Open Access Article
Open Access ArticleMultiple advanced logic gates made of DNA-Ag nanocluster and the application for intelligent detection of pathogenic bacterial genes†
Xiaodong
Lin
a,
Yaqing
Liu
 *a,
Jiankang
Deng
a,
Yanlong
Lyu
a,
Pengcheng
Qian
a,
Yunfei
Li
a and
Shuo
Wang
*b
*a,
Jiankang
Deng
a,
Yanlong
Lyu
a,
Pengcheng
Qian
a,
Yunfei
Li
a and
Shuo
Wang
*b
aKey Laboratory of Food Nutrition and Safety (Ministry of Education), College of Food Engineering and Biotechnology, Tianjin University of Science and Technology, Tianjin 300457, China. E-mail: yaqingliu@tust.edu.cn
bTianjin Key Laboratory of Food Science and Health, School of Medicine, Nankai University, Tianjin 300071, China. E-mail: wangshuo@nankai.edu.cn
First published on 8th January 2018
Abstract
The integration of multiple DNA logic gates on a universal platform to implement advance logic functions is a critical challenge for DNA computing. Herein, a straightforward and powerful strategy in which a guanine-rich DNA sequence lighting up a silver nanocluster and fluorophore was developed to construct a library of logic gates on a simple DNA-templated silver nanoclusters (DNA-AgNCs) platform. This library included basic logic gates, YES, AND, OR, INHIBIT, and XOR, which were further integrated into complex logic circuits to implement diverse advanced arithmetic/non-arithmetic functions including half-adder, half-subtractor, multiplexer, and demultiplexer. Under UV irradiation, all the logic functions could be instantly visualized, confirming an excellent repeatability. The logic operations were entirely based on DNA hybridization in an enzyme-free and label-free condition, avoiding waste accumulation and reducing cost consumption. Interestingly, a DNA-AgNCs-based multiplexer was, for the first time, used as an intelligent biosensor to identify pathogenic genes, E. coli and S. aureus genes, with a high sensitivity. The investigation provides a prototype for the wireless integration of multiple devices on even the simplest single-strand DNA platform to perform diverse complex functions in a straightforward and cost-effective way.
Introduction
To fulfil the demands of increased computational complexity, it is critical to integrate multiple logic gates into advanced logic circuits on a universal platform for the next generation of molecular computing.1–3 In electronics, higher-ordered logic circuits implement complex functional and information processing at varying degrees of complexity by electrically wiring multiple required logic gates, which is a significant challenge on a molecular scale.4 Thanks to its massive parallel computation ability, programmable sequence, and predictable configuration, DNA has been validated as an extremely powerful material for the engineering of DNA logic gates.5–8 To date, various DNA logic gates have been successfully constructed to implement versatile functions from data to information processing.9–18 According to the special catalysis of DNAzymes in many reactions, parallel gate arrays, multilayered gate cascades and fan-out gates have been successfully constructed.19–23 A toe-hold strategy based on entropy-driven DNA strand displacement provides an outstanding possibility for building enzyme-free DNA logic gates.24,25 Three DNA logic gates (one AND and two NOT) were integrated on a 2D platform via DNA crossover tile technology.3 The investigations significantly improve the development of DNA computing. However, it is worth noting that by-products are usually generated during logic operation, resulting in waste accumulation within the system. Meanwhile, cost-effective molecular logic gates constructed on a universal platform and featuring a simple operation is still a main challenge for the integration of molecular devices.26,27DNA-templated silver nanoclusters (DNA-AgNCs) with molecular-like properties have attracted increasing attention in extensive research fields due to their intrinsic merits such as excellent photostability, biocompatibility, and low cost.28 As signal transducers, DNA-AgNCs present advantages over organic dyes and quantum dots in that the use of DNA-AgNCs not only avoids the covalent label of DNA with a signal indicator, but also makes it easier to be introduced into a DNA logic system via the DNA template.29,30 The fluorescence emission of DNA-AgNCs exhibits size and sequence-dependent property31 and is highly sensitive to their micro-environment.32,33 These intrinsic properties endow DNA-AgNCs with promising applications in the development of parallel/cascade logic gates and computing circuits. Here, we demonstrated that a set of basic logic gates (YES, AND, OR, XOR, and INHIBIT (INH)) and advanced logic gates (half adder (HA), half subtractor (HS), multiplexer (MUX), and demultiplexer (DMUX)) could be integrated into a simple DNA-AgNCs platform to implement arithmetic and non-arithmetic logic functions.
Aside from the engineering of DNA computing, the construction of DNA logic gates is of scientific interesting for intelligent biosensing and diagnostics.34–37 To date, most of the logic-related analyses have been developed according to AND or OR logic gates with biomarkers as inputs.38,39 An AND gate based diagnosis system reports a high output signal in the coexistence of all biomarkers. For OR gate based detection, any one of the biomarkers can cause a high output signal response. To a certain degree, this provides a convincing way for intelligent diagnosis. However, AND or OR gate-based analysis suffers from the limitation of distinguishing biomarkers from each other, which might be overcome by a multiplexer gate-based biosensor. With two disease-related foodborne pathogen genes as inputs, here, a 2:1 multiplexer was demonstrated for the first time to act as intelligent biosensor to distinguish each of the genes with one output signal response under logic control.
Result and discussion
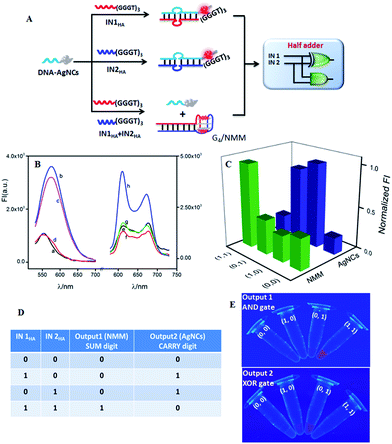
DNA-AgNCs were designed as a universal platform for the construction of multiple advanced logic gates, which contained two segments: a cytosine-rich (C-rich) segment for synthesizing AgNCs and an input-recognizing segment. A dim fluorescence of DNA-AgNCs could be lightened up by the approached guanine-rich (G-rich) DNA strand (see Fig. S1 in the ESI†), which was consistent with previous reports.40 This implemented the simplest YES gate function. Via simple modulation of the input sequences, a set of basic logic gates, including XOR, AND, OR, and INH, were successfully constructed on the same DNA-AgNCs platform (see Fig. S2–S8 in ESI†). Their combinations provide a powerful application to transfer a basic Boolean logic gate into advanced logic gates, thus implementing functions of almost the whole family of logic gates.41–43 Here, we demonstrated that arithmetic functions, such as half adder and half subtractor, and non-arithmetic functions, such as multiplexer and demultiplexer, could be reached by integrating the required basic logic gates on the universal platform of DNA-AgNCs.As important logic gates, half adder and half subtractor are highly demanded in numerical data and information processing.44 A half adder can implement an addition function on two binary digits by integrating an XOR gate and an AND gate in parallel to respectively code for a SUM and a CARRY digit, which requires two distinct output signals. Here, fluorescent AgNCs modulated by G-rich DNA sequences were used as signal indicators for the XOR logic gate. Interestingly, G-rich oligonucleotide sequences could form a G-quadruplex (G4) configuration to enhance the fluorescence of organic dyes.45,46 Such a combination of G-rich DNA modulating the fluorescence of AgNCs and fluorophore endows DNA-AgNCs as a powerful building block for the construction of advanced logic gates. To implement HA function, NMM (N-Methyl Mesoporphyrin IX) was selected as the signal indicator of the required AND gate since it has the properties of special structural selectivity for G4 rather than single, duplex, or triplex DNA strands.47 The resulting complex of G4/NMM can generate a pronounced fluorescent signal.48,49Fig. 1A outlines the operation principle of the developed HA. Both AgNCs and NMM emitted dim fluorescence signals in the absence of any input (Fig. 1B(a) and (e), respectively). Each of the two inputs, IN1HA and IN2HA, contained a G-rich segment of (GGGTGGGTGGGT) at the 3′-terminal to augment the fluorescence of the AgNCs by forming duplexes of DNA-AgNCs/IN1HA and DNA-AgNCs/IN2HA (Fig. 1B(b) and (c)). In contrast, the low fluorescence responses of NMM were monitored in both cases where the G-rich sequence of (GGGTGGGTGGGT) in IN1HA and IN2HA could not form a G4 configuration and thus had no significant influence on the fluorescence of NMM (Fig. 1B(f) and (g), respectively). When both inputs coexist, they preferred to hybridize together while they did not hybridize with DNA-AgNCs. Then, AgNCs reported a dim fluorescence signal (Fig. 1B(d)). However, the fluorescence of NMM was strongly augmented (Fig. 1B(h)) since the G-rich sequence of (GGGTGGGTGGGT) at the 3′-terminal of IN1HA formed a G4 configuration by hybridizing with the (TGGGT) sequence at the 5′-terminal of IN2HA, which further formed the G4/NMM complex. The formation of the G4 configuration was validated by circular dichroism (CD) experiments (Fig. S9 in ESI†). The DNA interactions were validated by native polyacrylamide gel electrophoresis (PAGE) experiment (Fig. S10 in ESI†).
To implement HA function, the normalized fluorescence intensities of the signal indicators at maximum peaks, FImax(NMM) and FImax(AgNCs), were plotted as a function of input combinations (Fig. 1C). Similar to the approach used in electronics, the output signal with an undefined range was defined as “1” or “0” when the normalized fluorescence intensity was higher than 0.45 or lower than 0.4, respectively. This definition was available for all logic operations in this work. The resulting truth table (Fig. 1D) clearly demonstrated that AND and XOR gates were successfully integrated in parallel to implement the HA arithmetic function. The entire logic system executes binary arithmetic operations of (0 + 0 = 0 0), (1 + 0 = 0 1), and (0 + 1 = 0 1). The case of (1 + 1 = 2) endowed the SUM digit as 0 and the CARRY digit as 1, yielding (1 + 1 = 1 0).50 Interestingly, the HA logic operation could be directly observed from the photos of the AND and XOR logic gates under UV irradiation (Fig. 1E). The fluorescence of NMM could be directly visualized in the coexistence of the two inputs, corresponding to the AND gate operation. For the XOR gate, the fluorescence of DNA-AgNCs could be clearly observed in the presence of either IN1HA or IN2HA. The results matched the fluorescence results quite well, validating the excellent reproducibility of the logic operations. Of note, the developed integration strategy of the molecular device presents advantages that the required AND and XOR gate share a common threshold value, a universal platform and the same set of inputs. Furthermore, the use of NMM as the second signal indicator by combining G-rich sequence modulation has the advantages of avoiding chemical labels and simplifying logic operations. This also provides an instructive strategy to wirelessly integrate molecular devices on a universal platform for untraditional molecular computing with a simple operation. This successful integration inspired us to construct other advanced logic gates on the DNA-AgNCs platform.
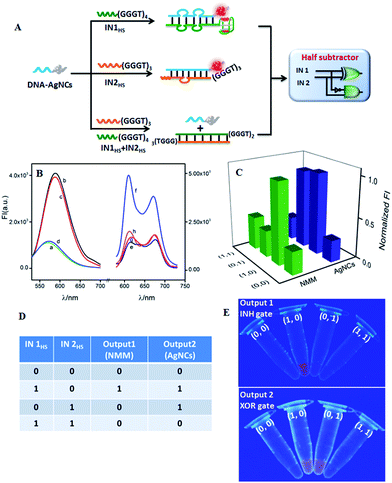
A half subtractor can implement a subtraction function of two bits by combining an XOR gate and an INH gate in parallel to produce a DIFFERENCE bit and a BORROW bit, respectively.51 Different from HA, an INH gate while not an AND gate is required by HS to integrate with an XOR gate. By modulating the input DNA sequences, an INH gate could feasibly be integrated with the developed XOR gate on the DNA-AgNCs platform. Fig. 2A outlines the operation principle of the as-developed HS. Both AgNCs and NMM reported a low fluorescence signal at the initial state (Fig. 2B(a) and (e), respectively). IN1HS containing the G-rich sequence (GGGTGGGTGGGTGGGT) at the 3′-terminal and IN2HS containing the G-rich sequence (GGGTGGGTGGGT) at the 3′-terminal could light up the fluorescence of the AgNCs by forming a duplex of DNA-AgNCs/IN1HS (Fig. 2B(b)) and DNA-AgNCs/IN2HS (Fig. 2B(c)), respectively. The coexistence of the two inputs produced a duplex of IN1HS/IN2HS due to the stronger affinity between IN1HS and IN2HS, leaving DNA-AgNCs alone with a low fluorescence response (Fig. 2B(d)). For the NMM-related INH gate, the G-rich sequence (GGGTGGGTGGGTGGGT) of INHS outstandingly enhanced the fluorescence of NMM by forming a G4/NMM complex (Fig. 2B(f)). The guanine base sequence (GGGTGGGTGGGT) at the 3′-terminal of IN2HS, however, was not sufficient to form a G4 configuration, producing a low fluorescence of NMM (Fig. 2B(g)). In the coexistence of the two inputs, the C-rich segment (CCCACCC) at the 5′-terminal of IN2HS hybridized with the G-rich segment at the 3′-terminal of IN1HS, inhibiting the formation of G4. Therefore, no enhanced fluorescence of NMM was monitored (Fig. 2B(h)). The modulation of the G4 configuration was validated by CD experiments (Fig. S11 in ESI†).
By plotting FImax(NMM) and FImax(AgNCs) against input combinations (Fig. 2C), the resulting truth table (Fig. 2D) indicates that the NMM-related INH logic gate and the AgNC-related XOR logic gate, respectively, performed the BORROW and DIFFERENT functions. The whole logic system implements binary arithmetic operations of (0 − 0 = 0 0), (1 − 0 = 0 1), and (1 − 1 = 0 0). The case of (0 − 1) is calculated via a high borrow to transfer it to (2 − 1 = 1), endowing both BORROW and DIFFERENT digits as “1” and finally getting the result of (0 − 1 = 1 1).50 Thus, the half subtractor was successfully achieved by integrating XOR and INH logic gates in parallel on a simple platform by hybridizing with two single strand DNA inputs in an enzyme-free and waste-free way. Fig. 2E exhibits the respective visions of INH and XOR logic operations under UV irradiation. The results further validate the reproducibility and reliability of the designed logic gate.
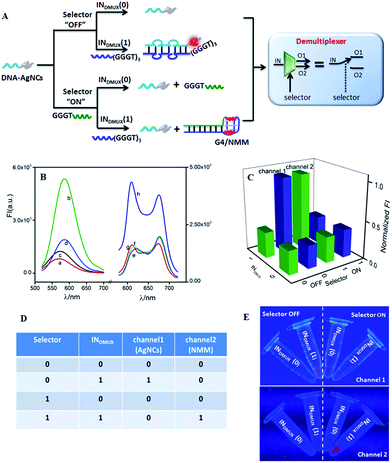
The developed half adder and half subtractor belong to the category of arithmetic logic gate. Molecular computing requires logic gates not only for arithmetic operations, but also for non-arithmetic operations, such as data transmission and compression/decompression, which are critical for information theory and computer science, while these have rarely been developed on a molecular scale.50 This inspired us to further construct non-arithmetic logic gates such as multiplexer (MUX) and demultiplexer (DMUX) on the DNA-AgNCs platform. Both multiplexer and demultiplexer consist of input, selector, and output, implementing data decompression and compression functions under selector control.52 A 1:2 DMUX can transmit one input (INDMUX) into two output channels via a selector (SDMUX) control, which could be realized by integrating an AND gate and an INH gate in parallel. As illustrated in Fig. 3A, the required AND logic gate with NMM as the signal indicator was integrated with the AgNC-related INH gate on the DNA-AgNCs platform to implement a demultiplexer function. Both the AgNCs and NMM exhibited a low fluorescence in the absence of INDMUX and SDMUX (Fig. 3B(a) and (e), respectively). The INDMUX could hybridize with the input-recognition segment of the DNA-AgNCs, forming a duplex of DNA-AgNCs/INDMUX. The G-rich sequence (GGGTGGGTGGGT) at the 3′-terminal of INDMUX was then approached to the AgNCs at the 5′-terminal of DNA-AgNCs, lighting up the fluorescence of AgNCs (Fig. 3B(b)). However, the G-rich sequence (GGGTGGGTGGGT) could not form G4 configuration, generating a low fluorescence of NMM (Fig. 3B(f)). The SDMUX contained the sequence of (GGGT) at the 5′-terminal, which was designed to neither hybridize with DNA-AgNCs nor form G4 configuration. Thus, neither the fluorescence of AgNCs (Fig. 3B(c)) nor the fluorescence of NMM (Fig. 3B(g)) was enhanced. In the coexistence of INDMUX and SDMUX, the duplex INDMUX/SDMUX rather than DNA-AgNCs/INDMUX was generated due to the higher affinity between INDMUX and SDMUX, resulting in a low fluorescence of DNA-AgNCs (Fig. 3B(d)). However, the formation of INDMUX/SDMUX gathered the G-rich sequence (GGGTGGGTGGGT) at the 3′-terminal of INDMUX and (GGGT) at the 5′-terminal of SDMUX together to generate a G4 configuration, leading to an outstanding fluorescence of NMM (Fig. 3B(h)). Therefore, the INDMUX was transmitted into channel 2 in the presence of SDMUX. By plotting the output channels of FImax(NMM) and FImax(AgNCs) against the input combinations (Fig. 3C), the demultiplexer operation could be conveniently obtained from the truth table (Fig. 3D). This can be instantly visualized from the photographs under UV irradiation (Fig. 3E). The input was transmitted into channel 1 with AgNCs as the signal indicator if the selector (SDMUX) was kept “OFF”. Otherwise, the INDMUX was transmitted into channel 2 with NMM as the signal indicator.
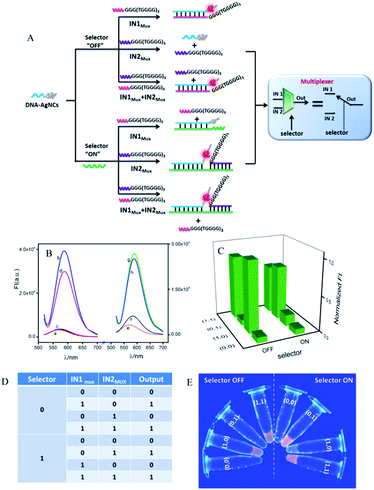
A 2:1 multiplexer (MUX) executes a reversed function to the 1:2 demultiplexer, which consists of two inputs, one selector and one output channel. By controlling the selector in the “ON” or “OFF” state, the two inputs can be selectively transmitted into a single output channel.53Fig. 4A depicts the operation principle of the developed 2:1 MUX with AgNCs as the signal reporter. Here, we first discussed the case in the absence of SMUX, i.e., when the selector is kept “OFF”. The original logic system reported a dim fluorescence signal in the absence of both inputs (IN1MUX and IN2MUX), Fig. 4B(a). Both IN1MUX and IN2MUX contained G-rich segments. The IN1MUX could hybridize with the DNA-AgNCs, thus transforming the AgNCs into bright-fluorescence-emitting clusters by the approaching G-rich segment, Fig. 4B(b). In contrast to IN1MUX, IN2MUX did not hybridize with DNA-AgNCs, resulting in a low output signal, Fig. 4B(c). In the coexistence of IN1MUX and IN2MUX, IN2MUX had no influence on the hybridization between IN1MUX and DNA-AgNCs. A high output signal was then monitored, which was similar to that in the presence of only IN1MUX, Fig. 4B(d). The results demonstrate that the multiplexer would report a high output signal of “1” in the presence of IN1MUX and a low output signal of “0” in the absence of IN1MUX, which was independent of the presence or absence of IN2MUX if the selector was absent. Considering the other case that the selector was in the state of “ON”, the added SMUX would associate with DNA-AgNCs to form a duplex of DNA-AgNCs/SMUX, which still reported a low output signal (Fig. 4B(e)). Here, DNA-AgNCs were designed to have a higher affinity to SMUX than to IN1MUX. Therefore, the addition of IN1MUX had no obvious influence on the duplex formation of SMUX/DNA-AgNCs. A low output signal would then be monitored (Fig. 4B(f)). With the addition of IN2MUX, the SMUX would act as linker, bringing IN2MUX and DNA-AgNCs together. The approaching G-rich segment of IN2MUX significantly enhanced the fluorescence of the AgNCs (Fig. 4B(g)). A similar augmented fluorescence signal was detected in the coexistence of SMUX, IN1MUX, and IN2MUX (Fig. 4B(h)), since IN1MUX also had no influence on the association among IN2MUX, SMUX, and DNA-AgNCs. Apparently, the multiplexer would report a high output signal in the presence of IN2MUX and a low output signal in the absence of IN2MUX no matter the absence or presence of IN1MUX if the selector was kept at “OFF”. The plotting of FIMAX against the input combinations (Fig. 4C) generated the corresponding truth table (Fig. 4D), which was further validated by the photographs of the multiplexer operation, triggered by UV irradiation (Fig. 4E). These results demonstrate that the multiplexer would transfer the state of IN1MUX into the output channel if the selector was kept “OFF”. It would transmit the state of the IN2MUX into the output channel if the selector was kept “ON”. This meets the feature of a 2:1 multiplexer.
Up to now, arithmetic logic gates of HA and HS and non-arithmetic logic gates of MUX and DMUX have been successfully constructed on the universal platform of DNA-AgNCs. Considering the reuse of the developed logic gates, resetting the logic system would be a possible way which could be realized by modifying the DNA-AgNCs on a magnetic nanoparticle surface (MNP/DNA-AgNCs). After finishing the logic operations, the logic system was heated at 90 °C for 10 min to de-hybridize the formed DNA duplexes. Under the assistance of a magnetic field, the MNP/DNA-AgNCs would be separated from the released DNA strands, which were then used to implement the next cycle of logic operations.
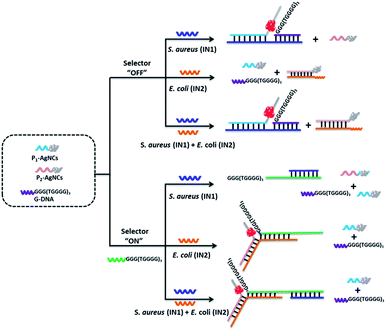
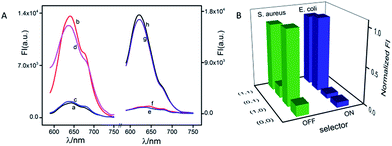
Except for data and information processing, the developed DNA logic system as an intelligent biosensor was further explored. As dangerous pathogenic bacteria, Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus) can cause foodborne infections and seriously threaten public health.54 Here, the E. coli gene and S. aureus gene were used as model targets to validate the logic-controlled detection via multiplexer function. To realize a 2:1 multiplexer-based intelligent detection, a G-rich DNA strand (G-DNA) and two DNA strand-templated AgNCs (P1-AgNCs and P2-AgNCs) were designed as the platform, which is illustrated in Fig. 5. Here, we first discuss the case in the absence of the selector DNA (S-DNA). The IN1 of the S. aureus gene acted as a linker to bridge P1-AgNCs and G-DNA together to form the complex IN1/P1-AgNCs/G-DNA. The dim fluorescence of the AgNCs (Fig. 6A(a)) was significantly enhanced by the approaching G-DNA (Fig. 6A(b)). In the presence of IN2, the E. coli gene, it would hybridize with P2-AgNCs and form a duplex of IN2/P2-AgNCs, which had no influence on the fluorescence of AgNCs (Fig. 6A(c)). In the coexistence of IN1 and IN2, IN1/P1-AgNCs/G-DNA and IN2/P2-AgNCs were generated, resulting in an enhanced fluorescence output signal (Fig. 6A(d)). The results demonstrate that the multiplexer would report a high output signal of “1” in the presence of the S. aureus gene and a low output signal of “0” in the absence of the S. aureus gene, which was independent of the presence or absence of the E. coli gene if the selector kept “OFF”.
With the addition of S-DNA, the IN1 of the S. aureus gene would hybridize with the S-DNA rather than with P1-AgNCs and G-DNA due to the higher affinity between IN1 and S-DNA. A low output signal similar to the original fluorescence (Fig. 6A(e)) was then monitored (Fig. 6A(f)). Different from that, IN2 of the E. coli gene would link P2-AgNCs and S-DNA together, forming a complex of IN2/P2-AgNCs/S-DNA. When the AgNCs of P2-AgNCs approach the G-rich sequence of S-DNA, an augmented fluorescence signal of AgNCs is produced (Fig. 6A(g)). In the coexistence of IN1 and IN2, IN1 would hybridize with S-DNA, which had no influence on the hybridization among IN2, P2-AgNCs and S-DNA, resulting in an enhanced output signal (Fig. 6A(h)). Obviously, the multiplexer would report a high output signal in the presence of the E. coli gene and a low output signal in the absence of the E. coli gene, which was independent of the presence or absence of the S. aureus gene if the selector was kept “ON”. By plotting the normalized fluorescence of the AgNCs against various input combinations (Fig. 6B), the results demonstrate that the multiplexer would transfer the state of the S. aureus gene into the output channel if the selector was kept “OFF” and would transmit the state of the E. coli gene into the output channel if the selector was switched “ON”. Meanwhile, the limit of detection reaches 100 fM for both E. coli and S. aureus gene detection due to the G-rich enhancement on the fluorescence of AgNCs (see Fig. S12 in ESI†).
Such a design enables us to identify multiple targets with a single signal indicator and also could help to make a rapid evaluation on various states of input in the sample.55 As illustrated in Table 1, the 2:1 multiplexer-based sensing system in the presence of the selector DNA was defined as detection kit A. The sensing system in the absence of the selector DNA was defined as detection kit B. For the decision making, the sample was divided into two parts and added into kit A and kit B, respectively. If both detection kits A and B reported a low output signal (“0”), then, neither S. aureus nor E. coli gene existed in the sample. If a high output (“1”) was detected from kit A, this indicated the existence of E. coli. The high output (“1”) of kit B suggested the presence of S. aureus. If both kits reported a high output (“1”), then, the sample contained both S. aureus and E. coli. The multiplexer-based biosensing system simplifies the operation effort and reduces the assay time, presenting a great potential application in high-throughput rapid detection.
| Output of detection kit A (with the S-DNA) | Output of detection kit B (without the S-DNA) | Rapid decision-making |
|---|---|---|
| 0 | 0 | Containing neither S. aureus nor E. coli |
| 1 | 0 | Containing E. coli |
| 0 | 1 | Containing S. aureus |
| 1 | 1 | Containing both S. aureus and E. coli |
Conclusions
We have successfully developed a novel strategy for the wireless integration of DNA logic gates into a simple and universal platform to implement multiple advanced arithmetic and non-arithmetic logic functions. To the best of our knowledge, this is the first time that a library of basic logic gates (YES, AND, OR, XOR and INH) and advanced logic gates (half adder, half subtractor, multiplexer and demultiplexer) have been constructed on the simplest single-strand DNA platform. The repeatability was further validated by instantly visualizing the logic operation under UV irradiation. All the logic gates shared a common threshold value, which would be beneficial for the integration of multiple molecular devices. The strategy of G-rich DNA sequence lighting up AgNCs and NMM for generating output signal avoids the chemical labeling of fluorophore and quencher, providing a cost-effective and powerful way for constructing complex logic gates. The enzyme-free logic operations were quite simple and entirely implemented on the basis of DNA strand-hybridization, thus eliminating waste accumulation in the system and reducing the overall time-consumption of DNA computing. Apart from a futuristic vision for molecular computing, the potential application of logic gates for intelligent detection has been further confirmed by distinguishing the E. coli and S. aureus genes with one output signal under multiplexer control. The developed strategy is straightforward and versatile, thus providing an instructive way for the integration of multiple molecular devices and intelligent diagnosis.Experimental details
The synthesis of DNA-AgNCs
The DNA-AgNCs were prepared according to a previous report.56 Briefly, AgNO3 solution (6 μM) was added into oligonucleotide solutions (1 μM) (phosphate buffer, 20 mM, pH 7.4) and incubated for 30 min at 4 °C. NaBH4 solution (6 μM) was then quickly added into the mixture under vigorous shaking. DNA-AgNCs with maximum emissions at 620 nm and 580 nm were obtained by modulating the mole ratio of DNA, Ag+ and NaBH4 with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 and 1
6 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, respectively. The resultant mixture was kept in the dark at 4 °C.
6, respectively. The resultant mixture was kept in the dark at 4 °C.
Native polyacrylamide gel electrophoresis
Polyacrylamide gels (20%) were prepared with 1× TBE buffer (pH 8.3). All the used DNAs were denatured by heating at 90 °C for 10 min and then slowly cooling down to about 25 °C. Each sample (9 μL) was mixed with loading buffer (1 μL) before loading onto the gel. The gel was run at a constant voltage of 100 V over a period of about 2.5 h. The gel was then immersed in SYBR@gold nucleic acid gel stain solution (1×) for about 30 min and then washed with water twice. The gel images were obtained from the Gel Doc™ XR+, Bio-RAD system (USA).UV photograph
The UV photographs of the as-prepared logic gates of YES, AND, OR, INH, XOR, HA, HS, MUX and DMUX were carried out under UV light irradiation with an excitation source of 365 nm. To take photographs of the advanced logic operations, HA, HS, and DMUX, the signal probes were added individually. NMM was absent when taking photographs for DNA-AgNCs-related logic operations. To directly observe NMM-related logic operations, only the DNA strand of the DNA-AgNCs, and not the DNA-AgNCs, was used.CD measurement
A Bio-Logic MOS450 spectropolarimeter (France) was utilized to collect the CD spectra of the oligonucleotides (4 μM) at room temperature. An average of three scans from 210 nm to 330 nm was recorded in an optical chamber (1 mm optical path length). The background of the buffer solution (PB buffer, 10 mM, pH 7.4, 10 mM K+) was subtracted from the CD data. Throughout the experiments, the lamp of the spectropolarimeter was always kept under a stable flow of dry purified nitrogen (99.99%) to avoid ozone production.Logic operation
DNA-AgNCs (maximum emission at 580 nm) were used as the platform for all the developed logic operations in this investigation, including YES, AND, OR, INH, XOR, HA, HS, MUX and DMUX. The required inputs with various combinations for each logic gate were added into the logic system according to the logic operation. After incubating for 1 h at room temperature, the fluorescence spectra of the DNA-AgNCs and NMM were recorded. The concentration of all the DNAs used was 500 nM for the YES, AND, OR, INH and XOR logic gates. The concentration of all the DNAs used was 200 nM for the HA, HS, MUX and DMUX logic gates. The concentration of NMM was 200 nM for the HA, HS and DMUX logic gates. All the used DNA strands cab be found in Table S1 in ESI.† All the chemicals and materials used can be found in ESI.†Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work is supported by the National Natural Science Foundation of China (No. 21575138 and No. 21775108). China International Science and Technology Cooperation Based of Food Nutrition/Safety and Medicinal Chemistry. The Tianjin Municipal Science and Technology Commission (Project No. 16PTSYJC00130). The International Science and Technology Cooperation Program of China (Project No. 2014DFR30350).Notes and references
- J. Macdonald, Y. Li, M. Sutovic, H. Lederman, K. Pendri, W. H. Lu, B. L. Andrews, D. Stefanovic and M. N. Stojanovic, Nano Lett., 2006, 6, 2598–2603 CrossRef CAS PubMed.
- R. Guliyev, S. Ozturk, Z. Kostereli and E. U. Akkaya, Angew. Chem., Int. Ed., 2011, 50, 9826–9831 CrossRef CAS PubMed.
- Y. V. Gerasimova and D. M. Kolpashchikov, Angew. Chem., Int. Ed., 2016, 55, 10244–10247 CrossRef CAS PubMed.
- K. S. Park, M. W. Seo, C. Jung, J. Y. Lee and H. G. Park, Small, 2012, 8, 2203–2212 CrossRef CAS PubMed.
- J. B. Zhu, L. B. Zhang, S. J. Dong and E. K. Wang, ACS Nano, 2013, 7, 10211–10217 CrossRef CAS PubMed.
- Y. H. Lin, Y. Tao, F. Pu, J. S. Ren and X. G. Qu, Adv. Funct. Mater., 2011, 21, 4565–4572 CrossRef CAS.
- J. Li, A. A. Green, H. Yand and C. H. Fan, Nat. Chem., 2017, 9, 1056–1067 CrossRef CAS PubMed.
- F. Pu, Z. Liu, J. S. Ren and X. G. Qu, Chem. Commun., 2013, 49, 2305–2307 RSC.
- F. Wang, C. H. Lu and I. Willner, Chem. Rev., 2014, 114, 2881–2941 CrossRef CAS PubMed.
- F. Pu, J. S. Ren and X. G. Qu, Adv. Mater., 2014, 26, 5742–5757 CrossRef CAS PubMed.
- M. N. Stojanovic, D. Stefanovic and S. Rudchenko, Acc. Chem. Res., 2014, 47, 1845–1852 CrossRef CAS PubMed.
- L. M. Adleman, Science, 1994, 266, 1021–1024 CAS.
- H. L. Li, W. Hong, S. J. Dong, Y. Q. Liu and E. K. Wang, ACS Nano, 2014, 8, 2796–2803 CrossRef CAS PubMed.
- L. Qian and E. Winfree, Science, 2011, 332, 1196–1201 CrossRef CAS PubMed.
- J. H. Chen, S. G. Zhou and J. L. Wen, Angew. Chem., Int. Ed., 2015, 54, 446–460 CAS.
- Y. Tam, Z. W. Dai, M. S. Chan, L. S. Liu, M. C. Cheung, F. Bolze, C. Tin and P. K. Lo, Angew. Chem., Int. Ed., 2016, 55, 164–168 CrossRef PubMed.
- J. Elbaz, F. Wang, F. Remacle and I. Willner, Nano Lett., 2012, 12, 6049–6054 CrossRef CAS PubMed.
- H. L. Li, S. J. Guo, Q. H. Liu, L. D. Qin, S. J. Dong, Y. Q. Liu and E. K. Wang, Adv. Sci., 2015, 2, 1500054 CrossRef PubMed.
- R. J. Pei, E. Matamoros, M. H. Liu, D. Stefanovic and M. Stojanovic, Nat. Nanotechnol., 2010, 5, 773–777 CrossRef CAS PubMed.
- R. Orbach, B. Willner and I. Willner, Chem. Commun., 2015, 51, 4144–4160 RSC.
- C. W. Brown III, M. R. Lakin, E. K. Horwitz, L. Fanning, H. E. West, D. Stefanovic and S. W. Graves, Angew. Chem., Int. Ed., 2014, 53, 7183–7187 CrossRef PubMed.
- J. Elbaz, O. Lioubashevski, F. Wang, F. Remacle, R. D. Levine and I. Willner, Nat. Nanotechnol., 2010, 5, 417–422 CrossRef CAS PubMed.
- R. Orbach, F. Wang, O. Lioubashevski, R. D. Levine and F. Remacle, Chem. Sci., 2014, 5, 3381–3387 RSC.
- B. M. Frezza, S. L. Cockroft and M. R. Ghadiri, J. Am. Chem. Soc., 2007, 129, 14875–14879 CrossRef CAS PubMed.
- J. B. Zhu, L. B. Zhang, T. Li, S. J. Dong and E. K. Wang, Adv. Mater., 2013, 25, 2440–2444 CrossRef CAS PubMed.
- H. Pei, L. Liang, G. B. Yao, J. Li, Q. Huang and C. H. Fan, Angew. Chem., Int. Ed., 2012, 51, 9020–9024 CrossRef CAS PubMed.
- A. J. Thubagere, C. Thachuk, J. Berleant, R. F. Johnson, D. A. Ardelean, K. M. Cherry and L. L. Qian, Nat. Commun., 2017, 8, 14373 CrossRef CAS PubMed.
- E. G. Gwinn, P. O'Neill, A. J. Guerrero, D. Bouwmeester and D. K. Fygenson, Adv. Mater., 2008, 20, 279–283 CrossRef CAS.
- D. Q. Fan, J. B. Zhu, Y. Q. Liu, E. K. Wang and S. J. Dong, Nanoscale, 2016, 8, 3834–3840 RSC.
- T. Li, L. B. Zhang, J. Ai, S. J. Dong and E. K. Wang, ACS Nano, 2011, 5, 6334–6338 CrossRef CAS PubMed.
- W. W. Guo, J. P. Yuan, Q. Z. Dong and E. K. Wang, J. Am. Chem. Soc., 2010, 132, 932–934 CrossRef CAS PubMed.
- H. C. Yeh, J. Sharma, J. J. Han and J. S. Martinez, Nano Lett., 2010, 10, 3106–3110 CrossRef CAS PubMed.
- N. Enkin, F. Wang, E. Sharon, H. B. Albada and I. Willner, ACS Nano, 2014, 8, 11666–11673 CrossRef CAS PubMed.
- M. X. You, L. Peng, N. Shao, L. Q. Zhang, L. P. Qiu, C. Cui and W. H. Tan, J. Am. Chem. Soc., 2014, 136, 1256–1259 CrossRef CAS PubMed.
- J. Hemphill and A. Deiters, J. Am. Chem. Soc., 2013, 135, 10512–10518 CrossRef CAS PubMed.
- Z. Xie, L. Wroblewska, L. Prochazka, R. Weiss and Y. Benenson, Science, 2011, 333, 1307–1311 CrossRef CAS PubMed.
- W. Liao, Y. Sohn, M. Riutin, A. Cecconello, W. J. Parak, R. Nechushtai and I. Willner, Adv. Funct. Mater., 2016, 26, 4262–4273 CrossRef CAS.
- C. C. Kloss, M. Condomines, M. Cartellieri, M. Bachmann and M. Sadelain, Nat. Biotechnol., 2013, 31, 71–75 CrossRef CAS PubMed.
- M. I. Shukoor, M. O. Altman, D. Han, A. T. Bayrac, I. Ocsoy, Z. Zhu and W. H. Tan, ACS Appl. Mater. Interfaces, 2012, 4, 3007–3011 CAS.
- H. C. Yeh, J. Sharma, I.-M. Shih, D. M. Vu, J. S. Martinez and J. H. Werner, J. Am. Chem. Soc., 2012, 134, 11550–11558 CrossRef CAS PubMed.
- K. Szaciłowski, Chem. Rev., 2008, 108, 3481–3548 CrossRef PubMed.
- F. Pu, E. Ju, J. S. Ren and X. G. Qu, Adv. Mater., 2014, 26, 1111–1117 CrossRef CAS PubMed.
- G. Seelig, D. Soloveichik, D. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 CrossRef CAS PubMed.
- S. Gibilisco, The Illustrated Dictionary of Electronics, ed. S. Gibilisco, McGraw-Hill, New York, 2001, p. 81 Search PubMed.
- Y. Q. Liu, J. T. Ren, Y. A. Qin, J. Li, J. Y. Liu and E. K. Wang, Chem. Commun., 2012, 48, 802–804 RSC.
- D. Q. Fan, K. Wang, J. B. Zhu, Y. Xia, Y. C. Han, Y. Q. Liu and E. K. Wang, Chem. Sci., 2015, 6, 1973–1978 RSC.
- S. S. Oh, K. Plakos, X. H. Lou, Y. Xiao and H. T. Soh, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14053–14058 CrossRef CAS PubMed.
- H. L. Li, J. Y. Liu, Y. X. Fang, Y. N. Qin, S. L. Xu, Y. Q. Liu and E. K. Wang, Biosens. Bioelectron., 2013, 41, 563–568 CrossRef CAS PubMed.
- S. L. Xu, H. L. Li, Y. Q. Miao, Y. Q. Liu and E. K. Wang, NPG Asia Mater., 2013, 5, e76 CrossRef CAS.
- J. Andréasson and U. Pischel, Chem. Soc. Rev., 2010, 39, 174–188 RSC.
- C. N. Yang, C. Y. Hsu and Y. C. Chuang, Chem. Commun., 2012, 48, 112–114 RSC.
- C. T. Wu, K. Wang, D. Q. Fan, C. Y. Zhou, Y. Q. Liu and E. K. Wang, Chem. Commun., 2015, 51, 15940–15943 RSC.
- J. Andréasson, S. D. Straight, S. Bandyopadhyay, R. H. Mitchell, T. A. Moore, A. L. Moore and D. Gust, Angew. Chem., Int. Ed., 2007, 46, 958–961 CrossRef PubMed.
- H. X. Yuan, Z. Liu, L. B. Liu, F. T. Lv, Y. L. Wang and S. Wang, Adv. Mater., 2014, 26, 4333–4338 CrossRef CAS PubMed.
- S. I. Stoeva, J. S. Lee, J. E. Smith, S. T. Rosen and C. A. Mirkin, J. Am. Chem. Soc., 2006, 128, 8378–8379 CrossRef CAS PubMed.
- J. T. Petty, J. Zheng, N. V. Hud and R. M. Dickson, J. Am. Chem. Soc., 2004, 126, 207–5212 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Chemicals, materials and DNA sequences used in the investigation, the construction of YES, AND, OR, XOR and INH logic gates, CD and PAGE experimental results. See DOI: 10.1039/c7sc05246d |
| This journal is © The Royal Society of Chemistry 2018 |