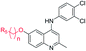Open Access Article
Open Access ArticleDesign, synthesis, and biological evaluation of AV6 derivatives as novel dual reactivators of latent HIV-1†
Mingtao Ao‡
a,
Zhenrui Pan‡a,
Yuqing Qian‡a,
Bowen Tanga,
Zeming Feng a,
Hua Fangc,
Zhen Wua,
Jingwei Chen*b,
Yuhua Xue*a and
Meijuan Fang
a,
Hua Fangc,
Zhen Wua,
Jingwei Chen*b,
Yuhua Xue*a and
Meijuan Fang *a
*a
aSchool of Pharmaceutical Sciences, Fujian Provincial Key Laboratory of Innovative Drug Target Research, Xiamen University, South Xiang-An Road, Xiamen, 361102, China. E-mail: xueyuhua@xmu.edu.cn; fangmj@xmu.edu.cn; Fax: +86-592-2189868; Tel: +86-592-2189868
bKey Laboratory for Chemical Biology of Fujian Province, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, China. E-mail: jwchen@xmu.edu.cn
cThe Third Institute of Oceanography of the State Oceanic Administration, Xiamen, 361005, China
First published on 11th May 2018
Abstract
The “shock and kill” strategy might be a promising therapeutic approach for HIV/AIDS due to the existence of latent viral reservoirs. A major challenge of the “shock and kill” strategy arises from the general lack of clinically effective latency-reversing agents (LRAs). The 2-methylquinoline derivative, antiviral 6 (AV6) has been reported to induce latent HIV-1 expression and act synergistically with a HDAC inhibitor VA to reverse HIV latency. We report herein the design and identification of AV6 analogues which possess the zinc-binding group of HDAC inhibitors and have dual acting mechanism for the reactivation of HIV-1 from latency. Evaluation of compounds for the reactivation of HIV-1 latency identified two excellent active compounds 12c and 12d. Further bioassays revealed that these two compounds reactivated latent HIV-1 through dual mechanism, the inhibition of HDACs and NFAT-required for early HIV-1 gene expression. Additionally, it was found that 12c and 12d could reactivate HIV-1 transcription by releasing P-TEFb from the inactive complex 7SK snRNP. At last, molecular docking identified their orientation and binding interactions at the active site of HDAC2. This experimental data suggests that 12c and 12d can be served as effective HIV-1 LRAs which can be taken up for further studies.
1. Introduction
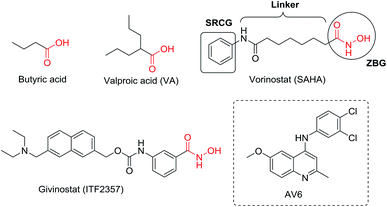
The latent HIV reservoirs in infected patients are the major barrier to eliminate HIV as reported.1,2 Recently, scientists have made efforts to use the “shock and kill” strategy to eradicate latently infected cells.3–10 In the “shock and kill” approach, latency-reversing agents (LRAs) should be used to force the replication and expression of latent HIV proviruses in the initial “shock” phase, and then the reactivated cells would be eliminated (“kill” phase) by a combination of viral cytopathic effects and the host immune response, as well as concomitant highly active anti-retroviral therapy (HAART) to prevent new infections.11–13 Currently, a wide range of small molecule have been explored as LRAs to reactivate latent provirus, including histone deacetylase (HDAC) inhibitors (Fig. 1), methyltransferase inhibitors, protein kinase C (PKC) activators, positive transcription elongation factor b (P-TEFb) activators and other unclassified HIV-1 latency-reversing agents.14–18 HDAC inhibitors, such as valproic acid (VA), vorinostat (also known as SAHA), and panobinostat have been used in clinical trials in HIV.19–23 However, preclinical and clinical studies suggest that these LRAs which target a single mechanism involved in latency may not be efficient enough to reactivate latent HIV-1 in vivo.24–27 Hence, there is an urgent requirement of developing more effective latency-reversing interventions with a unique structure and effective mode of action.Antiviral 6 (AV6, Fig. 1), a novel HIV-1 latency-reversing agent identified through high-throughput screening, has been reported to reproducibly activate latent HIV-1 in different cell-based latency model systems without causing general T cell proliferation or activation.28 Its activation is independent of the site of provirus integration, which complements the latency antagonist activity of HDAC inhibitor. For example, AV6 was demonstrated to act cooperatively with VA (a HDAC inhibitor) for enhanced induction of HIV-1 gene expression.28 This is a proof of concept showing that the combination of two or more inducers of HIV gene expression acting in accord via different mechanisms may produce a more potent reactivation effect.29,30 In this study, we design the AV6 analogues as LRAs with the goal of achieving HDACs inhibition. Key to the design is to identify the attachment appoint and the linker for the introduction of the essential active group of HDACs. Herein, we performed the chemical synthesis and biological evaluation of the first chemical series of AV6 analogues (series I) to determine the suitable attachment appoint for the introduction of the functional group of HDAC inhibitor. Then, a linker optimization strategy was pursued to obtain effective LRAs bearing AV6 scaffold structure and the functional group of HDAC inhibitors. These new AV6 derivatives (series II) were evaluated for their reactivation of latent HIV. The most potent reactivators were then taken up to do further bioassays.
2. Results and discussion
2.1. Design of the target compounds
Multi-target drugs have emerged as an innovative therapeutic approach for many diseases due to their complex etiology and their multifactorial progression.31–33 AV6 and HDAC inhibitors have been validated as two inducers of HIV gene expression acting in accord via two different mechanisms. The N-phenylquinoline nucleus of AV6 is considered as ‘Master Key Core’ in many compounds aimed at different targets to bring out varied pharmacological properties. Besides, many HDACIs have been designed and synthesized as dual- or multi-targeted inhibitors for the cancer therapy,34 and the synergistic effect on the activation of latent provirus between VA (a carboxylate HDACi) and AV6 has been confirmed.28 Thus, we developed an idea that the introduction of the function groups of HDACis in the N-phenylquinoline structural scaffold of AV6 might also result in new compounds both targeting HDACs and with AV6 related active mechanism to generate more potent LRAs.There are only three class I HDACs (HDAC 1, HDAC 2, HDAC 3) which are localized at the integrated HIV-1 LTR and are zinc-dependent deacetylasesas.14,35 The classical pharmacophore for their HDACi consists of a zinc-binding group (ZBG), an appropriate linker and a surface recognition cap group.36 Generally, ZBG plays a significant role in the binding efficiency between HDACi and enzyme. Therefore, we intend to incorporate the essential active group ZBG of HDACis into the N-phenylquinoline scaffold to generate new small molecule entities (AV6 analogues) which may display better potency to activate latent HIV-1. However, this is not a straight-forward procedure, and we must identify the appropriate common features, substitution sites, and then growing vectors.
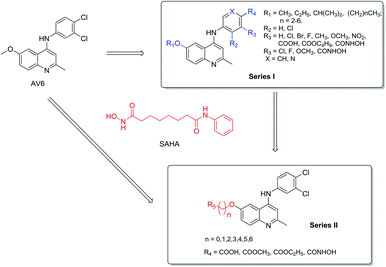
We hypothesized that the substituents on the benzene or quinoline rings contained in AV6 structure might modulate the activation activity of the compound. Therefore, we initially designed series I derivatives (Fig. 2, compounds 7b-7B) of AV6 to evaluate the influence of different substituents and various attachment points on the activation activity and figure out the best site for the introduction of the functional group ZBG of the HDACis. There are various ZBGs for HDACi, such as hydroxamic acids, aminobenzamides, carboxylates (short-chain fatty acids), electrophilic ketones, thiols, mercaptoacetamides, and 3-hydroxypyridin-2-thiones. Among all types of ZBGs, hydroxamic acid and carboxylate are common zinc ion chelating groups. So hydroxamic acid and carboxylates with different linker were subsequently incorporated into the suitable site of N-phenylquinoline nucleus in order to identify more potent novel LRAs (Fig. 2, series II). In this paper, we report the design, synthesis, and biological evaluation of AV6 analogues as novel LRAs with dual activated mechanism.
2.2. Syntheses of the designed compounds
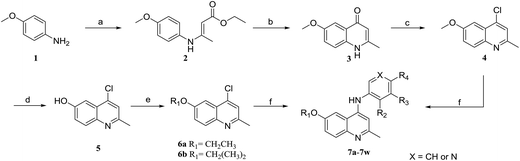
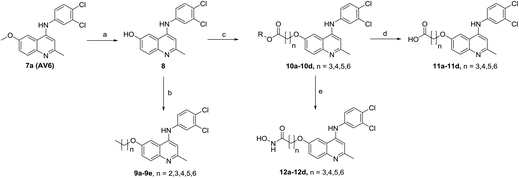
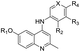
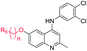
The synthetic approaches for the two different series of target compounds are outlined in Scheme 1 and 2. Compounds 7a–7w were prepared according to a four-step procedure described previously and shown in Scheme 1.37 p-Anisidine was condensed with ethyl acetoacetate to form compound 2, which was cyclized in diphenyl ether affording 6-methoxy-4-hydroxy-2-methylquinoline (3). The treatment of 3 with phosphorus oxychloride to provide the key intermediate 4-chloro-6-methoxy-2-methylquinoline (4).38–40 Compounds 7a–7e could be easily obtained through the reaction of 4 with different nucleophilic amine derivatives in the presence of catalytic amount of HCl. On the other hand, demethylation of 4 using BBr3 produced 4-chloro-2-methylquinolin-6-ol (5). Alkylation of 5 with bromoethane or 2-bromopropane provided 6a or 6b, and the reaction of halides (6a or 6b) with corresponding amine derivatives produced compounds 7f–7w (Scheme 1).41,42The designed products 9a–9e, 10a–10d, 11a–11d and 12a–12d (series II) were synthesized according to the pathway shown in Scheme 2. Demethylation on C-6 position of compound 7a gave corresponding C6–OH quinolone (8). Compound 8 converted to compounds 9a–9e and 10a–10d by alkylation of the C6–OH with appropriate alkyl halide and various lengths of ethyl or methyl bromoalkanoate, respectively. Carboxylic acid derivatives 11a–11d were finally obtained by the hydrolysis of esters 10a–10d. Conversion of esters 10a–10d to hydroxamic acids using freshly prepared hydroxylamine yielded the target compounds 12a–12d.43
The structures of the target compounds were confirmed with 1H-NMR, 13C-NMR, and high-resolution mass spectrometry (HRMS).
2.3. Biological evaluation
| Compound | X | R1 | R2 | R3 | R4 | % GFP(+) cells (10 μM) | % GFP(+) cells (5 μM) |
|---|---|---|---|---|---|---|---|
| a Data are presented as mean ± SD of at least three independent experiments performed in triplicate. Cells were treated with the indicated compounds at 5, 10 μM for 24 h. The expression of GFP were evaluated by FACS. | |||||||
| DMSO | 0.7 ± 0.1 | 0.7 ± 0.1 | |||||
| SAHA | 71.2 ± 4.2 | 56.8 ± 1.2 | |||||
| AV6/7a | CH | CH3 | H | Cl | Cl | 57.0 ± 1.2 | 3.8 ± 1.1 |
| 7b | CH | CH3 | H | Br | Cl | 8.3 ± 2.5 | 10.8 ± 0.3 |
| 7c | CH | CH3 | H | F | Cl | 1.8 ± 0.1 | 1.2 ± 0.1 |
| 7d | CH | CH3 | H | CH3 | Cl | 2.1 ± 0.3 | 0.9 ± 0.1 |
| 7e | CH | CH3 | H | OCH3 | Cl | 6.4 ± 0.5 | 1.9 ± 0.1 |
| 7f | CH | CH2CH3 | H | Cl | Cl | 47.3 ± 3.3 | 4.1 ± 0.1 |
| 7g | CH | CH2CH3 | H | Br | Cl | 39.9 ± 4.5 | 11.6 ± 2.3 |
| 7h | CH | CH2CH3 | H | F | Cl | 28.5 ± 1.6 | 15.3 ± 2.6 |
| 7i | CH | CH2CH3 | H | CH3 | Cl | 14.5 ± 0.5 | 8.5 ± 1.2 |
| 7j | CH | CH2CH3 | H | OCH3 | Cl | 5.1 ± 0.9 | 2.3 ± 0.6 |
| 7k | CH | CH2CH3 | H | NO2 | Cl | 1.3 ± 0.9 | 0.9 ± 1.9 |
| 7l | CH | CH2CH3 | H | COOH | Cl | 1.0 ± 0.1 | 0.9 ± 0.2 |
| 7m | CH | CH2CH3 | H | COOC4H9 | Cl | 5.2 ± 1.1 | 3.8 ± 0.9 |
| 7n | CH | CH2CH3 | H | CONHOH | Cl | 0.6 ± 0.1 | 0.6 ± 0.1 |
| 7o | CH | CH(CH3)2 | H | Cl | Cl | 0.9 ± 0.1 | 0.8 ± 0.1 |
| 7p | CH | CH(CH3)2 | H | Br | Cl | 0.9 ± 0.1 | 0.8 ± 0.1 |
| 7q | CH | CH(CH3)2 | H | F | Cl | 1.2 ± 0.2 | 0.8 ± 0.2 |
| 7r | CH | CH(CH3)2 | H | CH3 | Cl | 0.9 ± 0.2 | 0.8 ± 0.1 |
| 7s | CH | CH(CH3)2 | H | OCH3 | Cl | 1.3 ± 0.4 | 0.8 ± 0.3 |
| 7t | CH | CH(CH3)2 | H | NO2 | Cl | 1.5 ± 0.3 | 0.9 ± 0.3 |
| 7u | CH | CH(CH3)2 | H | COOH | Cl | 2.8 ± 0.1 | 1.5 ± 0.1 |
| 7v | CH | CH(CH3)2 | H | COOC4H9 | Cl | 1.4 ± 0.3 | 0.8 ± 0.2 |
| 7w | CH | CH3 | Cl | H | Cl | 0.7 ± 0.1 | 0.6 ± 0.1 |
| 7x | CH | CH2CH3 | CH3 | H | F | 1.0 ± 0.2 | 1.2 ± 0.2 |
| 7y | CH | CH2CH3 | H | H | CONHOH3 | 1.0 ± 0.2 | 0.9 ± 0.2 |
| 7z | CH | CH(CH3)2 | H | OCH3 | OCH3 | 0.9 ± 0.3 | 0.7 ± 0.1 |
| 7A | N | CH3 | H | Cl | Cl | 5.4 ± 0.4 | 5.1 ± 1.3 |
| 7B | N | CH2CH3 | H | Cl | Cl | 1.3 ± 0.3 | 0.8 ± 0.1 |
| Compound | n | R5 | % GFP(+) cells (μM) | % Cell viability (μM) | ||||
|---|---|---|---|---|---|---|---|---|
| 10 | 5 | 1 | 10 | 5 | 1 | |||
| a Data are presented as mean ± SD of at least three independent experiments performed in triplicate. Cells were treated with the indicated compounds at 1, 5, 10 μm for 24 h. The expression of GFP were evaluated by FACS. | ||||||||
| DMSO | 0.7 ± 0.1 | 0.7 ± 0.2 | 0.7 ± 0.2 | 98.6 ± 0.5 | 98.1 ± 1.9 | 98.8 ± 0.6 | ||
| SAHA | 71.2 ± 4.2 | 56.8 ± 1.2 | 20.1 ± 0.3 | 43.2 ± 3.2 | 50.9 ± 5.1 | 80.4 ± 2.4 | ||
| AV6/7a | 1 | H | 57.0 ± 1.2 | 3.8 ± 1.1 | 0.5 ± 0.5 | 47.6 ± 5.6 | 73.4 ± 3.5 | 80.7 ± 5.4 |
| 7f | 1 | CH3 | 43.3 ± 0.3 | 21.1 ± 1.2 | 2.4 ± 0.3 | 43.2 ± 3.2 | 50.9 ± 5.1 | 69.9 ± 2.4 |
| 7o | 0 | CH(CH3)2 | 0.9 ± 0.1 | 0.8 ± 0.1 | 0.8 ± 0.1 | 67.3 ± 3.3 | 76.7 ± 2.6 | 85.2 ± 2.1 |
| 9a | 2 | CH3 | 25.4 ± 4.1 | 18.6 ± 1.8 | 2.3 ± 0.3 | 17.7 ± 3.1 | 41.5 ± 1.6 | 56.1 ± 3.8 |
| 9b | 3 | CH3 | 19.9 ± 1.9 | 22.8 ± 1.0 | 5.7 ± 1.3 | 13.1 ± 2.3 | 34.1 ± 0.4 | 60.3 ± 0.5 |
| 9c | 4 | CH3 | 19.7 ± 0.7 | 21.6 ± 0.9 | 3.1 ± 0.5 | 19.6 ± 3.9 | 39.6 ± 0.8 | 57.6 ± 4.3 |
| 9d | 5 | CH3 | 24.5 ± 2.8 | 29.1 ± 2.2 | 5.6 ± 1.2 | 17.2 ± 3.1 | 37.6 ± 3.1 | 58.2 ± 3.8 |
| 9e | 6 | CH3 | 32.4 ± 2.9 | 11.1 ± 1.8 | 1.4 ± 0.1 | 20.4 ± 2.5 | 52.3 ± 1.0 | 72.9 ± 3.3 |
| Compound | n | R5 | % GFP(+) cells (μM) | % Cell viability (μM) | ||||
|---|---|---|---|---|---|---|---|---|
| 10 | 5 | 1 | 10 | 5 | 1 | |||
| a Data are presented as mean ± SD of at least three independent experiments performed in triplicate. Cells were treated with the indicated compounds at 1, 5, 10 μm for 24 h. The expression of GFP were evaluated by FACS. | ||||||||
| DMSO | 0.7 ± 0.1 | 0.7 ± 0.2 | 0.7 ± 0.2 | 98.6 ± 0.5 | 98.1 ± 1.9 | 98.8 ± 0.6 | ||
| SAHA | 71.2 ± 4.2 | 56.8 ± 1.2 | 20.1 ± 0.3 | 43.2 ± 3.2 | 50.9 ± 5.1 | 80.4 ± 2.4 | ||
| AV6/7a | 1 | H | 57.0 ± 1.2 | 3.8 ± 1.1 | 0.5 ± 0.5 | 47.6 ± 5.6 | 73.4 ± 3.5 | 80.7 ± 5.4 |
| 10a | 3 | COOC2H5 | 3.3 ± 0.2 | 3.2 ± 0.5 | 0.8 ± 0.1 | 94.3 ± 2.5 | 97.1 ± 2.9 | 86.3 ± 3.4 |
| 10b | 4 | COOCH3 | 1.6 ± 0.7 | 1.4 ± 0.4 | 0.9 ± 0.2 | 92.6 ± 3.4 | 95.6 ± 3.7 | 85.8 ± 4.7 |
| 10c | 5 | COOC2H5 | 0.7 ± 0.2 | 1.5 ± 0.5 | 0.9 ± 0.2 | 95.4 ± 3.3 | 97.3 ± 2.9 | 87.0 ± 3.1 |
| 10d | 6 | COOCH3 | 0.9 ± 0.2 | 1.7 ± 0.3 | 0.8 ± 0.1 | 96.6 ± 3.2 | 96.7 ± 2.5 | 86.1 ± 2.3 |
| 11a | 3 | COOH | 0.6 ± 0.1 | 0.7 ± 0.2 | 0.9 ± 0.1 | 97.6 ± 2.8 | 97.1 ± 1.9 | 76.8 ± 2.6 |
| 11b | 4 | COOH | 1.1 ± 0.1 | 1.2 ± 0.1 | 0.7 ± 0.1 | 95.6 ± 2.4 | 97.2 ± 3.0 | 83.7 ± 3.5 |
| 11c | 5 | COOH | 0.8 ± 0.1 | 1.1 ± 0.1 | 0.7 ± 0.2 | 94.6 ± 2.8 | 95.4 ± 2.9 | 94.9 ± 0.6 |
| 11d | 6 | COOH | 1.0 ± 0.1 | 1.1 ± 0.1 | 0.9 ± 0.2 | 97.6 ± 3.5 | 96.3 ± 1.9 | 97.8 ± 1.6 |
| 12a | 3 | CONHOH | 2.7 ± 0.1 | 1.3 ± 0.9 | 1.1 ± 0.3 | 85.7 ± 4.2 | 94.2 ± 1.1 | 95.3 ± 3.1 |
| 12b | 4 | CONHOH | 19.7 ± 2.1 | 2.8 ± 1.7 | 0.7 ± 0.6 | 54.1 ± 3.7 | 89.5 ± 4.2 | 89.3 ± 4.8 |
| 12c | 5 | CONHOH | 87.2 ± 2.0 | 81.5 ± 1.5 | 39.7 ± 4.1 | 44.1 ± 3.4 | 49.8 ± 3.9 | 44.2 ± 3.6 |
| 12d | 6 | CONHOH | 84.9 ± 3.5 | 77.1 ± 2.4 | 49.9 ± 3.5 | 38.6 ± 3.1 | 32.7 ± 3.4 | 39.1 ± 2.9 |
At the first initial exploration, a variety of substituents and different attachment points at the benzene or quinoline rings contained in AV6 structure were examined. As shown in Table 1, compound 7a (AV6) with 6-methoxy and 3′,4′-dichloro substitutions induced 57.0 ± 1.2 percent GFP production in J-Lat A2 cells at 10 μM. Upon comparison of the activities of 6-ethoxy derivatives 7f–7j and 6-isopropoxy derivatives 7o–7s with 6-methoxy derivatives 7a–7e, we found that the reactivation potency of the derivatives for latent HIV-1 is very sensitive to the type of R1 substituent. Compounds 7o–7s have no effect on HIV-1 gene expression in J-Lat A2 cells, which suggest that the replacement of the methoxy group with the isoproyloxy group at C-6 position of quinoline ring is very detrimental for the reactivation potency of latent HIV-1. While in 6-ethoxy derivatives 7f–7j, 7f exhibits good activity of latent HIV-1 reversing (47.3 ± 3.3% at 10 μM), and compounds 7g–7i produce enhanced potency (14.5–39.9% of the GFP expression at 10 μM) compared to corresponding 6-methoxy derivatives 7b–7d (with GFP% values ranged from 2.1–8.3% at 10 μM). These results imply that the C-6 position (R1) of the quinoline ring may be a promising attachment point for the introducing of the ZBG groups by using the linear alkyl linker.
On the other hand, the replacement of Cl with linker groups (CH3 (7d and 7r) and OCH3 (7e, 7j, 7s, and 7z)), ZBG groups (COOH (7l and 7u), COOR (7m and 7v), and CONHOH (7n and 7y)), or other groups (NO2 (7k, and 7t)) at the C-3′ (R3) and C-4′ (R4) positions of phenyl ring leads to the disappearance of activity as shown in Table 1. It indicates that 3′,4′-dichloro substitutions at the phenyl ring are essential for potent reactivation of latent HIV-1 gene expression. Moreover, compounds 7A (5.4 ± 0.4%) and 7B (1.3 ± 0.3%) in which the phenyl ring is changed to the pyridine ring display weak or no induction of GFP expression in J-Lat A2 cells at 10 μM in comparison with AV6/7a (57.0 ± 1.2%) and 7f (47.3 ± 3.3%), respectively. It implies that the phenyl ring is also essential for the activity. Additionally, 7b, 7f–7j, 7m, and 7A also showed certain degree of activation effect (with GFP% values > 2.0%) at 5 μM. Among them, 6-ethoxy derivatives 7f–7j could reactivated latent HIV-1 in a dose-dependent manner better than 6-methoxy derivatives 7a–7e. Above all, the position C-6 (R1) of quinoline ring might be better suited for the introduction of ZBG group than the positions R3 and R4 of benzene ring, and the ZBG group would like to be introduced in R1 position by using linear linker.
To explore the effect of chain length of linear linker on activity, a series of ether analogues of AV6 (9a–9e) with 3–7 carbons of n-alkyl chain at the C-6 position of the quinolone ring was generated and their reactivation of latent HIV-1 gene expression was evaluated. As shown in Table 2, all ether analogues of AV6 (9a–9e) with 3–7 carbons of linear alkyl chain retained potent activities in the induction of GFP expression in J-Lat A2 cells at 10 μM (19.7 ± 0.7%–32.4 ± 2.9%) and 5 μM (11.1 ± 1.8%–29.1 ± 2.2%). It was further confirmed that together with the oxygen connection, the straight alkyl chain could be used as linear linker for the introduction of ZBG groups at 6-position of quinolone ring.
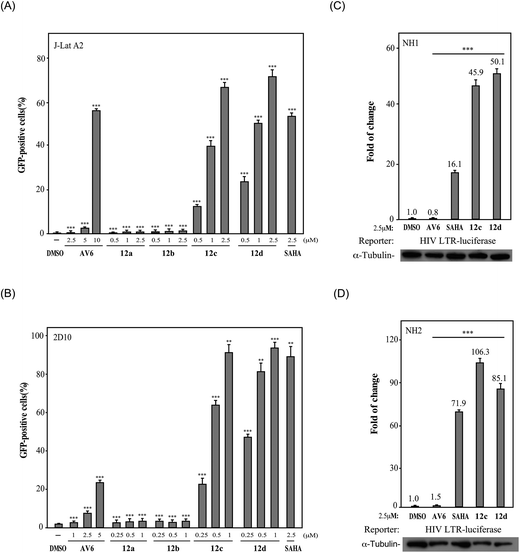
Finally, we investigated the effect of various ZBG groups with linear alky linker connected to the C-6 position of the quinoline ring on latent HIV-1 reactivation. Different ZBG groups (COOC2H5, COOH, and CONHOH) were introduced at the terminal of the straight alkyl chain on 6-position of quinolone ring, and another three series of AV6 analogues (10a–10d, 11a–11d, and 12a–12d) were generated and analyzed. Among these new derivatives with various ZBG groups, compounds 10a–10d (carboxylates) and 11a–11d (carboxylic acids) were either weakly active or completely inactive at tested concentrations (Table 3). Compounds with hydroxamic acid side chain showed varying degrees of potency to reverse HIV latency (12b–12d), except for compound 12a. Compounds 12c and 12d exerted a remarkable improvement in the latent HIV-1 reactivation activity, inducing 39.7 ± 4.1% and 49.9 ± 3.5% GFP production in J-Lat A2 cells at 1 μM, respectively. However, they displayed slightly higher toxicity than SAHA and AV6 according to the cell viability (Table 2). In this experiment, we also observed that the length of the alkyl linker with oxygen bonded connection at the C-6 position had an important effect on the activity. For example, for compounds that had the same terminal basic group –CONHOH, the latency-reversing activity of 12c (n = 5) and 12d (n = 6) were much better than that of 12a (n = 3) and 12b (n = 4). These results indicate that the length of the linear alkyl linker (n) and its terminal basic group (R5) have important impacts on the compounds regarding their binding activities.
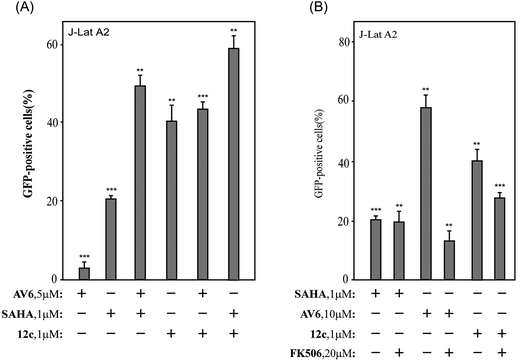
Among all these AV6 analogues, 12c and 12d showed the most optimal properties in terms of its efficient latency-reversing activity and displayed relatively mild cytotoxicity, thus they were selected for further bioassays as outlined below.
2.3.3.1 Activate latent HIV-1 via the NFAT-required mechanism. It was reported that AV6 requires nuclear factor of activated T-cells (NFAT) for inducing latent HIV-1 expression, but not SAHA. The function of NFAT is regulated by calcineurin (CaN), a calcium and calmodulin dependent serine/threonine protein phosphatase, which activates the T cells of the immune system and can be blocked by its inhibitors.48 To determine whether NFAT is involved in the reversal of HIV latency by 12c, J-Lat A2 cells were pre-treated with the calcineurin inhibitor FK506 before the incubation with 12c. As illustrated in Fig. 4(B), the calcineurin inhibitor FK506 could block AV6-dependent viral expression but did not affect the reactivation of HIV latency induced by SAHA. The reactivation of HIV latency induced by 12c was decreased after pre-treated with FK506, so FK506 also did block 12c-dependent viral gene expression to some extent. These results suggest that 12c retains the reactivation mechanism of AV6, which requires NFAT for early HIV-1 gene expression. While compound 12c is not as sensitive to FK506 as AV6 is, it indicates that 12c may activate latent HIV-1 from latent reservoirs as a HDACs inhibitor.
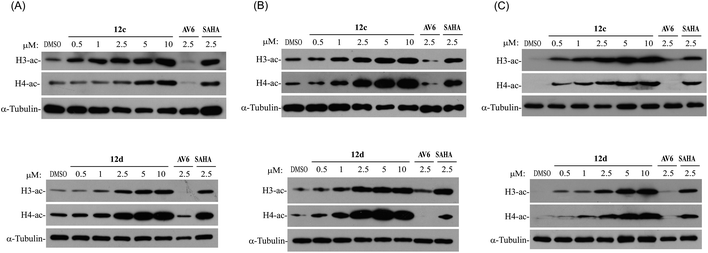
2.3.3.2 Activate latent HIV-1 as histone deacetylase (HDAC) inhibitors. It is well known that inhibition of HDACs is sufficient to reactivate a fraction of latent HIV in a variety of experimental systems. Histone acetylation consists of the addition of an acetyl group to the ε-amino group of lysine residues. Histone lysine acetylation, on lysine 9 or 14 of histone H3 (H3K9, H3K14) or on lysine 16 of histone H4 (H4K16), is generally associated with active gene transcription.15,21 Thus, we examined whether compounds 12c and 12d with the functional group ZBG of HDACs inhibitors also inhibited intracellular even nuclear HDACs by monitoring the acetylation levels of histones H3 and H4 in HeLa and A549, and J-Lat A2 cell lines. In this assay, SAHA (a clinically approved pan-HDAC inhibitor) was used as positive control, DMSO and AV6 were used as negative controls. It was found that after the treatment of compounds 12c and 12d, the acetylation of histone H3 and H4 was significantly up regulated in a dose-dependent manner, whereas AV6 which lacked HDAC inhibition activity was not effect on the acetylation levels of histones H3 and H4 (Fig. 5). It indicated that the right introduction of a ZBG (–CONHOH) in the AV6 scaffold generated two excellent LRAs 12c and 12d, which possessed strong activity on HDACs inhibition to induce HIV-1 latency reversal.
2.4. Molecular docking study of compound 12c and 12d to HDAC2
SAHA achieves broad HDAC inhibitor activity through recognition of conserved structural features within this family of enzymes. We performed molecular docking study in the HDAC2 to determine the binding mode for AV6 analogues and study the key interactions between AV6 analogues and HDAC2. The X-ray protein structure of the human HDAC2 co-crystallized with SAHA in the ligand binding pocket (PDB code: 4LXZ) was selected to run the docking calculations. Re-docking was first carried out with the co-crystal ligand (SAHA) into the binding site of HDAC2 and compared with the co-crystal pose (4LXZ) to assess the docking protocol in this study. The RMSD value of re-docked SAHA refereeing with the co-crystal pose is 0.773 Å (see Fig. S1 of the ESI†), which indicated our docking results were reliable as the success rate of docking is usually judged using the RMSD value lesser than 2.0 Å. Then, we performed the docking analysis of 12c binding to HDAC2. Molecular docking study indicated that the docking score of 12c (−5.949 kcal mol−1) beyond 2-fold to SAHA (−2.759 kcal mol−1). Comparison of the binding poses of SAHA and 12c in the active site of HDAC2 (Fig. 7) shows that the ZBGs of 12c and SAHA both form interactions with the zinc ion and TYR308, but only the OH group of 12c's ZBG establishes an additional hydrogen bond which interacted with the side chain of residue HID145 (distance: 1.96 Å). In addition, the interaction between the surface of binding site and their cap groups was apparently different. The cap group of SAHA phenyl group has π–π stacking interaction with residue HID33, while for 12c binding mode the quinolone core formed a key π–π stacking interaction with residue TYR209. These findings might lead to the better docking score for 12c than SAHA. In this work, molecular model of HDAC2 in complex with 12d was also constructed. It was found that compound 12d showed a similar binding mode with 12c (see Fig. S2 of the ESI†).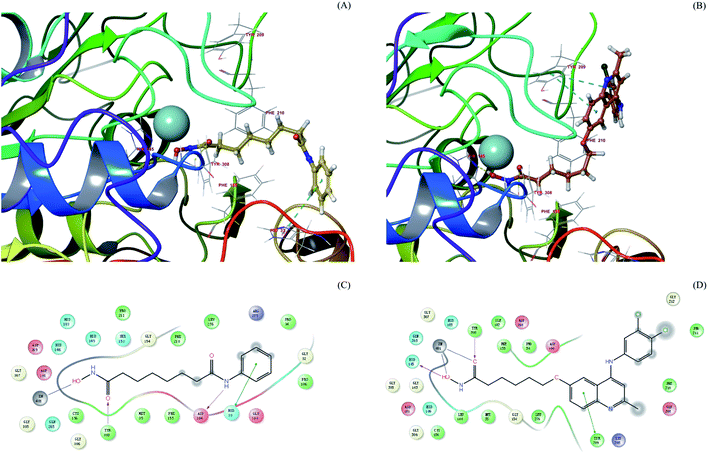 | ||
| Fig. 7 SAHA (A) and 12c (B) docked into the active site of HDAC2 (PDB entry 4LXZ). SAHA and 12c are shown in stick with yellow and brown carbon, respectively. The catalytic zinc in magenta CPK representation. The HDAC2 binding site is presented with a coloured ribbon. Neighbouring amino acids were displayed in lines within a distance of 5 Å approximately to ligand. 2D binding models of SAHA (C) and 12c (D) with HDAC2. Hydrogen bonds are indicated with solid arrows, colour lines around ligands (SAHA and 12c) stand for the binding pocket and the residues in colours nearby established the pocket. The green colour denotes the hydrophobic nature of amino acids, the red colour denotes the acid amino acids, the purple denotes the alkalinity of amino acids, the cyan denotes the polar amino acids, and the grey points of ligand atoms denotes the solvent accessibility. | ||
3. Conclusion
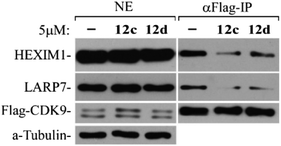
In summary, series of 2-methylquinolin-4-amine compound were synthesized and their ability to activate latent HIV-1 were evaluated in J-Lat A2 cells. Among all synthesized compounds, compound 12c and 12d exhibited excellent potency to reverse HIV latency. Compared with the obviously a lot of synergy between SAHA and AV6, no cooperative effect was observed using the combination of 12c and AV6 for the reactivation of latent HIV-1 gene expression, and to some extent the 12c-dependent viral gene expression was blocked by the calcineurin inhibitor FK506 which did block AV6-dependent mRNA expression.28 These data indicate that 12c retains the reactivation mechanism of AV6, which requires NFAT for early HIV-1 gene expression. On the other hand, the western blotting clearly showed that both in the HepG2 cells and A549 cells treated with 12c and 12d, the lysine acetylation of histone H3 and histone H4 increased in a dose-dependent manner. This study implies that 12c and 12d also act as HDAC inhibitors to reactivate HIV-1 transcription. Positive transcription elongation factorb (P-TEFb) is a critical component of the cellular transcriptional machinery that is required for the elongation of HIV transcription. In this work, the co-immunoprecipitation assay confirmed that 12c and 12d were able to release P-TEFb from the inactive complex 7SK snRNP to activate HIV transcription. Molecular docking yielded binding modes for compounds 12c and 12d with the similar metal coordination with ZN2+ as well as SAHA, and its larger cap group quinoline core increased the interactions with HDAC2 and turned in the opposite direction from the cap group of SAHA. All these results proved the potential of 12c and 12d as novel dual reactivator of latent HIV-1. However, further studies are required to determine their roles in reactivating latent HIV-1.4. Experimental section
4.1. Chemistry
All reagents were commercial available and used without further purification, unless otherwise indicated. Reactions, which required the use of anhydrous, inert atmosphere techniques, were carried out under an atmosphere of nitrogen. Reactions were magnetically stirred and monitored by thin-layer chromatography (TLC) on Merck silica gel 60F-254 by fluorescence. 1H-NMR and 13C-NMR spectra were obtained using a Bruker (Bruker Biospin, Zug, Switzerland) AV2600 Ultrashield spectrometer at 600 and 150 MHz, respectively. Chemical shifts were reported in parts per million (ppm) relative to tetramethylsilane (TMS) as an internal standard. Multiplicities were abbreviated as follows: single (s), doublet (d), doublet of doublets (dd), doublet–triplet (dt), triplet (t), triplet–triplet (tt), triplet–doublet (td), quartet (q), quartet–doublet (qd), multiplet (m), and broad signal (brs). High-resolution mass spectral (HRMS) data were acquired on a Q Exactive. Melting points were measured on a SGW X-4 micro-melting point spectrometer and were uncorrected. The purities of synthesized compounds were assessed by high performance liquid chromatography (HPLC) with a COSMOSIL C18-MS-II column (250 mm × 4.6 mm i.d., 5 μm). The HPLC analysis was performed on an Agilent Technologies 1100 Series HPLC system. The mobile phase was methanol (A) and water (B) in a linear gradient mode as follows: (A) from 5% to 100% and (B) from 95% to 0% during 0–30 min. The flow rate was 1 mL min−1, and the detection wavelengths were 254 nm and 280 nm.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 4.04 (q, J = 7.0 Hz, 2H, OCH2CH3), 3.75 (s, 3H, OCH3), 1.89 (s, 3H, CH3), 1.19 (t, J = 7.1 Hz, 3H, OCH2CH3). 13C NMR (150 MHz, DMSO-d6): δ 169.9 (C
CH), 4.04 (q, J = 7.0 Hz, 2H, OCH2CH3), 3.75 (s, 3H, OCH3), 1.89 (s, 3H, CH3), 1.19 (t, J = 7.1 Hz, 3H, OCH2CH3). 13C NMR (150 MHz, DMSO-d6): δ 169.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 160.3 (NHCH3–C
O), 160.3 (NHCH3–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 157.4 (C–OCH3), 132.0 (Ph-C), 126.7 (s, 2C, Ph-C), 114.7 (s, 2C, Ph-C), 84.8 (C
CH), 157.4 (C–OCH3), 132.0 (Ph-C), 126.7 (s, 2C, Ph-C), 114.7 (s, 2C, Ph-C), 84.8 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 58.5 (OCH2CH3), 55.7 (OCH3), 20.2 (OCH2CH3), 15.0 (CH3). ESI-HRMS (+): m/z [M + H]+ calculated for C13H18NO3+, 236.1128, found, 236.1281.
CH), 58.5 (OCH2CH3), 55.7 (OCH3), 20.2 (OCH2CH3), 15.0 (CH3). ESI-HRMS (+): m/z [M + H]+ calculated for C13H18NO3+, 236.1128, found, 236.1281.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 156.2 (C-6), 150.5 (C-2), 135.2 (C-9), 124.7 (C-10), 123.1 (C-7), 120.4 (C-8), 107.4 (C-3), 104.1 (C-5), 55.9 (OCH3), 19.9 (CH3). ESI-HRMS (+): m/z [M + H]+ calculated for C11H12NO2+, 190.0863, found, 190.0862.
O), 156.2 (C-6), 150.5 (C-2), 135.2 (C-9), 124.7 (C-10), 123.1 (C-7), 120.4 (C-8), 107.4 (C-3), 104.1 (C-5), 55.9 (OCH3), 19.9 (CH3). ESI-HRMS (+): m/z [M + H]+ calculated for C11H12NO2+, 190.0863, found, 190.0862.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 6a as a white solid (1.2 g, 93.6%). 1H NMR (600 MHz, CDCl3): δ 8.02 (d, J = 9.5 Hz, 1H, H-8), 7.39–7.42 (m, 2H, H-5 and H-7), 7.38 (s, 1H, H-3), 4.19 (q, J = 7.0 Hz, 2H, OCH2CH3), 2.72 (s, 3H, CH3), 1.51 (t, J = 7.0 Hz, 3H, OCH2CH3). 13C NMR (150 MHz, CDCl3): δ 157.6 (C-6), 155.7 (C-2), 144.0 (C-9), 141.4 (C-4), 130.1 (C-8), 125.7 (C-7), 123.5 (C-10), 122.1 (C-3), 102.4 (C-5), 64.0 (OCH2CH3), 24.5 (CH3), 14.7 (OCH2CH3). ESI-HRMS (+): m/z [M + H]+ calculated for C12H13ClNO+, 222.0680, found, 222.0681.
1) to afford 6a as a white solid (1.2 g, 93.6%). 1H NMR (600 MHz, CDCl3): δ 8.02 (d, J = 9.5 Hz, 1H, H-8), 7.39–7.42 (m, 2H, H-5 and H-7), 7.38 (s, 1H, H-3), 4.19 (q, J = 7.0 Hz, 2H, OCH2CH3), 2.72 (s, 3H, CH3), 1.51 (t, J = 7.0 Hz, 3H, OCH2CH3). 13C NMR (150 MHz, CDCl3): δ 157.6 (C-6), 155.7 (C-2), 144.0 (C-9), 141.4 (C-4), 130.1 (C-8), 125.7 (C-7), 123.5 (C-10), 122.1 (C-3), 102.4 (C-5), 64.0 (OCH2CH3), 24.5 (CH3), 14.7 (OCH2CH3). ESI-HRMS (+): m/z [M + H]+ calculated for C12H13ClNO+, 222.0680, found, 222.0681.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 6b as a white solid (2.9 g, 59.6%). 1H NMR (600 MHz, CDCl3): δ 7.91 (d, J = 9.2 Hz, 1H, H-8), 7.43 (d, J = 2.8 Hz, 1H, H-5), 7.32–7.36 (m, 2H, H-3 and H-7), 4.71–4.78 (m, 1H, OCH(CH3)2), 2.67 (s, 3H, CH3), 1.43 (s, 3H, OCH(CH3)2), 1.42 (s, 3H, OCH(CH3)2). 13C NMR (150 MHz, CDCl3): δ 156.4 (C-6), 155.7 (C-2), 143.9 (C-9), 141.3 (C-4), 130.2 (C-8), 125.7 (C-7), 124.1 (C-10), 122.1 (C-3), 104.0 (C-5), 70.4 (OCH(CH3)2), 24.5 (CH3), 21.9 (s, 2C, OCH(CH3)2). ESI-HRMS (+): m/z [M + H]+ calculated for C13H15ClNO+, 236.0837, found, 236.0837.
1) to afford 6b as a white solid (2.9 g, 59.6%). 1H NMR (600 MHz, CDCl3): δ 7.91 (d, J = 9.2 Hz, 1H, H-8), 7.43 (d, J = 2.8 Hz, 1H, H-5), 7.32–7.36 (m, 2H, H-3 and H-7), 4.71–4.78 (m, 1H, OCH(CH3)2), 2.67 (s, 3H, CH3), 1.43 (s, 3H, OCH(CH3)2), 1.42 (s, 3H, OCH(CH3)2). 13C NMR (150 MHz, CDCl3): δ 156.4 (C-6), 155.7 (C-2), 143.9 (C-9), 141.3 (C-4), 130.2 (C-8), 125.7 (C-7), 124.1 (C-10), 122.1 (C-3), 104.0 (C-5), 70.4 (OCH(CH3)2), 24.5 (CH3), 21.9 (s, 2C, OCH(CH3)2). ESI-HRMS (+): m/z [M + H]+ calculated for C13H15ClNO+, 236.0837, found, 236.0837.4.1.7.1 N-(3,4-Dichlorophenyl)-6-methoxy-2-methylquinolin-4-amine (7a). White solid; yield: 35.8%; HPLC purity: 98.9% (tR = 21.92 min); mp: 266–267 °C. 1H NMR (600 MHz, DMSO-d6): δ 8.08 (s, 1H, H-5), 7.96 (d, J = 9.2 Hz, 1H, H-8), 7.77 (d, J = 4.2 Hz, 1H, Ph-H), 7.76 (d, J = 1.7 Hz, 1H, Ph-H), 7.58 (dd, J = 2.6, 9.2 Hz, 1H, H-7), 7.50 (dd, J = 2.4, 8.6 Hz, 1H, Ph-H), 6.89 (s, 1H, H-3), 3.97 (s, 3H, OCH3), 2.59 (s, 3H, CH3); 13C NMR (150 MHz, DMSO-d6): δ 157.9 (C-2), 153.9 (C-6), 151.9 (C-4), 151.8 (C-9), 139.2 (C-1′), 132.4 (Ph-C), 132.0 (C-8), 128.3 (Ph-C), 126.6 (Ph-C), 125.0 (C-7), 124.7 (Ph-C), 123.7 (Ph-C), 118.5 (C-10), 103.0 (C-3), 101.9 (C-5), 56.8 (OCH3), 21.1 (CH3). ESI-HRMS (+): m/z [M + H]+ calculated for C17H15Cl2N2O+, 333.0556, found, 333.0552.
Other target compounds (7b-7B) were synthesized following the general procedure as described above with the yield of 25.0–86.0%. Their structures were assigned by the spectroscopic data including 1H NMR, 13C NMR, and ESI-HRMS (see ‘ESI†’).
4.1.9.1 N-(3,4-Dichlorophenyl)-2-methyl-6-propoxyquinolin-4-amine (9a). White solid; yield: 30.0%; HPLC purity: 96.1% (tR = 23.96 min); mp: 266–267 °C. 1H NMR (600 MHz, DMSO-d6): δ 7.77 (d, J = 9.2 Hz, 1H, H-8), 7.69 (d, J = 2.0 Hz, 1H, H-5), 7.63 (d, J = 8.6 Hz, 1H, Ph-H), 7.58 (d, J = 2.4 Hz, 1H, Ph-H), 7.36–7.40 (m, 2H), 6.94 (s, 1H, H-3), 4.08 (t, J = 6.5 Hz, 2H, OCH2), 2.49 (s, 3H, CH3), 1.81 (sxt, J = 7.0 Hz, 2H, OCH2CH2CH3), 1.03 (t, J = 7.3 Hz, 3H, OCH2CH2CH3); 13C NMR (150 MHz, DMSO-d6): δ 156.2 (C-2), 155.9 (C-6), 147.1 (C-4), 143.0 (C-9), 141.7 (C-1′), 132.1 (Ph-C), 131.6 (C-8), 129.1 (Ph-C), 125.0 (C-7), 123.5 (Ph-C), 122.3 (Ph-C), 121.9 (Ph-C), 119.6 (C-10), 103.7 (C-3), 102.5 (C-5), 70.0 (OCH2), 24.4 (OCH2CH2CH3), 22.5 (C-11), 11.0 (OCH2CH2CH3). ESI-HRMS (+): m/z [M + H]+ calculated for C19H19Cl2N2O+, 361.0869, found, 361.0865.
Other target compounds (9b–9e) were synthesized following the general procedure as described above with the yield of 30.0–55.0%. Their structures were assigned by the spectroscopic data including 1H NMR, 13C NMR, and ESI-HRMS (see ‘ESI†’).
4.1.10.1 Methyl 6-((4-((3,4-dichlorophenyl)amino)-2-methylquinolin-6-yl)oxy)hexanoate (10c). White solid; yield: 64.2%; HPLC purity: 93.3% (tR = 24.35 min); mp: 129–131 °C. 1H NMR (600 MHz, CDCl3): δ 7.90 (d, J = 9.2 Hz, 1H, H-8), 7.44 (d, J = 8.6 Hz, 1H, Ar-H), 7.38 (d, J = 2.6 Hz, 1H, H-5), 7.30–7.33 (m, 1H, H-7), 7.13–7.17 (m, 2H, Ar-H), 6.92 (s, 1H, H-3), 3.96–4.04 (m, 2H, 12-CH2), 3.68 (s, 3H, COO–CH3), 2.58 (s, 3H, 11-CH3), 2.34–2.41 (m, 2H), 1.81–1.85 (m, 2H), 1.69–1.73 (m, 2H), 1.47–1.54 (m, 2H); 13C NMR (150 MHz, CDCl3): δ 173.3 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 155.7 (C-2), 155.3 (C-6), 145.0 (C-4), 143.4 (C-9), 139.7 (C-1′), 132.2 (Ph-C), 130.0 (C-8), 129.2 (Ph-C), 125.5 (C-7), 121.9 (Ph-C), 120.9 (Ph-C), 119.7 (C-10), 118.5 (Ph-C), 103.5 (C-3), 99.4 (C-5), 67.0 (C-12), 50.6 (COO–CH3), 32.9, 27.7, 24.5, 23.5 (C-11). ESI-HRMS (+): m/z [M + H]+ calculated for C23H25Cl2N2O3+, 447.1237, found, 447.1236.
O), 155.7 (C-2), 155.3 (C-6), 145.0 (C-4), 143.4 (C-9), 139.7 (C-1′), 132.2 (Ph-C), 130.0 (C-8), 129.2 (Ph-C), 125.5 (C-7), 121.9 (Ph-C), 120.9 (Ph-C), 119.7 (C-10), 118.5 (Ph-C), 103.5 (C-3), 99.4 (C-5), 67.0 (C-12), 50.6 (COO–CH3), 32.9, 27.7, 24.5, 23.5 (C-11). ESI-HRMS (+): m/z [M + H]+ calculated for C23H25Cl2N2O3+, 447.1237, found, 447.1236.
4.1.10.2 Ethyl 7-((4-((3,4-dichlorophenyl)amino)-2-methylquinolin-6-yl)oxy)heptanoate (10d). White solid; yield: 59.1%; HPLC purity: 96.4% (tR = 25.76 min); mp: 129–132 °C. 1H NMR (600 MHz, CDCl3): δ 7.90 (d, J = 9.2 Hz, 1H, H-8), 7.43 (d, J = 8.6 Hz, 1H, Ph-H), 7.37 (d, J = 2.4 Hz, 1H, H-5), 7.31 (dd, J = 2.6, 9.2 Hz, 1H, H-7), 7.17–7.20 (m, 1H, Ph-H), 7.14 (dd, J = 2.5, 8.5 Hz, 1H, Ph-H), 6.91 (s, 1H, H-3), 4.12 (q, J = 7.0 Hz, 2H, COO–CH2CH3), 3.98 (t, J = 6.4 Hz, 2H, 12-CH2), 2.58 (s, 3H, 11-CH3), 2.28–2.35 (m, 2H), 1.79–1.82 (m, 2H), 1.66 (quin, J = 7.5 Hz, 2H), 1.43–1.51 (m, 2H), 1.35–1.42 (m, 2H), 1.25 (t, J = 7.2 Hz, 1H, COO–CH2CH3); 13C NMR (150 MHz, CDCl3): δ 174.0 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 156.7 (C-2), 156.5 (C-6), 146.0 (C-4), 144.4 (C-9), 140.7 (C-1′), 133.3 (Ph-C), 131.1 (C-8), 130.3 (Ph-C), 126.6 (C-7), 123.0 (Ph-C), 121.8 (Ph-C), 120.8 (Ph-C), 119.5 (C-10), 104.5 (C-3), 100.2 (C-5), 68.2 (C-12), 60.4 (COO–CH2CH3), 34.2, 28.8, 28.7, 25.6, 25.1, 24.8 (C-11), 14.2 (COO–CH2CH3). ESI-HRMS (+): m/z [M + H]+ calculated for C25H29Cl2N2O3+, 475.1550, found, 475.1549.
O), 156.7 (C-2), 156.5 (C-6), 146.0 (C-4), 144.4 (C-9), 140.7 (C-1′), 133.3 (Ph-C), 131.1 (C-8), 130.3 (Ph-C), 126.6 (C-7), 123.0 (Ph-C), 121.8 (Ph-C), 120.8 (Ph-C), 119.5 (C-10), 104.5 (C-3), 100.2 (C-5), 68.2 (C-12), 60.4 (COO–CH2CH3), 34.2, 28.8, 28.7, 25.6, 25.1, 24.8 (C-11), 14.2 (COO–CH2CH3). ESI-HRMS (+): m/z [M + H]+ calculated for C25H29Cl2N2O3+, 475.1550, found, 475.1549.Other target compounds (10a-10b) were synthesized following the general procedure as described above with the yield of 34.0–64.0%. Their structures were assigned by the spectroscopic data including 1H NMR, 13C NMR, and ESI-HRMS (see ‘ESI†’).
4.1.11.1 4-((4-((3,4-dichlorophenyl)amino)-2-methylquinolin-6-yl)oxy) butanoic acid (11a). White solid; yield: 70.0%; HPLC purity: 97.0% (tR = 21.99 min); mp: 266–267 °C. 1H NMR (600 MHz, DMSO-d6): δ 7.79 (d, J = 9.2 Hz, 1H, H-8), 7.72 (d, J = 2.4 Hz, 1H, H-5), 7.65 (d, J = 8.8 Hz, 1H, Ph-H), 7.59 (d, J = 2.6 Hz, 1H, Ph -H), 7.39 (td, J = 2.2, 8.8 Hz, 2H, H-7 and H-6′), 6.93 (s, 1H, H-3), 4.14 (t, J = 6.4 Hz, 2H, 12-CH2), 2.49 (s, 3H, CH3), 2.45 (t, J = 7.2 Hz, 2H, 14-CH2), 2.03 (quin, J = 6.8 Hz, 2H, 13-CH2); 13C NMR (150 MHz, DMSO-d6): δ 174.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 156.2 (C-2), 155.8 (C-6), 147.7 (C-4), 142.1 (C-9), 141.3 (C-1′), 132.1 (Ph-C), 131.7 (C-8), 128.4 (Ph-C), 125.5 (C-7), 124.0 (Ph-C), 122.7 (Ph-C), 122.4 (Ph-C), 119.3 (C-10), 103.3 (C-3), 102.5 (C-5), 67.7 (OCH2), 30.6 (CH2–COOH), 24.7 (OCH2CH2), 24.0 (C-11). ESI-HRMS (−): m/z [M − H]− calculated for C20H17Cl2N2O3−, 403.0622, found, 403.0618.
O), 156.2 (C-2), 155.8 (C-6), 147.7 (C-4), 142.1 (C-9), 141.3 (C-1′), 132.1 (Ph-C), 131.7 (C-8), 128.4 (Ph-C), 125.5 (C-7), 124.0 (Ph-C), 122.7 (Ph-C), 122.4 (Ph-C), 119.3 (C-10), 103.3 (C-3), 102.5 (C-5), 67.7 (OCH2), 30.6 (CH2–COOH), 24.7 (OCH2CH2), 24.0 (C-11). ESI-HRMS (−): m/z [M − H]− calculated for C20H17Cl2N2O3−, 403.0622, found, 403.0618.Other target compounds (11b–11d) were synthesized following the general procedure as described above with the yield of 70.0–91.0%. Their structures were assigned by the spectroscopic data including 1H NMR, 13C NMR, and ESI-HRMS (see ‘ESI†’).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (2.0 mL/4.0 mL) at 0 °C. The reaction mixture was warmed to room temperature and stirred at 25 °C for 24 h. After completion of the reaction (monitored by TLC), the mixture was neutralized with acetic acid. The formed precipitate was collected by filtration, washed with water and recrystallized from ethanol to give the crude product. The crude was purified by column chromatography using appropriate mixtures of dichloromethane and methanol to yield the titled compound.
2 (2.0 mL/4.0 mL) at 0 °C. The reaction mixture was warmed to room temperature and stirred at 25 °C for 24 h. After completion of the reaction (monitored by TLC), the mixture was neutralized with acetic acid. The formed precipitate was collected by filtration, washed with water and recrystallized from ethanol to give the crude product. The crude was purified by column chromatography using appropriate mixtures of dichloromethane and methanol to yield the titled compound.
4.1.12.1 6-((4-((3,4-dichlorophenyl)amino)-2-methylquinolin-6-yl)oxy)-N-hydroxy-hexanamide (12c). White solid; yield: 50.1%; HPLC purity: 97.9% (tR = 22.09 min); mp: 266–267 °C. 1H NMR (600 MHz, CD3OD): δ 7.70 (d, J = 9.2 Hz, 1H, H-8), 7.65 (d, J = 2.4 Hz, 1H, H-5), 7.54 (d, J = 8.6 Hz, 1H, Ph-H), 7.52 (d, J = 2.6 Hz, 1H, Ph-H), 7.41 (dd, J = 2.5, 9.3 Hz, 1H, H-7), 7.30 (dd, J = 2.4, 8.6 Hz, 1H, Ph-H), 6.75 (s, 1H, H-3), 4.07 (t, J = 6.3 Hz, 2H, 12-CH2), 2.48 (s, 3H, CH3), 2.05 (t, J = 7.3 Hz, 2H, 16-CH2), 1.74–1.85 (m, 2H, 13-CH2), 1.62 (quin, J = 7.6 Hz, 2H, 15-CH2), 1.43–1.51 (m, 2H, 14-CH2); 13C NMR (150 MHz, CD3OD): δ 171.4 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 157.7 (C-2), 153.9 (C-6), 151.8 (C-4), 138.7 (C-9), 136.9 (C-1′), 133.0 (Ph-C), 131.3 (C-8), 129.1 (Ph-C), 125.8 (C-7), 124.4 (Ph-C), 123.8 (Ph-C), 123.4 (Ph-C), 118.3 (C-10), 101.7 (C-3), 101.2 (C-5), 68.3 (C-12), 32.3 (C-16), 28.4 (C-13), 25.2 (C-15), 25.0 (C-14), 20.1 (C-11). ESI-HRMS (−): m/z [M − H]− calculated for C22H22Cl2N3O3−, 446.1044, found, 446.1040.
O), 157.7 (C-2), 153.9 (C-6), 151.8 (C-4), 138.7 (C-9), 136.9 (C-1′), 133.0 (Ph-C), 131.3 (C-8), 129.1 (Ph-C), 125.8 (C-7), 124.4 (Ph-C), 123.8 (Ph-C), 123.4 (Ph-C), 118.3 (C-10), 101.7 (C-3), 101.2 (C-5), 68.3 (C-12), 32.3 (C-16), 28.4 (C-13), 25.2 (C-15), 25.0 (C-14), 20.1 (C-11). ESI-HRMS (−): m/z [M − H]− calculated for C22H22Cl2N3O3−, 446.1044, found, 446.1040.
4.1.12.2 7-((4-((3,4-Dichlorophenyl)amino)-2-methylquinolin-6-yl)oxy)-N-hydroxy-heptanamide (12d). White solid; yield: 64.9%; HPLC purity: 98.4% (tR = 22.76 min); mp: 266–267 °C. 1H NMR (600 MHz, DMSO-d6): δ 7.72 (d, J = 9.0 Hz, 1H, H-8), 7.68 (d, J = 2.6 Hz, 1H, H-5), 7.59 (d, J = 0.9 Hz, 1H, Ph-H), 7.58 (d, J = 4.8 Hz, 1H, Ph-H), 7.38 (dd, J = 2.5, 8.7 Hz, 1H, Ph-H), 7.30 (dd, J = 2.6, 9.2 Hz, 1H, H-7), 6.96 (s, 1H, H-3), 4.11 (t, J = 6.4 Hz, 2H, H-12), 2.46 (s, 3H, CH3), 2.17 (t, J = 7.2 Hz, 2H, H-17), 1.74–1.82 (m, 2H, H-13), 1.52 (quin, J = 7.4 Hz, 2H, H-16), 1.45 (quin, J = 7.5 Hz, 2H, H-14), 1.31–1.39 (m, 2H, H-13); 13C NMR (150 MHz, DMSO-d6): δ 169.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 157.1 (C-2), 154.1 (C-6), 153.0 (C-4), 139.7 (C-9), 138.2 (C-1′), 134.3 (Ph-C), 131.8 (C-8), 130.3 (Ph-C), 125.6 (C-7), 125.1 (Ph-C), 124.4 (Ph-C), 122.7 (Ph-C), 118.6 (C-10), 103.4 (C-3), 100.3 (C-5), 69.0 (C-12), 49.1 (C-17), 32.7 (C-13), 28.8 (C-15), 25.8 (C-16), 25.5 (C-14), 20.6 (C-11). ESI-HRMS (−): m/z [M − H]− calculated for C23H24Cl2N3O3−, 426.1200, found, 426.1197.
O), 157.1 (C-2), 154.1 (C-6), 153.0 (C-4), 139.7 (C-9), 138.2 (C-1′), 134.3 (Ph-C), 131.8 (C-8), 130.3 (Ph-C), 125.6 (C-7), 125.1 (Ph-C), 124.4 (Ph-C), 122.7 (Ph-C), 118.6 (C-10), 103.4 (C-3), 100.3 (C-5), 69.0 (C-12), 49.1 (C-17), 32.7 (C-13), 28.8 (C-15), 25.8 (C-16), 25.5 (C-14), 20.6 (C-11). ESI-HRMS (−): m/z [M − H]− calculated for C23H24Cl2N3O3−, 426.1200, found, 426.1197.Other target compounds (12a-12b) were synthesized following the general procedure as described above with the yield of 35.0–65.0%. Their structures were assigned by the spectroscopic data including 1H NMR, 13C NMR, and ESI-HRMS (see ‘ESI†’).
4.2. Biological assays
4.3. Molecular modeling
X-ray protein structure of human HDAC2 (PDB code: 4LXZ) was download from Protein Data Bank (PDB) and prepared with Protein Preparation Wizard panel as implemented in Maestro 10.5 (Schrödinger Release 2016–1: Maestro, Schrödinger, LLC, New York, NY, 2016.). Missing hydrogens and residues were added, bond orders were assigned, and water molecules beyond 5 Å to inhibitor were removed. Both the native ligand SAHA and compound 12c were prepared with LigPrep panel with OPLS2005 force field, generating the protonation and tautomeric states at 7.0 ± 2.0 pH units. The glide grid center was setting according the geometrical center of original inhibitor (SAHA) and the grid size was 20 × 20 × 20 Å3, in which the metal coordination was set for ZN2+ in the binding site. Prior to docking compound 12c, a re-docking run was performed to evaluate the docking performance of Glide extra precision (Glide XP) mode. Glide XP mode (Schrödinger Release 2016-1: Glide, Schrödinger, LLC, New York, NY, 2017.) was successful in reproducing the binding mode of native ligand SAHA with a RMSD value of 0.773 Å. Then based on this docking grid, all inhibitors including the co-crystal ligand SAHA were docked flexibly by Glide 7.0 in extra precision with default options.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81773600, 81672955 and 81302652), the Natural Science Foundation of Fujian Province of China (No. 2018J01132 and 2016J011172), and the Fundamental Research Funds for the Central Universities (No. 20720180051 and 20720160062) to Y. X. This research was also sponsored by the Scientific Research Foundation of Third Institute of Oceanography, SOA (Grant No. 2016042) and the Educational Department of Fujian Province (JA15017).References
- D. Finzi, J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant and R. F. Siliciano, Nat Med, 1999, 5, 512–517 CrossRef CAS PubMed.
- J. D. Siliciano, J. Kajdas, D. Finzi, T. C. Quinn, K. Chadwick, J. B. Margolick, C. Kovacs, S. J. Gange and R. F. Siliciano, Nat Med, 2003, 9, 727–728 CrossRef CAS PubMed.
- A. R. Cillo and J. W. Mellors, Curr. Opin. Virol., 2016, 18, 14–19 CrossRef PubMed.
- A. S. S. W. G. o. H. I. V. C. International, S. G. Deeks, B. Autran, B. Berkhout, M. Benkirane, S. Cairns, N. Chomont, T. W. Chun, M. Churchill, M. Di Mascio, C. Katlama, A. Lafeuillade, A. Landay, M. Lederman, S. R. Lewin, F. Maldarelli, D. Margolis, M. Markowitz, J. Martinez-Picado, J. I. Mullins, J. Mellors, S. Moreno, U. O'Doherty, S. Palmer, M. C. Penicaud, M. Peterlin, G. Poli, J. P. Routy, C. Rouzioux, G. Silvestri, M. Stevenson, A. Telenti, C. Van Lint, E. Verdin, A. Woolfrey, J. Zaia and F. Barre-Sinoussi, Nat. Rev. Immunol., 2012, 12, 607–614 CrossRef PubMed.
- J. D. Siliciano and R. F. Siliciano, J. Clin. Invest., 2016, 126, 409–414 CrossRef PubMed.
- K. M. Barton, B. D. Burch, N. Soriano-Sarabia and D. M. Margolis, Clin. Pharmacol. Ther., 2013, 93, 46–56 CrossRef CAS PubMed.
- C. Schwartz, S. Bouchat, C. Marban, V. Gautier, C. Van Lint, O. Rohr and V. Le Douce, Biochem. Pharmacol., 2017, 146, 10–22 CrossRef CAS PubMed.
- S. G. Deeks, Nature, 2012, 487, 439–440 CrossRef CAS PubMed.
- G. Darcis, B. Van Driessche and C. Van Lint, Trends Immunol., 2017, 38, 217–228 CrossRef CAS PubMed.
- P. K. Datta, R. Kaminski, W. Hu, V. Pirrone, N. T. Sullivan, M. R. Nonnemacher, W. Dampier, B. Wigdahl and K. Khalili, Curr. HIV Res., 2016, 14, 431–441 CrossRef CAS PubMed.
- D. M. Margolis, J. V. Garcia, D. J. Hazuda and B. F. Haynes, Science, 2016, 353, aaf6517 CrossRef PubMed.
- N. M. Archin and D. M. Margolis, Curr. Opin. Infect. Dis., 2014, 27, 29–35 CrossRef CAS PubMed.
- J. T. Kimata, A. P. Rice and J. Wang, Curr. Opin. Immunol., 2016, 42, 65–70 CrossRef CAS PubMed.
- S. Xing and R. F. Siliciano, Drug discovery today, 2013, 18, 541–551 CrossRef CAS PubMed.
- D. M. Margolis, Curr. Opin. HIV AIDS, 2011, 6, 25–29 CrossRef PubMed.
- X. Contreras, M. Barboric, T. Lenasi and B. M. Peterlin, PLoS Pathog., 2007, 3, 1459–1469 CAS.
- J. Kulkosky, D. M. Culnan, J. Roman, G. Dornadula, M. Schnell, M. R. Boyd and R. J. Pomerantz, Blood, 2001, 98, 3006–3015 CrossRef CAS PubMed.
- M. Stoszko, E. De Crignis, C. Rokx, M. M. Khalid, C. Lungu, R. J. Palstra, T. W. Kan, C. Boucher, A. Verbon, E. C. Dykhuizen and T. Mahmoudi, EBioMedicine, 2016, 3, 108–121 CrossRef PubMed.
- N. M. Archin, A. Espeseth, D. Parker, M. Cheema, D. Hazuda and D. M. Margolis, AIDS Res. Hum. Retroviruses, 2009, 25, 207–212 CrossRef CAS PubMed.
- (a) M. Dokmanovic and P. A. Marks, J. Cell. Biochem., 2005, 96, 293 CrossRef CAS PubMed; (b) D. Demonte, V. Quivy, Y. Colette and C. Van Lint, Biochem. Pharmacol., 2004, 68, 1231–1238 CrossRef CAS PubMed; (c) K. Shirakawa, L. Chavez, S. Hakre, V. Calvanese and E. Verdin, Trends Microbiol., 2013, 21, 277–285 CrossRef CAS PubMed.
- X. Contreras, M. Schweneker, C. S. Chen, J. M. McCune, S. G. Deeks, J. Martin and B. M. Peterlin, J. Biol. Chem., 2009, 284, 6782–6789 CrossRef CAS PubMed.
- N. M. Archin, J. L. Kirchherr, J. A. M. Sung, G. Clutton, K. Sholtis, Y. Y. Xu, B. Allard, E. Stuelke, A. D. Kashuba, J. D. Kuruc, J. Eron, C. L. Gay, N. Goonetilleke and D. M. Margolis, J. Clin. Invest., 2017, 127, 3126–3135 Search PubMed.
- T. D. Zaikos, M. M. Painter, N. T. Sebastian, V. H. Terry and K. L. Collins, J. Virol., 2018, 02110–02117 Search PubMed.
- N. Sagotlerolle, A. Lamine, M. L. Chaix, F. Boufassa, J. P. Aboulker, D. Costagliola, C. Goujard, C. Pallier, C. Paller and J. F. Delfraissy, AIDS, 2008, 22, 1125–1129 CrossRef CAS PubMed.
- J. P. Routy, C. L. Tremblay, J. B. Angel, B. Trottier, D. Rouleau, J. G. Baril, M. Harris, S. Trottier, J. Singer, N. Chomont, R. P. Sekaly and M. R. Boulassel, HIV Med., 2012, 13, 291–296 CrossRef CAS PubMed.
- N. M. Archin, R. Bateson, M. K. Tripathy, A. M. Crooks, K. H. Yang, N. P. Dahl, M. F. Kearney, E. M. Anderson, J. M. Coffin, M. C. Strain, D. D. Richman, K. R. Robertson, A. D. Kashuba, R. J. Bosch, D. J. Hazuda, J. D. Kuruc, J. J. Eron and D. M. Margolis, J. Infect. Dis., 2014, 210, 728–735 CrossRef CAS PubMed.
- N. M. Archin, M. Cheema, D. Parker, A. Wiegand, R. J. Bosch, J. M. Coffin, J. Eron, M. Cohen and D. M. Margolis, PLoS One, 2010, 5, e9390 Search PubMed.
- S. Micheva-Viteva, Y. Kobayashi, L. C. Edelstein, A. L. Pacchia, H. L. Lee, J. D. Graci, J. Breslin, B. D. Phelan, L. K. Miller, J. M. Colacino, Z. Gu, Y. Ron, S. W. Peltz and J. P. Dougherty, J. Biol. Chem., 2011, 286, 21083–21091 CrossRef CAS PubMed.
- G. M. Laird, C. K. Bullen, D. I. Rosenbloom, A. R. Martin, A. L. Hill, C. M. Durand, J. D. Siliciano and R. F. Siliciano, J. Clin. Invest., 2015, 125, 1901–1912 Search PubMed.
- E. D. Crignis and T. Mahmoudi, Viruses, 2014, 6, 4581–4608 CrossRef PubMed.
- M. Purwin, J. Hernández-Toribio, C. Coderch, R. Panchuk, N. Skorokhyd, K. Filipiak, B. de Pascual-Teresa and A. Ramos, RSC Adv., 2016, 6, 66595–66608 RSC.
- X. Chen, S. Zhao, Y. Wu, Y. Chen, T. Lu and Y. Zhu, RSC Adv., 2016, 6, 103178–103184 RSC.
- (a) A. K. Halder, S. Mallick, D. Shikha, A. Saha, K. D. Saha and T. Jha, RSC Adv., 2015, 5, 72373–72386 RSC; (b) X. Zhang, J. Zhang, M. Su, Y. Zhou, Y. Chen, J. Li and W. Lu, RSC Adv., 2014, 4, 40444–40448 RSC.
- A. Petrelli and G. Valabrega, Expert Opin. Pharmacother., 2009, 10, 589–600 CrossRef CAS PubMed.
- K. S. Keedy, N. M. Archin, A. T. Gates, A. Espeseth, D. J. Hazuda and D. M. Margolis, J. Virol., 2009, 83, 4749–4756 CrossRef CAS PubMed.
- E. Bourguet, K. Ozdarska, G. Moroy, J. Jeanblanc and M. Naassila, J. Med. Chem., 2018, 61, 1745–1766 CrossRef CAS PubMed.
- K. Abouzid and S. Shouman, Bioorg. Med. Chem., 2008, 16, 7543–7551 CrossRef CAS PubMed.
- J. A. Joule, K. Mills and G. F. Smith, Quinolines and isoquinolines: reactions and synthesis, Heterocyclic Chemistry, 1995, pp. 120–145 Search PubMed.
- X. Q. Wang, C. L. Xia, S. B. Chen, J. H. Tan, T. M. Ou, S. L. Huang, D. Li, L. Q. Gu and Z. S. Huang, Eur. J. Med. Chem., 2015, 89, 349–361 CrossRef CAS PubMed.
- E. Pitta, M. K. Rogacki, O. Balabon, S. Huss, F. Cunningham, E. M. Lopez-Roman, J. Joossens, K. Augustyns, L. Ballell, R. H. Bates and P. Van der Veken, J. Med. Chem., 2016, 59, 6709–6728 CrossRef CAS PubMed.
- V. S. Gopinath, J. Pinjari, R. T. Dere, A. Verma, P. Vishwakarma, R. Shivahare, M. Moger, P. S. Kumar Goud, V. Ramanathan, P. Bose, M. V. Rao, S. Gupta, S. K. Puri, D. Launay and D. Martin, Eur. J. Med. Chem., 2013, 69, 527–536 CrossRef CAS PubMed.
- H. Cope, R. Mutter, W. Heal, C. Pascoe, P. Brown, S. Pratt and B. Chen, Eur. J. Med. Chem., 2006, 41, 1124–1143 CrossRef CAS PubMed.
- Y. Wu, W. Dai, X. Chen, A. Geng, Y. Chen, T. Lu and Y. Zhu, RSC Adv., 2017, 7, 52180–52186 RSC.
- A. Jordan, D. Bisgrove and E. Verdin, EMBO J., 2003, 22, 1868–1877 CrossRef CAS PubMed.
- Y. K. K., R. Pearson, J. Hokello, K. Lassen, J. Friedman, M. Tyagi and J. Karn, J. Virol., 2008, 82, 12291–12303 CrossRef PubMed.
- V. T. Nguyen, T. Kiss, A. A. Michels and O. Bensaude, Nature, 2001, 414, 322–325 CrossRef CAS PubMed.
- Z. Li, J. Guo, Y. Wu and Q. Zhou, Nucleic Acids Res., 2013, 41, 277–287 CrossRef CAS PubMed.
- P. G. Mccaffrey, C. Luo, T. K. Kerppola, J. Jain, T. M. Badalian, A. M. Ho, E. Burgeon, W. S. Lane, J. N. Lambert and T. Curran, Science, 1993, 262, 750–754 CAS.
- D. D. Richman, D. M. Margolis, M. Delaney, W. C. Greene, D. Hazuda and R. J. Pomerantz, Science, 2009, 323, 1304–1307 CrossRef CAS PubMed.
- J. H. N. Yik, R. C. Chen, R. Nishimura, J. L. Jennings, A. J. Link and Q. Zhou, Mol. Cell, 2003, 12, 971–982 CrossRef CAS PubMed.
- Z. Yang, Q. Zhu, K. Luo and Q. Zhou, Nature, 2001, 414, 317–322 CrossRef CAS PubMed.
- C. Jeronimo, D. Forget, A. Bouchard, Q. Li, G. Chua, C. Poitras, C. Thérien, D. Bergeron, S. Bourassa and J. Greenblatt, Mol. Cell, 2007, 27, 262–274 CrossRef CAS PubMed.
- B. J. Krueger, C. Jeronimo, B. B. Roy, A. Bouchard, C. Barrandon, S. A. Byers, C. E. Searcey, J. J. Cooper, O. Bensaude and É. A. Cohen, Nucleic Acids Res., 2008, 36, 2219–2229 CrossRef CAS PubMed.
- B. Sobhian, N. Laguette, A. Yatim, M. Nakamura, Y. Levy, R. Kiernan and M. Benkirane, Mol. Cell, 2010, 38, 439–451 CrossRef CAS PubMed.
- J. E. Brogie and D. H. Price, Nucleic Acids Res., 2017, 45, 6864–6880 CrossRef CAS PubMed.
- J. Wu, M. T. Ao, R. Shao, H. R. Wang, D. Yu, M. J. Fang, X. Gao, Z. Wu, Q. Zhou and Y. H. Xue, Sci. Rep., 2017, 7 Search PubMed.
- N. He, N. S. Jahchan, E. Hong, Q. Li, M. A. Bayfield, R. J. Maraia, K. Luo and Q. Zhou, Mol. Cell, 2008, 29, 588–599 CrossRef CAS PubMed.
- Y. H. Xue, Z. Y. Yang, R. C. Chen and Q. Zhou, Nucleic Acids Res., 2010, 38, 360–369 CrossRef CAS PubMed.
- Z. Y. Yang, J. H. N. Yik, R. C. Chen, N. H. He, M. K. Jang, K. Ozato and Q. Zhou, Mol. Cell, 2005, 19, 535–545 CrossRef CAS PubMed.
- N. H. He, M. Liu, J. Hsu, Y. H. Xue, S. Chou, A. Burlingame, N. J. Krogan, T. Alber and Q. Zhou, Mol. Cell, 2010, 38, 428–438 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra01216d |
| ‡ These authors contribute equally to this paper. |
| This journal is © The Royal Society of Chemistry 2018 |