results were summarized in Fig. S4B and C.† A total of 1519 candidate N-homocysteinylated targets were quantified across three biological replicates (RH/L ≥ 2, HTL-treated proteome versus the untreated control) (Table S3†). In addition to the in vitro profiling, we also applied the AT-3 probe to identify N-homocysteinylation from HeLa cells treated with HTL in situ. The results of cell viability assays indicated no measurable cytotoxicity for HeLa cells after incubation with HTL (0.5 mM) for 24 h at 37 °C (Fig. S5A†). Under this condition, HTL-dependent labeling signals were again detected with AT-3 (Fig. S5B†) and quantitative chemoproteomic experiments identified more than 1000 potential targets of N-homocysteinylation (Fig. 3B and S5C, Table S4†). We further verified the enrichment of N-homocysteinylation by AT-3 on four candidate proteins, ATP5A1, GAPDH, PKM and ACTIN, by affinity purification and immunoblotting (Fig. 3C and S6†). Finally, among the 1087 potential targets identified in three biological replicates, 885 targets were overlapped with the in vitro data (Fig. 3D, Table S4†). Gene ontology analysis by DAVID (Database for Annotation, Visualization and Integrated Discovery)46 revealed that the N-homocysteinylated protein targets were enriched in important biological processes such as cell–cell adhesion (annexin A2; filamin B), translational initiation (eukaryotic translation initiation factors; ribosomal proteins) as well as protein folding (peptidylprolyl isomerases; heat shock proteins) (Fig. 3E and Table S4†). Comparison with the whole proteome data of HeLa cells47 reveals that the majority of N-homocysteinylated targets were of high abundance (Fig. 3F). The results were consistent with a previous report that human plasma proteins were N-homocysteinylated proportionally to its abundance.48We finally sought to identify exact sites of N-homocysteinylation using a tandem orthogonal proteolysis strategy.49 As illustrated in Fig. 4A, proteomes of HTL-induced HeLa cells (0.5 mM) were labeled with AT-3, conjugated with a photo-cleavable biotin-azide tag via CuAAC, and subjected to streptavidin enrichment and on-beads trypsin digestion. The AT-3 adducted peptides were released from streptavidin beads upon irradiation with ultraviolet (UV) light (365 nm, 0.5 J cm−2, 60 min) and analyzed by LC-MS/MS for site identification. In total, 304 unique sites of N-homocysteinylation were identified across two biological replicates (Fig. 4B and Table S5†). For example, the MS/MS spectrum generated by higher-energy collisional dissociation (HCD) fragmentation unambiguously supports the adduct of N-homocysteinylation on Lys54 of Profilin-1 (TFVNITPAEVGVLVGK*DR) (Fig. 4C and S7†). We also identified two sites of N-homocysteinylation, K57 (Fig. S8†) and K80 (Fig. S9†), from Histone H3, the former of which has been previously identified from purified histones of HEK293T cells.50 The identified sites of N-homocysteinylation belong to 168 proteins, most of which (≥97%) were quantified from HeLa cells with in situ treatment of HTL (Fig. 4D and Table S5†). Among them, 25 proteins were with the corresponding ratios, RH/L, less than 2, suggesting possible non-specific labeling of lysine residues within the same protein. The sequence analysis did not reveal any obvious conserved motifs surrounding N-homocysteinylated lysine residues (Fig. S10A†). In addition, most of the N-homocysteinylated lysines do not have heightened intrinsic reactivity according to a recent ABPP profiling study23 (Fig. S10B and Table S5†). Interestingly, 10 proteins reported with thioester-reactive lysine residues by Meier and colleagues43 are also found in our list (Fig. S10C and Table S5†), including GAPDH that has three lysine residues (K66, K84 and K219) modified by both malonyl-CoA and HTL due to similar reactivity. Finally, 46 sites of N-homocysteinylation have been annotated as sites of acetylation according to the Uniprot database, suggesting a potential crosstalk between N-homocysteinylation and acetylation as previously revealed on purified histones50 (Fig. S10D and Table S5†).
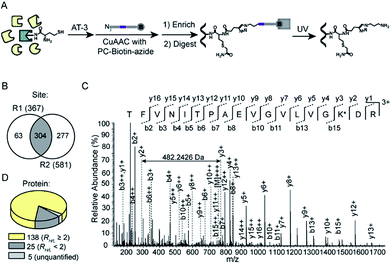 | ||
| Fig. 4 Identification of sites of N-homocysteinylation from HeLa cells induced with HTL in situ. (A) Schematic workflow for MS-based identification of residue sites of N-homocysteinylation using a tandem orthogonal proteolysis strategy. (B) Venn diagram showing the number of sites of N-homocysteinylation identified across two biological replicates. (C) MS/MS spectrum of TFVNITPAEVGVLVGK*DR, a peptide from Profilin-1. All b and y ions are labeled. The m/z difference of 482.2426 between y2+ and y3+ supports the expected modification on the lysine residue (see Fig. S7† for the exact structure) as denoted by K*. (D) Categorization of 168 proteins with identified sites of N-homocysteinylation based on distribution of their quantification ratios, RH/L. | ||
Conclusions
In summary, we have developed a chemoselective strategy inspired by NCL to label protein N-homocysteinylation. Although NCL was developed more than two decades ago, its applications have been mainly limited to protein synthesis. Our method exploited the unique structure of N-homocysteinylation and extended, for the first time, the usage of thioester probes to the profiling of this disease-related protein post-translational modification. We further applied the thioester probe in combination with MS-based quantitative proteomics to generate a global portrait of protein N-homocysteinylation in mammalian cells. Particularly, over 800 N-homocysteinylated proteins and 300 sites of N-homocysteinylation were identified from HeLa cells induced under HHcy-mimicking conditions. Our data suggested that the non-enzymatic process is tightly associated with physical accessibility of lysine residues towards the reactive intermediate and might be functionally implicated in crosstalk with other lysine modification. This powerful chemoproteomic platform will provide valuable information to help decipher molecular mechanisms of HHcy-implicated diseases.Conflicts of interest
The authors declare that there is no conflict of interest regarding the publication of this article.Acknowledgements
We thank Prof. Suwei Dong for discussions. We thank the Computing Platform of the Center for Life Science for supporting the proteomic data analysis. This work was supported by Ministry of Science and Technology of China (2016YFA0501500), National Science Foundation of China (21472008 and 81490741), and a “1000 Talents Plan” Young Investigator Award (C. W.).Notes and references
- J. Selhub, Annu. Rev. Nutr., 1999, 19, 217–246 CrossRef CAS PubMed.
- J. Perla-Kajan, T. Twardowski and H. Jakubowski, Amino Acids, 2007, 32, 561–572 CrossRef CAS PubMed.
- S. Seshadri, A. Beiser, J. Selhub, P. F. Jacques, I. H. Rosenberg, R. B. D'Agostino, P. W. Wilson and P. A. Wolf, N. Engl. J. Med., 2002, 346, 476–483 CrossRef CAS PubMed.
- G. J. Hankey and J. W. Eikelboom, Lancet, 1999, 354, 407–413 CrossRef CAS.
- G. S. Sharma, T. Kumar, T. A. Dar and L. R. Singh, Biochim. Biophys. Acta, 2015, 1850, 2239–2245 CrossRef CAS PubMed.
- P. Paoli, F. Sbrana, B. Tiribilli, A. Caselli, B. Pantera, P. Cirri, A. De Donatis, L. Formigli, D. Nosi, G. Manao, G. Camici and G. Ramponi, J. Mol. Biol., 2010, 400, 889–907 CrossRef CAS PubMed.
- H. Jakubowski, L. Zhang, A. Bardeguez and A. Aviv, Circ. Res., 2000, 87, 45–51 CrossRef CAS PubMed.
- H. Jakubowski, Cell. Mol. Life Sci., 2004, 61, 470–487 CrossRef CAS PubMed.
- H. Jakubowski, FASEB J., 1999, 13, 2277–2283 CrossRef CAS PubMed.
- H. Jakubowski and R. Glowacki, Adv. Clin. Chem., 2011, 55, 81–103 CAS.
- J. Perla-Kajan, L. Marczak, L. Kajan, P. Skowronek, T. Twardowski and H. Jakubowski, Biochemistry, 2007, 46, 6225–6231 CrossRef CAS PubMed.
- D. L. Sauls, E. Lockhart, M. E. Warren, A. Lenkowski, S. E. Wilhelm and M. Hoffman, Biochemistry, 2006, 45, 2480–2487 CrossRef CAS PubMed.
- R. Glowacki and H. Jakubowski, J. Biol. Chem., 2004, 279, 10864–10871 CrossRef CAS PubMed.
- H. Jakubowski, Clin. Chem. Lab. Med., 2005, 43, 1011–1014 CrossRef CAS PubMed.
- A. Undas, J. Perla, M. Lacinski, W. Trzeciak, R. Kazmierski and H. Jakubowski, Stroke, 2004, 35, 1299–1304 CrossRef CAS PubMed.
- H. Jakubowski, Anal. Biochem., 2002, 308, 112–119 CrossRef CAS PubMed.
- J. Perla, A. Undas, T. Twardowski and H. Jakubowski, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2004, 807, 257–261 CrossRef CAS PubMed.
- M. Sikora, L. Marczak, J. Kubalska, A. Graban and H. Jakubowski, Amino Acids, 2014, 46, 235–244 CrossRef CAS PubMed.
- L. Marczak, M. Sikora, M. Stobiecki and H. Jakubowski, J. Proteomics, 2011, 74, 967–974 CrossRef CAS PubMed.
- M. Sikora, L. Marczak, T. Twardowski, M. Stobiecki and H. Jakubowski, Anal. Biochem., 2010, 405, 132–134 CrossRef CAS PubMed.
- T. Zang, S. Dai, D. Chen, B. W. Lee, S. Liu, B. L. Karger and Z. S. Zhou, Anal. Chem., 2009, 81, 9065–9071 CrossRef CAS PubMed.
- C. Wang and N. Chen, Huaxue Xuebao, 2015, 73, 657–668 CrossRef CAS.
- S. M. Hacker, K. M. Backus, M. R. Lazear, S. Forli, B. E. Correia and B. F. Cravatt, Nat. Chem., 2017, 9, 1181–1190 CrossRef CAS PubMed.
- E. Weerapana, C. Wang, G. M. Simon, F. Richter, S. Khare, M. B. Dillon, D. A. Bachovchin, K. Mowen, D. Baker and B. F. Cravatt, Nature, 2010, 468, 790–795 CrossRef CAS PubMed.
- M. Abo and E. Weerapana, J. Am. Chem. Soc., 2015, 137, 7087–7090 CrossRef CAS PubMed.
- S. Lin, X. Yang, S. Jia, A. M. Weeks, M. Hornsby, P. S. Lee, R. V. Nichiporuk, A. T. Iavarone, J. A. Wells, F. D. Toste and C. J. Chang, Science, 2017, 355, 597–602 CrossRef CAS PubMed.
- M. M. Blewett, J. Xie, B. W. Zaro, K. M. Backus, A. Altman, J. R. Teijaro and B. F. Cravatt, Sci. Signaling, 2016, 9, rs10 CrossRef PubMed.
- S. Zhuang, Q. Li, L. Cai, C. Wang and X. Lei, ACS Cent. Sci., 2017, 3, 501–509 CrossRef CAS PubMed.
- T. Kambe, B. E. Correia, M. J. Niphakis and B. F. Cravatt, J. Am. Chem. Soc., 2014, 136, 10777–10782 CrossRef CAS PubMed.
- B. D. Horning, R. M. Suciu, D. A. Ghadiri, O. A. Ulanovskaya, M. L. Matthews, K. M. Lum, K. M. Backus, S. J. Brown, H. Rosen and B. F. Cravatt, J. Am. Chem. Soc., 2016, 138, 13335–13343 CrossRef CAS PubMed.
- Y. Yang, H. Song, D. He, S. Zhang, S. Dai, S. Lin, R. Meng, C. Wang and P. R. Chen, Nat. Commun., 2016, 7, 12299 CrossRef CAS PubMed.
- D. Tan, Q. Li, M. J. Zhang, C. Liu, C. Ma, P. Zhang, Y. H. Ding, S. B. Fan, L. Tao, B. Yang, X. Li, S. Ma, J. Liu, B. Feng, X. Liu, H. W. Wang, S. M. He, N. Gao, K. Ye, M. Q. Dong and X. Lei, eLife, 2016, 5, e12509 Search PubMed.
- X. H. Gao, D. Krokowski, B. J. Guan, I. Bederman, M. Majumder, M. Parisien, L. Diatchenko, O. Kabil, B. Willard, R. Banerjee, B. Wang, G. Bebek, C. R. Evans, P. L. Fox, S. L. Gerson, C. L. Hoppel, M. Liu, P. Arvan and M. Hatzoglou, eLife, 2015, 4, e10067 Search PubMed.
- J. D. Majmudar, A. M. Konopko, K. J. Labby, C. T. Tom, J. E. Crellin, A. Prakash and B. R. Martin, J. Am. Chem. Soc., 2016, 138, 1852–1859 CrossRef CAS PubMed.
- M. Broncel, R. A. Serwa, T. D. Bunney, M. Katan and E. W. Tate, Mol. Cell. Proteomics, 2016, 15, 715–725 CAS.
- G. Xu, J. S. Paige and S. R. Jaffrey, Nat. Biotechnol., 2010, 28, 868–873 CrossRef CAS PubMed.
- P. E. Dawson, T. W. Muir, I. Clark-Lewis and S. B. Kent, Science, 1994, 266, 776–779 CAS.
- A. Dirksen and P. E. Dawson, Curr. Opin. Chem. Biol., 2008, 12, 760–766 CrossRef CAS PubMed.
- J. P. Tam and Q. Yu, Biopolymers, 1998, 46, 319–327 CrossRef CAS PubMed.
- P. Wang, S. Dong, J. H. Shieh, E. Peguero, R. Hendrickson, M. A. S. Moore and S. J. Danishefsky, Science, 2013, 342, 1357–1360 CrossRef CAS PubMed.
- E. C. Johnson and S. B. Kent, J. Am. Chem. Soc., 2006, 128, 6640–6646 CrossRef CAS PubMed.
- Y. Yasueda, T. Tamura, A. Fujisawa, K. Kuwata, S. Tsukiji, S. Kiyonaka and I. Hamachi, J. Am. Chem. Soc., 2016, 138, 7592–7602 CrossRef CAS PubMed.
- R. A. Kulkarni, A. J. Worth, T. T. Zengeya, J. H. Shrimp, J. M. Garlick, A. M. Roberts, D. C. Montgomery, C. Sourbier, B. K. Gibbs, C. Mesaros, Y. C. Tsai, S. Das, K. C. Chan, M. Zhou, T. Andresson, A. M. Weissman, W. M. Linehan, I. A. Blair, N. W. Snyder and J. L. Meier, Cell Chem. Biol., 2017, 24, 231–242 CrossRef CAS PubMed.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef CAS PubMed.
- P. J. Boersema, R. Raijmakers, S. Lemeer, S. Mohammed and A. J. Heck, Nat. Protoc., 2009, 4, 484–494 CrossRef CAS PubMed.
- G. Dennis Jr, B. T. Sherman, D. A. Hosack, J. Yang, W. Gao, H. C. Lane and R. A. Lempicki, Genome Biol., 2003, 4, P3 CrossRef.
- T. Geiger, A. Wehner, C. Schaab, J. Cox and M. Mann, Mol. Cell. Proteomics, 2012, 11, M111.014050 Search PubMed.
- H. Jakubowski, J. Biol. Chem., 2002, 277, 30425–30428 CrossRef CAS PubMed.
- E. Weerapana, A. E. Speers and B. F. Cravatt, Nat. Protoc., 2007, 2, 1414–1425 CrossRef CAS PubMed.
- L. Xu, J. Chen, J. Gao, H. Yu and P. Yang, Analyst, 2015, 140, 3057–3063 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, synthetic procedures, characterization data for reaction products and additional figures. See DOI: 10.1039/c8sc00221e |
| This journal is © The Royal Society of Chemistry 2018 |