A facile atom economic one pot multicomponent synthesis of bioactive spiro-indenoquinoxaline pyrrolizines as potent antioxidants and anti-cancer agents†
Kailasam
Saravana Mani
 a,
Werner
Kaminsky
a,
Werner
Kaminsky
 b and
Subramaniam Parameswaran
Rajendran
*a
b and
Subramaniam Parameswaran
Rajendran
*a
aDepartment of Chemistry, Bharathiar University, Tamil Nadu, India. E-mail: chemsaran.mani@gmail.com; rajendransp@yahoo.com
bDepartment of Chemistry, University of Washington, Seattle, WA 98195, USA
First published on 14th November 2017
Abstract
A competent and highly discriminating one-pot method for the synthesis of biologically active novel spiro-indenoquinoxaline pyrrolizines accumulating three pharmocophoric cores, heterocyclic quinoline scaffold, pyrrolizines and indeno-quinoxaline, in a single molecular framework by means of a four-component reaction between ninhydrin, o-phenylenediamine, L-proline and quinolinyl chalcones in methanol via [3+2] cycloaddition was developed. The structures of the compounds were well characterized by FT-IR, NMR, ESI-MS, XRD and elemental analysis. The synthesized compounds were screened for in vitro antioxidant activity using DPPH, nitric oxide, super oxide radical and in vitro cytotoxic activity against MCF-7 and A-549 cancer cell lines and visualized using the fluorescent microscopic technique. The newly synthesized compounds exhibited excellent antioxidant activity compared to the standard molecule, i.e., BHT. In terms of cytotoxic activity, compounds 5c, 5e, 6c and 6e were found to exhibit significant activity, when compared to standard doxorubicin against MCF-7 and A-549 cancer cells using the MTT assay method. The cell morphology analysis clearly indicates cell death was induced by apoptosis and necrosis pathways. Molecular docking studies were performed using an EGFR inhibitor to determine the molecular interactions between the compounds and proteins.
1. Introduction
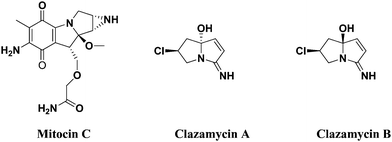
At present, it is still a great challenge for synthetic chemists to develop novel bioactive molecules for the treatment of cancer, which are safe and have fewer side effects. Chemotherapy is the leading cancer treatment approach; however, its drawbacks including death of normal cells and long time-duration or dose level limit its clinical application.1 Nitrogen containing heterocycles constructed through 1,3-dipolar cycloaddition of azomethine ylides2–5 have been documented as an important cluster of significant bioactive compounds. It is a major class of heterocyclic systems and plays an essential role in the field of anticancer drug enlargement. In addition, it finds several applications in medicinal chemistry, including antibacterial,6 antimalarial,7 anti-inflammatory,8 apoptosis,9 DNA repair and tyrosine kinases (TK) inhibition.10 Tetracyclic indeno quinoxalines are also an another important class of heterocycles used as intermediates in the synthesis of several types of spiro compounds,11,12 which possess potent α-glycosidase inhibitors.Pyrrolizine derivatives represent a class of novel heterocycles that serve as promising scaffolds for anticancer drugs. The unique antitumor properties of mitocin C inspired chemists to develop different pyrrolizine systems and assess their potential antitumor activities against a wide variety of cancer types.13
Mitocin C isolated from Streptomyces caespitosus or Streptomyces laevendulae is used to treat upper esophageal carcinoma, anal, breast cancers and superficial bladder tumors.14–16 Further, it is believed to act as a good DNA alkylator, which can cross-link DNA with high efficiency and specificity.17,18 clazamycin A and clazamycin B are naturally occurring pyrrolizines with very interesting anti-tumor properties19 (Fig. 1). Recently, multicomponent reactions have become a powerful tool to construct molecules with structural diversity and complexity with minimum waste generation and are widely applicable in both combinatorial and medicinal chemistry arenas.20 The abovementioned tremendous biological evaluation of quinolines, indeno quinoxaline and pyrrolizines prompted us to develop a cluster of all three heterocyclic scaffolds together by a multicomponent reaction and perform their biological evaluation.
In this paper, we report a new synthetic methodology for the synthesis of novel bioactive spiro-indenoquinoxaline pyrrolizine molecules through multicomponent reactions. Herein, we would like to report an efficient, atom economic, regioselective and high yielding MCR protocol for one-pot facile synthesis of bioactive functionalized spiro-indenoquinoxaline pyrrolizine heterocyclic scaffolds by the reaction of ninhydrin, o-phenylenediamine, proline, quinoline bearing chalcones on refluxing in methanol and the evaluation of their in vitro antioxidant and in vitro anti tumor activity.
2. Results and discussion
2.1. Chemistry
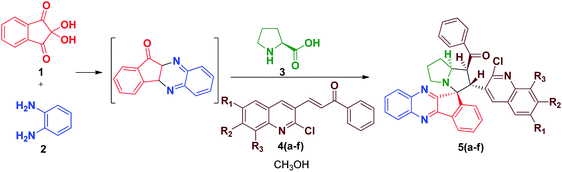
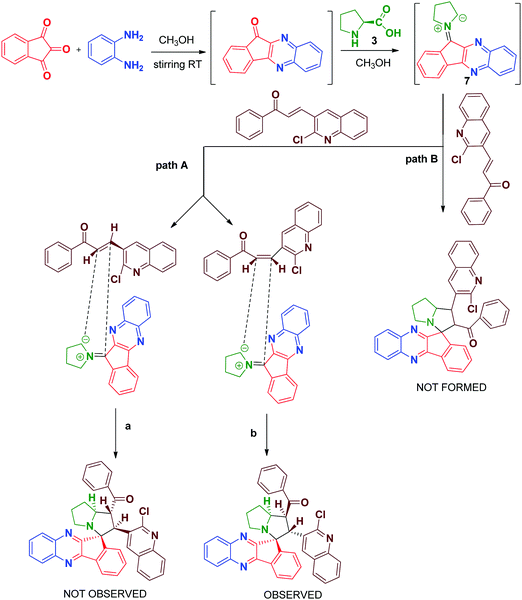
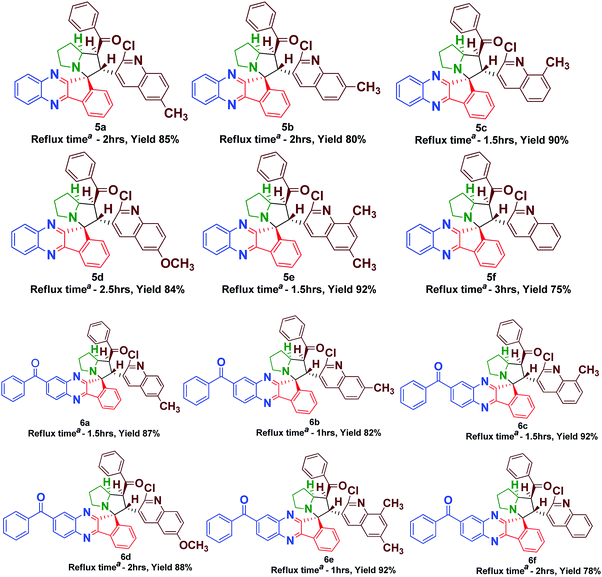
The synthesis of spiro [indeno[1,2-b] quinoxaline] pyrrolizines 5(a–f) and 6(a–f) was achieved through a 1,3-dipolar cycloaddition reaction protocol.21 Initially, quinoline based dipolarophiles 4(a–f) were synthesized by the Claizen Schmidt condensation reaction between 2-chloro-3-formyl quinoline derivatives and acetophenone under alcoholic KOH conditions. An equimolar mixture of ninhydrin 1, o-phenylenediamine 2, L-proline 3 and dipolarophile 4a in methanol was refluxed for 2 to 3 h (Scheme 1). After monitoring the completion of the reaction by TLC, a single product, spiro-indenoquinoxaline pyrrolizines 5a, was separated by evaporation of the solvent in good yields. The optimized reaction condition, time and yields of 5(a–f) are mentioned in Table 1. The FT-IR spectrum of 5a displayed characteristic bands at 1680 cm−1, 1588 cm−1 and 759 cm−1 corresponding to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, –C
O, –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C–Cl in quinoline dipolarophile moiety, respectively. The 1H NMR spectrum of product 5a, showed the characteristic multiplets at δ 1.89–2.64 corresponding to the pyrrolizine ring.
N and C–Cl in quinoline dipolarophile moiety, respectively. The 1H NMR spectrum of product 5a, showed the characteristic multiplets at δ 1.89–2.64 corresponding to the pyrrolizine ring.
| a Isolated yield, reaction condition: ninhydrin 1 (1 mmol), o-phenylenediamine 2 or 3,4-diamino benzophenone 2a (1mmol), L-proline 3 (1 mmol) quinoline chalcones 4 (1 mmol), methanol (10 ml). |
|---|
|
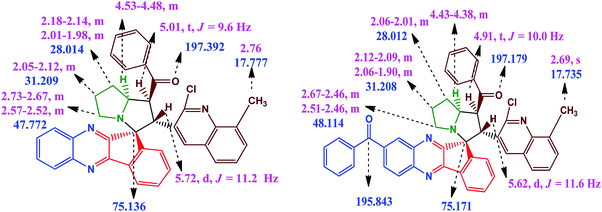
The proton attached to the benzoyl group appeared at δ 4.99 as a triplet with a coupling constant (J = 9.6 Hz) and a pyrrolizine ring proton attached to the quinoline moiety appeared at δ 5.70 as a doublet with a coupling constant (J = 11.6 Hz). The proton attached to the benzoyl moiety is consistent with trans disposition with the proton attached to the quinoline moiety. Similarly, the –NCH proton attached to the pyrrolizine ring is consistent with the cis position to the proton attached to the benzoyl group as shown in the XRD structure in Fig. 2, while the signals for all other protons in the aromatic region are in good agreement with the proposed structure (Fig. 3).
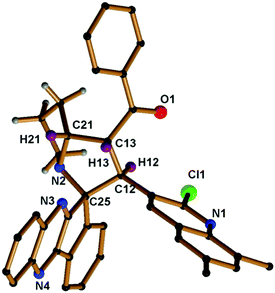 | ||
| Fig. 2 Molecular structure of compound 5e; hydrogen atoms in the pyrrolizine ring are omitted for clarity (CCDC 1569306).† | ||
In the 13C NMR spectrum, the quaternary spiro carbon appears at δ 75.12 and the carbonyl carbon attached to the phenyl ring appears at δ 197.34. Moreover, the signals for all other carbons located at appropriate chemical shifts were in perfect agreement with the proposed structure. In DEPT-135 NMR, the appearance of three negative signals corresponding to the methylene protons in the pyrrolizine core further strongly attested the formation of the product. The observed mass spectrum of the product HRMS (TOF ES+) m/z 593.2092 further confirmed the formation of compound 5a.
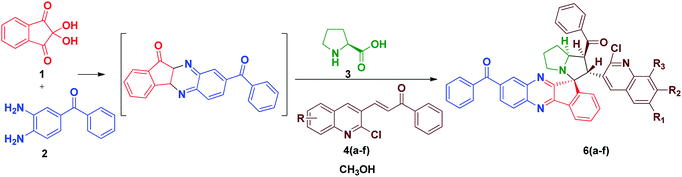
In order to introduce the electron withdrawing substituent benzoyl group in the quinoxaline core, we used in 3,4-diaminobenzophenone instead of o-phenylenediamine. Using the optimized conditions, we then explored the scope of this cycloaddition reaction using ninhydrin 1, 3, 4-diaminobenzophenone 2a, L-proline 3 and quinoline dipolarophile 4(a–f) in refluxing methanol (Scheme 2). After the completion of reaction, monitored by TLC, the precipitate obtained was filtered and washed with water to afford the single product 6(a–f). In the IR spectrum of compound 6a, characteristic peaks appeared at 1659 cm−1, 1586 cm−1 and 701 cm−1, which are attributed to the carbonyl stretching, quinoline –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C–Cl, respectively. The yield of compounds 6(a–f) would be slightly high when compared to compounds 5(a–f).
N and C–Cl, respectively. The yield of compounds 6(a–f) would be slightly high when compared to compounds 5(a–f).
In the 1H NMR spectrum of compound 6a, a sharp singlet appeared at δ 2.50, attributed to the –CH3 group in the quinoline scaffold. Multiplets of the pyrrolizine ring proton appeared in the range δ 1.97–δ 2.71. The proton attached to the quinoline moiety would be slightly deshielded to δ 5.69 as the doublet (J = 11.6 Hz) and the proton attached to benzoyl group resonated at δ 4.96 as the triplet (J = 9.6 Hz). The chemical shifts at aromatic regions are in good agreement with the proposed structure. The observed mass spectrum of the product HRMS (TOF ES+) m/z 697.2482 further confirmed the formation of compound 6b.
In the 13C NMR spectrum, the peaks at δ 197.116 and δ 195.863 are attributed to two carbonyl carbons and the peak at δ 75.181 is attributed to the quaternary spiro carbon. These observed chemical shift values are in good agreement with the structure of the compound 6a. In the DEPT-135 NMR spectrum, the negative signals correspond to three –CH2 protons in the pyrrolizine core. Furthermore, the presence of a molecular ion peak at m/z 697.2476 in the mass spectrum confirmed the formation of product 6c. The formation of the spiro[indeno[1,2-b]quinoxaline]-11,3′-pyrrolizines scaffold probably involves a complex multistep sequence. Initially, the cyclocondensation reaction takes place between ninhydrin 1 and o-phenylenediamine 2 forms indeno-quinoxaline-11-one, which further condenses with proline 3 that undergoes decaboxylation to give azomethine ylide 7 subsequently through cycloaddition reaction with dipolarophile 4 to afford the final product (Scheme 3).
3. Biological evaluation of synthesized compounds
3.1. In vitro antioxidant activity
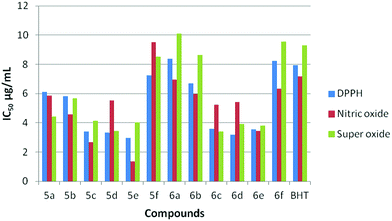
The antioxidant effectiveness primarily involves scavenging radicals and inhibiting oxidations of biological species. A number of quinoline derivatives were tested for anti oxidant activity as reflected by the ability to inhibit lipid per-oxidation in rat brain and kidney homogenates. Free radical scavenging capacity can be investigated by the neutralization of electrons or hydrogens. Compounds having low IC50 values and high hydrogen donating ability exhibit enhanced antioxidant activity. The radical scavenging capacity of compounds 5(a–f) and 6(a–f) along with the standard BHT were tested against DPPH, nitric oxide and super oxide radicals and the results are summarized in (Table 2).| Compounds | DPPH | Nitric oxide | Super oxide |
|---|---|---|---|
| 5a | 6.13 | 5.87 | 4.43 |
| 5b | 5.82 | 4.56 | 5.69 |
| 5c | 3.42 | 2.68 | 4.12 |
| 5d | 3.32 | 5.51 | 3.45 |
| 5e | 2.96 | 1.34 | 4.01 |
| 5f | 7.23 | 9.52 | 8.54 |
| 6a | 8.4 | 6.95 | 10.11 |
| 6b | 6.71 | 5.98 | 8.65 |
| 6c | 3.6 | 5.24 | 3.4 |
| 6d | 3.2 | 5.43 | 3.9 |
| 6e | 3.56 | 3.43 | 3.8 |
| 6f | 8.24 | 6.32 | 9.54 |
| BHT | 7.93 | 7.19 | 9.30 |
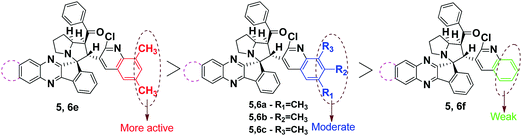
From the results obtained, all the synthesized compounds (Fig. 4) exhibited excellent activity when compared to that of the standard BHT. Interestingly, compound 5e exhibits the most promising activity with the IC50 values of 2.96, 1.34 and 4.01 against DPPH, Nitric oxide and super oxide radicals, which outperformed the standard BHT with the IC50 values of 7.93, 7.19 and 9.30, respectively. Next in the series, compounds 5c, 5d, 6c, 6d and 6e showed excellent activity in all three radical scavenging assays. The compounds 5a, 5b, 5f, 6a, 6b and 6f displayed equipotent activity with the standard molecule BHT. The reason for high scavenging capacity of the compounds can be the nature of substituents in the quinoline core. The results clearly suggest that compounds possessing electron donating groups22,23 at the 6th and 8th positions of the quinoline scaffold would be significantly more active than compounds possessing an electron donating group at the 7th position. The presence of –OCH324 at the 6th position would be more active when compared to the –CH3 group at the 6th position. From the structural analysis, the absence of an electron donating group in the quinoline moiety led to decrease in activity when compared to rest of the compounds (Scheme 4).
3.2. In vitro cytotoxic activity
The use of drugs in different targets, often discrete with multiple biological targets, with the expectation of synergistic effects and low toxicity, safety becomes an ideal approach to increase interest in the present research scenario. The anticancer properties of the synthesized compounds 5(a–f) and 6(a–f) were screened against MCF-7 and A-549 cancer cell lines by the MTT [3-(4,5-dimethylthiazo-2-yl)-2,5-diphenyltetrazolium bromide] assay method. Doxorubicin can act as a positive control for this study. Inhibition of cells was measured in terms of IC50 values after 48 h of cell exposure. The results obtained are summarized in Table 3.| Samples | MCF-7a | A-549b |
|---|---|---|
| a MCF-7 (breast cancer cell line). b A-549(lung cancer cell line). | ||
| 5a | 33 ± 1.4 | 24 ± 1.0 |
| 5b | 23 ± 0.5 | 36 ± 1.5 |
| 5c | 17 ± 1.6 | 19 ± 0.2 |
| 5d | 28 ± 1.5 | 24 ± 1.5 |
| 5e | 15 ± 1.9 | 16 ± 1.8 |
| 5f | 35 ± 1.5 | 37 ± 1.7 |
| 6a | 25 ± 1.4 | 32 ± 1.0 |
| 6b | 24 ± 1.5 | 32 ± 0.9 |
| 6c | 18 ± 1.3 | 20 ± 1.5 |
| 6d | 26 ± 1.3 | 24 ± 0.5 |
| 6e | 16 ± 0.5 | 17 ± 1.8 |
| 6f | 30 ± 0.7 | 33 ± 1.8 |
| Doxorubucin | 14 ± 1.0 | 15 ± 1.4 |
As listed in Table 3, all the synthesized compounds exhibited good to moderate activity against two cell lines when compared to the Doxorubicin standard. Among the synthesized compounds, compounds 5e and 6e exhibited equipotent activity with IC50 values of 15 ± 1.6 and 16 ± 0.5 against the MCF-7 cell line and 16 ± 1.8 and 17 ± 1.8 against the A-549 cell lines, compared to that of positive control Doxorubicin 14 ± 1.0 (MCF-7) and 15 ± 1.4 (A-549), respectively. Increase in the cytotoxic values of 5e and 6e suggests that the electron donating methyl group can be favorable at the 6th and 8th positions of the quinoline core.
Upon further investigation of the methyl group in the position of the quinoline core, it was revealed that surprisingly the methyl group at the 8th position possess good activity with IC50 values of 17 ± 1.6 and 18 ± 1.3 (compound 5c) against MCF-7 and 19 ± 0.2 and 20 ± 1.8 (compound 6c) against A-549 cancer cell lines. Moreover, rest of the compounds with electron donating groups at the 6th and 7th position exhibited moderate activity against both cancer cell lines. This clearly indicates that an electron donating group at the 8th position increases the anticancer potency rather than other positions. The absence of electron donating groups in any of the positions in the quinoline core displayed weak activity towards MCF-7 and A-549 cell lines (Scheme 4).
3.3. Cell morphology analysis
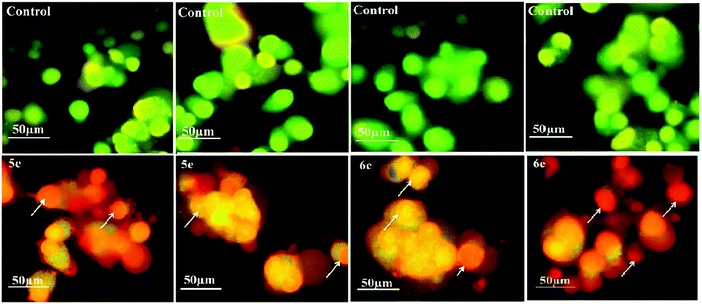
To monitor the effect of synthesized compounds on cell morphology, treated cancer cells were examined by inverted light microscopy and compared with untreated cells. The treated cells showed significant changes compared with untreated cells. Cytological investigations elucidate the anticancer effect routed through membrane blebbing, membrane instability and disturbing the cytoskeleton of the cells by the compounds.Fig. 5 and 6 reveal the morphological changes in MCF-7 and A-549 after treatment with compounds with their respective inhibitory concentrations for 24 h. Phase contrast micrographs revealed that the compounds 5c, 5e, 6c and 6e induced increased cell shrinkage, membrane blebbing and form floating cells in a dose independent manner.
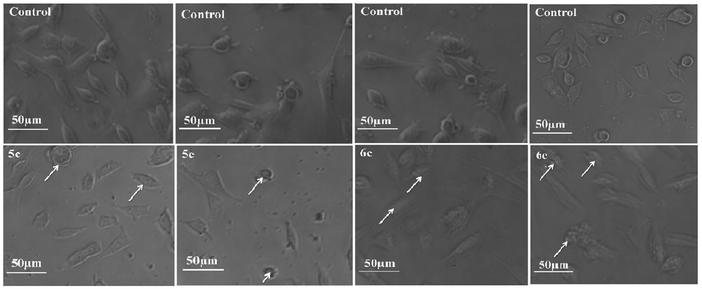 | ||
| Fig. 5 Morphological analysis of treated MCF-7 cancer cells and the arrow mark indicates the formation of floating cells and appearance of membrane blebbing. | ||
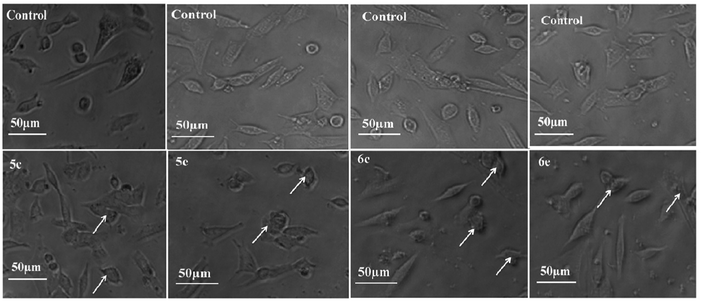 | ||
| Fig. 6 Morphological analysis of treated A-549 cells and the arrow mark indicates the formation of floating cells and appearance of membrane blebbing. | ||
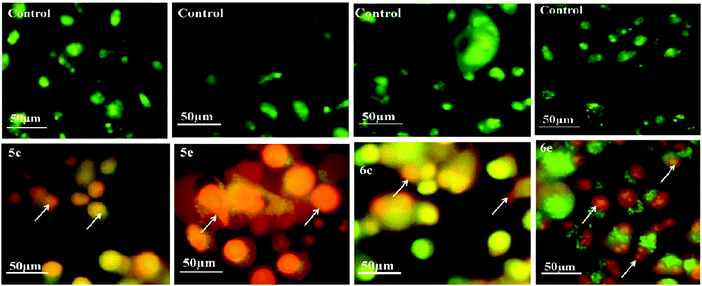
3.4. Acridine orange–ethidium bromide (AO–EB) staining
Acridine orange/ethidium bromide (AO/EB) fluorescent staining assay were performed to distinguish the pathway, i.e. apoptotic, necrotic and live cells.25 AO permeates the intact cell membrane and stains the nuclei green, while EB permeates only cells with loss in membrane integrity and stains the nucleus orange. MCF-7 and A-549 cells untreated and treated with compounds 5c, 5e, 6c and 6e were stained with AO–EB and analyzed under a fluorescent microscope. Fig. 7 and 8 show that the untreated cancer cells (control) did not exhibit any significant adverse effects compared to the compounds-treated cancer cells. It can be observed that with the addition of compounds to the cancer cells, the green color cells are converted into orange/red color cells, which is due to induced apoptosis and the nuclear condensation effects on the cells. Fig. 7 and 8 also show that the compounds were significantly inducing apoptosis in the investigated cancer lines.3.5. DAPI straining method
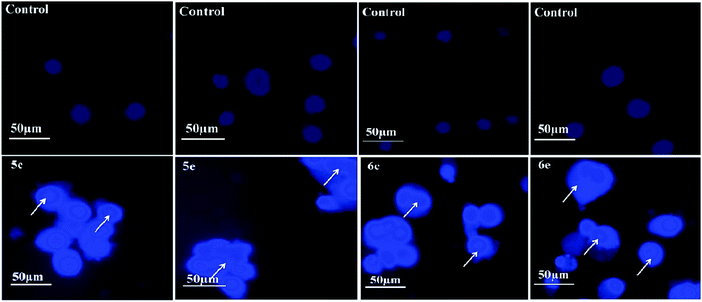
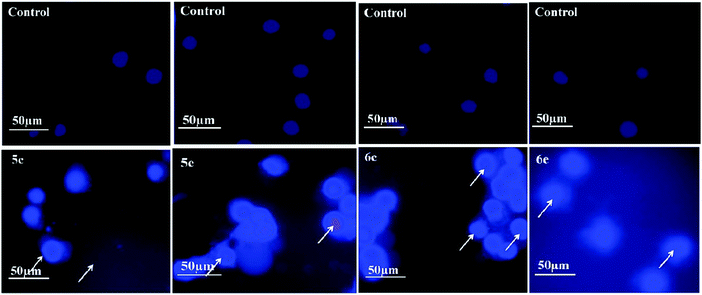
4′,6-Diamidino-2-phenylindole (DAPI) is a nuclear stain which strongly binds to the adenine–thymine clusters of the minor groove of double-stranded DNA and detect the chromatin condensation or nuclear damage. Binding of DAPI to DNA produces a ∼20-fold enhanced fluorescence apparently due to the displacement of water molecules from both DAPI and the minor groove.26 DAPI stains the apoptotic cells as bright colored due to the condensed nucleus, which is a typical apoptotic feature. Fluorescence microscopy images of lung and breast cancer cells after 24 h stained with DAPI in the absence and presence of compounds are shown in Fig. 9 and 10, respectively, which reveal that the untreated cells did not show any significant changes, while cancer cells treated with compounds show bright fetches, which indicates the presence of condensed chromatins and nuclear fragmentations in the cancer cells. Thus, from the results of MTT assay and Fluorescence microscopy analysis, we confidently report herein that the compounds can be used as potent therapeutic agents.3.6. Molecular docking
Docking studies predicted the interaction of inhibitors with protein and residues involved in this complex. The orientation and conformation of inhibitors bound in the active site of the protein and the formed protein–ligand complex with the highest binding energy value are the most important requirements for docking studies. The inhibition potency of the newly synthesized compounds, 5e and 6e, was subjected to further docking studies to explore the binding pattern against the Epidermal growth factor receptor (EGFR) (PDB code: 1M17)27 using Autodock 4.2 software.28 As depicted from Fig. 11, compound 5e is surrounded by hydrophobic amino acids such as GLY 695, CYS 773, TYR 777, GLY 772, LEU 694, VAL 693, LYS 704 and LYS 692. The key amino acid CYS 773 is considered to play a crucial role in the stabilization of the receptor by compound 5e, which can be observed in the hydrophobic enclosure, in which the nitrogen atom in the quinoxaline ring is mounted towards the CYS 773 hydrophobic amino acid (1.8 Å) with a glide score (−5.54 kcal mol−1). Similarly, the compound 6e is surrounded by hydrophobic amino acids ARG 817, LEU 820, GLY 772, CYS 773, LYS 692, PHE 699 etc. Similar to compound 5e, it was also observed in case of compound 6e that the key amino acid CYS 773 is considered to play a crucial role in the stabilization of the receptor through hydrophobic enclosure where the nitrogen atom in the quinoxaline ring is hydrogen bonded with CYS 773 hydrophobic amino acid (2.2 Å) with glide score (−6.05 kcal mol−1). Therefore, it is assumed that hydrophobicity is one of the important physicochemical properties, which impart lipophilicity to the molecule as well as membrane permeability and their absorption in biological systems.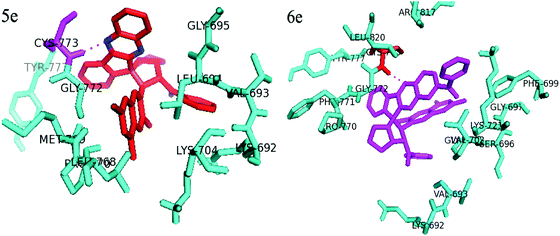 | ||
| Fig. 11 Docking studies on compounds 5e and 6e with EGF receptor and dotted lines shows hydrogen bond. | ||
4. Conclusion
In summary, we have developed a highly atom economic method for the synthesis of spiro indeno-quinoxaline derivatives via one pot four component 1,3-dipolar cycloaddition reactions of azomethine ylide and quinoline derived dipolarophile in a successful manner. The pure products were isolated by recrystallization. The structures of the synthesized compounds were well characterized using single crystal XRD, spectral and elemental analysis. All the synthesized compounds were successfully screened against antioxidants and cytotoxic activity for two cancer cell lines (MCF-7 and A-549). Interestingly, all the compounds exhibited excellent activity against DPPH, nitric oxide and super oxide free radicals. Compounds 5c, 5e, 6c and 6e were found to be significantly active in this series and also induced cell death by apoptosis and necrosis. The substituents of electron donating groups in the 6th and 8th positions turned out to be the best candidates in this screened series.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank SAIF, CUSAT, Cochin for NMR spectral analysis and University of Washington, Seattle, US for Single crystal XRD data. We thank the Department of Botany and Zoology, Bharathiar University for evaluating antioxidant and anticancer studies.References
- S. Dhar, N. Kolishetti, S. J. Lippard and O. C. Farokhzad, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1850 CrossRef CAS PubMed.
- A. M. Akondi, S. Mekala, M. L. Kantam, R. Trivedi, L. Raju Chowhane and A. Dasbc, New J. Chem., 2017, 41, 873 RSC.
- R. Sakhuja, S. S. Panda, L. Khanna, S. Khurana and S. C. Jain, Bioorg. Med. Chem., 2011, 21, 5465 CrossRef CAS PubMed.
- A. A. Raj, R. Raghunathan, M. R. Sridevi Kumari and N. Raman, Bioorg. Med. Chem., 2003, 11, 407 CrossRef CAS PubMed.
- B. V. Subba Reddy, G. Karthik, T. Rajasekaran, A. Antony and B. Sridhar, Tetrahedron Lett., 2012, 53, 2396 CrossRef CAS.
- N. Sun, R. L. Du, Y. Y. Zheng, B. H. Huang, Q. Guo, R. F. Zhang, K. Y. Wong and Y. J. Lu, Eur. J. Med. Chem., 2017, 135, 1 CrossRef CAS PubMed.
- K. Kaur, M. Jain, R. P. Reddy and R. Jain, Eur. J. Med. Chem., 2010, 45, 8 Search PubMed.
- E. Rajanarendar, M. Nagi Reddy, S. Rama Krishna, K. Rama Murthy, Y. N. Reddy and M. V. Rajam, Eur. J. Med. Chem., 2012, 55, 273 CrossRef CAS PubMed.
- O. Afzal, S. Kumar, M. R. Haider, M. R. Ali, R. Kumar, M. Jaggi and S. Bawa, Eur. J. Med. Chem., 2015, 97, 871 CrossRef CAS PubMed.
- K. M. Amin, H. H. Georgey and F. M. Awadallah, Med. Chem. Res., 2011, 20, 1042 CrossRef CAS.
- M. T. Maghsoodlu, S. M. Habibi-Khorassani, A. Morani, N. Hazeri, A. Davodi and S. S. Sajadikhah, Tetrahedron, 2011, 67, 8492 CrossRef.
- M. Sahar Kahn, M. A. Munawar and M. Ashraf, Bioorg. Med. Chem. Lett., 2014, 22, 1195 CrossRef PubMed.
- A. Belal, B. El-Dien and M. El-Gendy, Bioorg. Med. Chem., 2014, 22, 46 CrossRef CAS PubMed.
- S. K. Carter and S. T. Crooke, Mitocin C: Current status and new developments, Academic Press, 1979 Search PubMed.
- H. Schwartz, J. E. Sodergren and F. S. Philips, Science, 1963, 142, 1181 CAS.
- W. T. Bradner, Cancer Treat. Rev., 2001, 27, 35 CrossRef CAS PubMed.
- M. Tomasz, R. Lipman, D. Chowdary, J. Pawlak, G. L. Verdine and K. Nakanishi, Science, 1987, 235, 1204 CAS.
- M. Tomasz, Chem. Biol., 1995, 2, 575 CrossRef CAS.
- D. D. Buechter and D. E. Thurston, J. Nat. Prod., 1987, 50, 360 CrossRef CAS.
- (a) A. Domling and I. Ugi, Angew. Chem., Int. Ed., 2000, 39, 3168 CrossRef CAS; (b) A. Domling and I. Ugi, Chem. Rev., 2006, 106, 17 CrossRef PubMed; (c) R. V. A. Orru and M. de Greef, Synthesis, 2003, 1471 CrossRef CAS; (d) J. D. Sunderhaus and S. F. Martin, Chem. – Eur. J., 2009, 15, 1300 CrossRef CAS PubMed; (e) N. Isambert and R. Lavilla, Chem. – Eur. J., 2008, 14, 8444 CrossRef CAS PubMed; (f) D. J. Ramon and M. Yus, Angew. Chem., Int. Ed., 2005, 44, 1602 CrossRef CAS PubMed.
- (a) A. Dandia, V. Parewa, S. Kumari, S. Bansal and A. Sharma, Green Chem., 2016, 18, 2488 RSC; (b) A. Dandia, A. K. Jain, A. K. Laxkar and D. S. Bhati, Tetrahedron Lett., 2013, 54(12), 3180 CrossRef CAS; (c) A. Dandia, A. K. Jain and A. K. Laxkar, RSC Adv., 2013, 3, 8422 RSC; (d) A. Dandia, A. K. Jain, A. K. Laxkar and D. S. Bhati, Tetrahedron, 2013, 69(8), 2062 CrossRef CAS.
- H. Y. Zhang, Curr. Comput. Aided Drugs Des., 2005, 1, 257 CrossRef CAS.
- M. Sankaran, K. Chandraprakash, C. Uvarani and P. S. Mohan, Bioorg. Med. Chem. Lett., 2010, 20, 7147 CrossRef CAS PubMed.
- K. P. Rakesh, H. M. Manukumar and D. C. Gowda, Bioorg. Med. Chem. Lett., 2015, 25, 1072 CrossRef CAS PubMed.
- Y. Chang, Y. Li, N. Ye, X. Guo, Z. Li, G. Sun and Y. Sun, Apoptosis, 2016, 21, 977 CrossRef CAS PubMed.
- T. S. Reddy, H. Kulhari, V. G. Reddy, A. V. S. Rao, V. Bansal, A. Kamal and R. Shukla, Org. Biomol. Chem., 2015, 13, 10136 Search PubMed.
- J. Stamos, M. X. Sliwkowski and C. Eigenbrot, J. Biol. Chem., 2002, 277, 46265 CrossRef CAS PubMed.
- E. M. Pique and R. Huey, The Autodock4.2 molecular graphics system, http://autodock.scripps.edu/downloads/autodock-registration/autodock-4-2-download-page, 2010.
Footnote |
| † Electronic supplementary information (ESI) available: X-ray crystal data and Spectral data of all the compounds associated with article will be available. CCDC 1569306. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c7nj02993d |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2018 |