Nanoscale membrane architecture of healthy and pathological red blood cells†
Andra C.
Dumitru‡
a,
Mégane A.
Poncin‡
a,
Louise
Conrard
b,
Yves F.
Dufrêne
ac,
Donatienne
Tyteca
b and
David
Alsteens
 *a
*a
aUniversité catholique de Louvain, Institute of Life Sciences, Croix du Sud 4-5, bte L7.07.06, B-1348 Louvain-la-Neuve, Belgium. E-mail: david.alsteens@uclouvain.be
bUniversité catholique de Louvain, de Duve Institute, Avenue Hippocrate 75/B1.75.05, B-1200 Woluwe-Saint-Lambert, Belgium
cWalloon Excellence in Life sciences and Biotechnology (WELBIO), Belgium
First published on 10th March 2018
Abstract
Red blood cells feature remarkable mechanical properties while navigating through microcirculation vessels and during spleen filtration. An unusual combination of plasma membrane and cytoskeleton physical properties allows red blood cells to undergo extensive deformation. Here we used atomic force microscopy multiparametric imaging to probe how cellular organization influences nanoscale and global mechanical properties of cells in both physiological and pathological conditions. Our data obtained in native conditions confirmed that, compared to healthy cells, cells from patients with hereditary spherocytosis are stiffer. Through vertical segmentation of the cell elasticity, we found that healthy and pathological cells display nanoscale architecture with an increasing stiffness along the direction of the applied force. By decoupling the mechanical response of the plasma membrane from its underlying cytoskeleton, we find that both components show altered properties in pathological conditions. Nanoscale multiparametric imaging also revealed lipid domains that exhibit differential mechanical properties than the bulk membrane in both healthy and pathological conditions. Thanks to correlated AFM-fluorescence imaging, we identified submicrometric sphingomyelin-enriched lipid domains of variable stiffness at the red blood cell surface. Our experiments provide novel insights into the interplay between nanoscale organization of red blood cell plasma membrane and their nanomechanical properties. Overall, this work contributes to a better understanding of the complex relationship between cellular nanoscale organization, cellular nanomechanics and how this 3D organization is altered in pathological conditions.
Conceptual insightsRed blood cells (RBCs) feature remarkable mechanical properties while navigating through microcirculation vessels and during spleen filtration. We investigated the architecture of RBCs at different scale, from the cellular level to the nanoscale and determined the biological properties by linking the nanoscale architecture with nanomechanical properties. Here, we report the use of force–distance curve-based atomic force microscopy (AFM) to study the three-dimensional architecture and nanomechanical response of the red blood cells in physiological and pathological conditions. The study was carried out in parallel on RBCs from healthy volunteers and from patients with hereditary spherocytosis. Mechanical analysis of both patient types confirms previous studies showing that RBCs from patients with hereditary spherocytosis are stiffer. Hence our results demonstrate a novel correlation between the lipid composition, organization in lipid domains and cell surface nanomechanics. We suggest that these domains contribute to the membrane homeostasis and that their mechanobiological function is directly involved in local cellular deformations. Our nanoscale observations provide direct links with RBC pathologies (spherocytosis, elliptocytosis,…) and contribute to a better understanding of these RBC diseases. In nanomedicine, this research demonstrates the power of AFM to decipher new pathological mechanisms, thus opening new avenues for therapies. |
The unique structural organization of the human red blood cell (RBC) enables it to undergo large reversible deformations while maintaining constant volume and membrane surface area.1–3 Lacking the actin–myosin–microtubule cytoskeleton that is responsible for shape changes in nucleated cells, RBCs maintain their structural integrity and can display dynamic local deformations thanks to a dynamic network of spectrin filaments tethered to the cytosolic side of the plasma membrane (PM) via multiprotein complexes centered on ankyrin and 4.1R protein.4 The surface tension of the lipid bilayer and the constant remodeling of the flexible spectrin network are two key elements governing the elastic response of the RBC membrane skeleton. Currently, there is increasing evidence that submicrometric lipid domains exist at the RBC surface and could contribute to PM tension regulation, being involved in local deformations, such as local budding and vesicle formation.5,6 Submicrometric domains may also serve as recruitment or exclusion platforms for membrane proteins, participating in the regulation of dynamic cellular processes such as signal transduction. For example, we have recently shown that sphingomyelin (SM)-enriched submicrometric domains increase in abundance along with secondary calcium (Ca2+) efflux, a process involved in shape and volume restoration after deformation.6 However, whether such submicrometric domains coexist with nanoscale structural or physical heterogeneities at the RBC surface and whether they contribute with the cytoskeleton to RBC deformation remain to be determined.
Disorders due to mutations in various membrane or skeletal proteins, such as in hereditary spherocytosis (HS), induce loss of membrane surface and reduced RBC deformability, in turn altering cell function and leading to splenic trapping and ultimately anemia. Dysregulation of lipid loss and the presence of altered membrane proteins have been alluded as key components altering RBC plasticity and biconcavity in pioneering biochemical studies of HS PMs several decades ago.7–9 Though the aforementioned studies shed light on the link between cellular composition and pathology, direct evidence for a correlation between specific lipid distribution in submicrometric domains or PM-cytoskeleton anchorage and their role in PM deformability at the nanoscale is missing. While these domains have been suggested to contribute to the membrane homeostasis,6 we believe that PM lateral organization also contributes to the cell mechano-function, being directly involved in local cellular deformations.
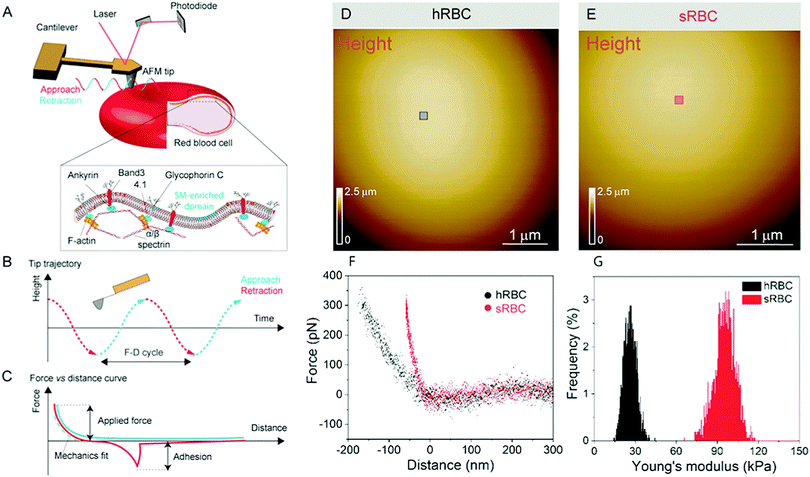
Numerous techniques have been used to study the changes accompanying RBCs during mechanical deformation, including micropipette aspiration, optical tweezers, atomic force microscopy (AFM), electric field deformation, membrane flickering analysis and vital confocal imaging.2,6,10–12 Among these, AFM stands out as an ideal tool, which can integrate high-resolution, vital imaging and quantitative mechanical characterization of living cells. In the past, AFM has been applied to the investigation of whole RBCs or ghost cells in order to establish a link between RBCs in healthy or pathological conditions, focusing on their nanomechanical properties, morphology or roughness.12–18 However, RBCs underwent different treatments in most of those AFM studies, including chemical crosslinking of lipids and proteins or air-drying, which alter their mechanical and morphological properties and makes it difficult to relate these observations with the behavior of cells in physiological conditions.19–21 We use force distance (FD) curve-based AFM, which has been developed over the past 20 years and can now be applied to simultaneously image the topography of complex biological systems while mapping quantitative sample properties, such as physical, chemical and biological interactions.22 This mode consists in recording the force while approaching and retracting the AFM tip from the surface (Fig. 1A). Recent advances pushed the limits of both spatial and temporal resolution, leading to so-called multiparametric imaging, thus opening the way to explore highly dynamic cellular processes occurring at the molecular scale.23–25 In living cells, FD curve-based AFM was used to obtain topography and mechanical properties maps of cytoskeletal structures at unprecedented resolution in physiological conditions.26–29 The operating principle of FD-based AFM is based on a cantilever being oscillated well below its resonance frequency, while using the tip–sample interaction force (peak force) as feedback (Fig. 1A–C). The tip is continuously approached and retracted from the sample and from each FD cycle the sample height is determined and tip–sample interactions are analyzed. Quantitative parameters, such as Young's modulus, stiffness, energy dissipation and adhesion, can then be extracted, mapped pixel-by-pixel and directly correlated to the sample topography.
Here, we report the use of FD-based AFM to study the nanoscale architecture and nanomechanical response of RBCs in physiological and pathological conditions. The study was carried out in parallel on RBCs from healthy volunteers (hRBCs) and from patients with HS (sRBCs). Vertical segmentation of the cell mechanics and use of cytoskeleton-disrupting drugs allowed us to estimate the relative contributions of PM and cytoskeleton in controlling cellular nanomechanics. Combined AFM and fluorescence microscopy imaging measurements enabled us to identify SM-enriched domains on the surface of RBC PM while extracting their heterogeneous nanomechanical properties. The current study sheds new light into the relation between membrane nanoscale organization, nanomechanical properties and RBC biological functions. Moreover, the present results unravel several key features of submicrometric lipid domains, i.e. their real existence, exact size, mechanical properties as well as interplay with the cytoskeleton in the control of RBC deformation. Indeed, we here reveal on RBCs in native conditions that submicrometric domains with differential nanomechanical properties as compared to the bulk membrane coexist with smaller domains and that both the cytoskeleton and the local membrane composition influence the local mechanical properties of the RBC.
Results and discussion
Probing the structure and mechanics of RBCs by AFM
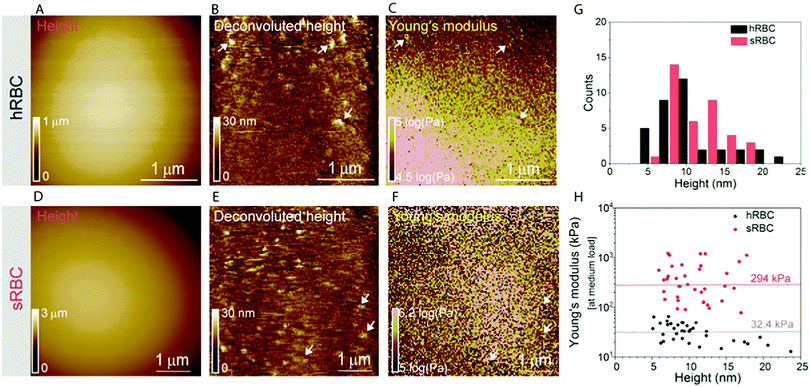
RBCs are typically found in a biconcave shape when in suspension, but they partially lose their biconcavity during the first minutes of interaction with the poly-L-lysine (PLL)-coated substrate, probably due to the strong interaction forces between the PM and the positively charged surface.6,12 Immobilization on PLL enables to avoid the most worrisome echinocyte conversion that could be generated by the repulsion between negative charges from the glycocalyx at the RBC surface and the silicates from the glass. We first evaluated the morphology, roughness and nanomechanical properties of healthy (hRBCs) and spherocytotic (sRBCs) RBCs. After immobilization on PLL-coated surfaces, RBCs in both physiological and pathological conditions feature a rounded shape (Fig. 1D and E). Due to the large curvature of the cells, fine structures are difficult to observe in raw height images. However, roughness analysis revealed a relatively smooth surface for both cell types, the root mean square (rms) roughness on height images being 3.34 ± 1.3 nm and 5.63 ± 2.6 nm for hRBCS and sRBCs respectively (from 20 cells for each condition, measured on 500 × 500 nm areas after a 2nd order polynomial fitting of the height image). The height of the cells varied between 1–3.5 μm, sRBCs being higher (2.5 ± 0.9 μm) than hRBCs (1.7 ± 0.5 μm, from 20 cells for each condition).Recording single FD curves on the surface of the cells allowed us to evaluate the mechanical properties (Young's modulus) of RBCs from healthy donors and HS patients (Fig. S1A–C, ESI†). A 2 × 2 μm area in the central region of the cell, where the height reaches maximum values (>2–3 μm), was selected and the indentation range used for the analysis was chosen in such manner (<10% of the height) to avoid any substrate effect.30 As can be directly observed from individual FD curves, hRBCs typically present a shallower slope than the one observed for sRBCs (Fig. S1C, ESI†). Young's moduli were extracted using a Hertz fit (Fig. S1C, ESI†) and the values for hRBCs and sRBCs are presented in Fig. S1D (ESI†). hRBCs have a mean Young's modulus value of 1.45 ± 0.66 kPa (N = 980 curves, from 10 different cells), while spherocytotic RBCs are stiffer at 2.04 ± 0.76 kPa (N = 561 curves, from 9 different cells), as can be directly observed from the FD curves. The increased stiffness of sRBCs lowers their ability to spread on the sample substrate and explains the differences in height observed.
We suggest that the structural and mechanical properties of sRBCs and hRBCs are of biological relevance. Mechanically stressed erythrocytes have been previously described as displaying surfaces with increased roughness,31 while ageing was linked to decreased roughness values.32 The correlation we observed between membrane roughness and stiffness for sRBCs as compared with hRBCs is in good agreement with these previous reports. These observations support previous observations in the literature that less deformable sRBCs are subjected to increased mechanical stresses during their passage through narrow capillaries in the microcirculation, which in turn decreases their life span.
Vertical segmentation of the RBC mechanical properties
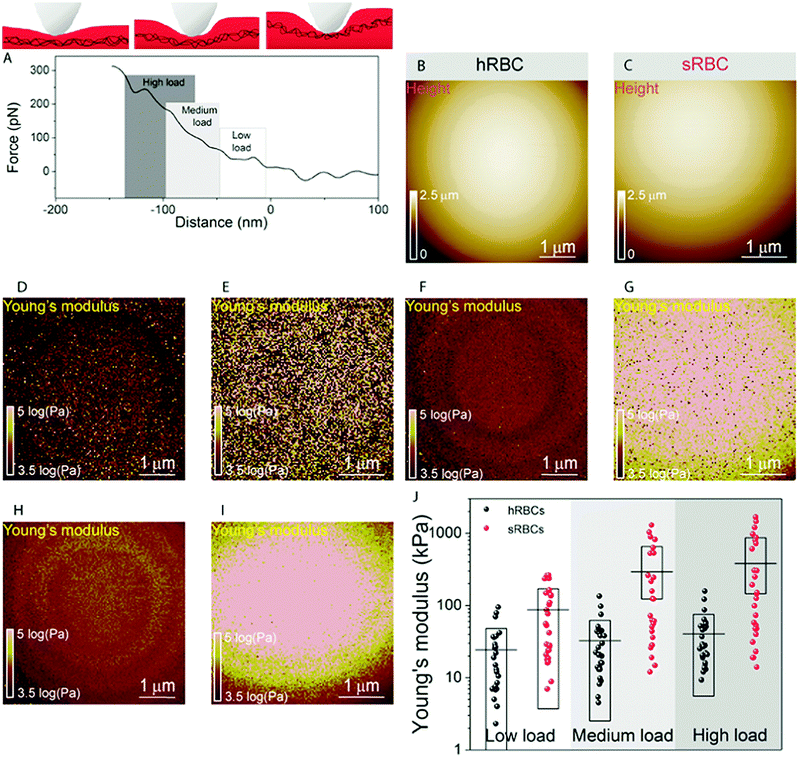
To gain further insight into the complex mechanical architecture of living RBCs, we used multiparametric FD-based AFM imaging to simultaneously investigate at high-resolution the topography of individual RBCs and locally map their Young's modulus. hRBCs and sRBCs from patients were imaged in similar conditions, by acquiring simultaneous topography and Young's modulus maps. We observed that FD curves recorded on sRBCs have a steeper slope as compared to the ones recorded on hRBCs (Fig. 1F), similarly to single FD curves (Fig. S1C, ESI†). We selected 2 × 2 μm areas in the central region of the hRBC and sRBC cells in Fig. 1D, E and calculated the corresponding Young's modulus values (∼2500 points). Histograms showing a comparison of the mechanical properties of hRBCs and sRBCs as measured by FD-based AFM are shown in Fig. 1G. A 3-fold increase in Young's modulus is observed on sRBCs (91 ± 15 kPa) as compared to hRBCs (29 ± 9 kPa). In comparison with the indentation experiments performed in single FD curves mode (Fig. S1C and D, ESI†), the indentation velocities in FD-based AFM are about two orders of magnitude higher (∼300 μm s−1vs. 1 μm s−1 range). Given the fact that RBCs are viscoelastic bodies,33,34 their mechanical response will depend on the frequency at which they are probed.26,35–37 Our results are in good agreement with Young's moduli observed on other living mammalian cells, including different types of cancer cells, fibroblasts or cardiomyocytes.26,38–40The architecture of the erythrocyte membrane includes lipids and proteins as well as an underlying actin–spectrin network.4 To better understand how individual components contribute to cell mechanics, we fitted the different regions of the retract FD curve in the contact area using the Hertz model (see Methods and Fig. S2, ESI†). We selected three fitting ranges within our applied overall load (300 pN) (Fig. 2A), as follows: low load (5–20%), which could mainly comprise the contribution of the lipid PM; medium load (20–60%), which could correspond to a contribution of both the PM and the underlying actin–spectrin cytoskeleton and high load (30–90%), which could be attributed mostly to the cytoskeleton (as well as previous components). Topography images (Fig. 2B and C) were recorded simultaneously with the corresponding elasticity maps (Fig. 2D, F, H and Fig. 2E, G, I). Remarkably, we observed that Young's modulus progressively increased with the applied load for both hRBCs and sRBCs suggesting that the actin–spectrin cytoskeleton plays the major role in mechanical properties of erythrocytes.
At first sight, the spherocytotic cell shows a brighter contrast throughout the three indentation ranges meaning that sRBCs are less deformable than the hRBCs. We next quantified the mechanical properties of both physiological and pathological conditions at low, medium and high load (Fig. 2J). The in-depth characterization of the hRBCs membrane's mechanical behavior revealed that Young's modulus increases from 24 ± 23.9 kPa at low load to 32 ± 29.9 kPa at medium load and finally reaching 40 ± 34.9 kPa at high load. Contrastingly, sRBCs show a rapid increase of the Young's modulus with the applied load, i.e. from 87 ± 83.4 kPa at low load to 298 ± 362.7 kPa at medium load and 380 ± 482.9 kPa at high load. A vertical segmentation of the mechanical properties has been previously observed on nucleated cells with a 3D cytoskeleton.41–43 However, RBCs have a less complex structure, lacking internal organelles, a nucleus and the actin–myosin–microtubule cytoskeleton found in nucleated cells. Their cytoskeleton is a 2D network with quasi-hexagonal symmetry. The main components are ∼200 nm long flexible strands of (α1β1)2 spectrin tetramers interconnected to short actin filaments at junctional complexes centered on the protein 4.1R (horizontal linkages).44 Hence, spectrin filaments are connected to the PM through 4.1R and ankyrin complexes (vertical linkages).4 Hereditary spherocytosis is generally attributed to defects in proteins of the ankyrin complex, such as ankyrin, band-3 or β-spectrin, resulting in RBC morphology and deformability changes. In this study, we examined RBCs from three splenectomized patients with mutations in the gene coding for β-spectrin. Erythrocytes from these patients show a slightly reduced spectrin density compared to healthy donors. This could affect the distribution of anchorage points between the membrane and the underlying cytoskeleton, resulting in the impairment of its dynamic remodeling and contributing to a stiffer cytoskeleton. Although this remains to be determined, this hypothesis is actually in good agreement with theoretical studies showing that the loss of the spectrin:membrane anchorage stiffens the cytoskeleton45,46 and with our observations of an almost 10-fold difference between Young's modulus of hRBCs and sRBCs at high loads. A recent study of the elastic properties of RBCs during morphological changes has shown that spectrin can be found in two independent conformations: a soft and easily deformable one in hRBCs, while a transition towards a stiff conformation and a bimodal distribution of spectrin conformations is observed in sRBCs.47 Our AFM measurements suggest, for the first time directly in physiological conditions and on intact RBCs, the importance of the cytoskeleton in controlling the RBC mechanical properties. Surprisingly, our approach revealed that at low indentation load the difference between hRBCs and sRBCs is also present, indicating that besides the cytoskeleton, PM lipid content and its nanoscale organization should also influence the nanomechanics of erythrocytes.
Role of cytoskeleton in RBC mechanics
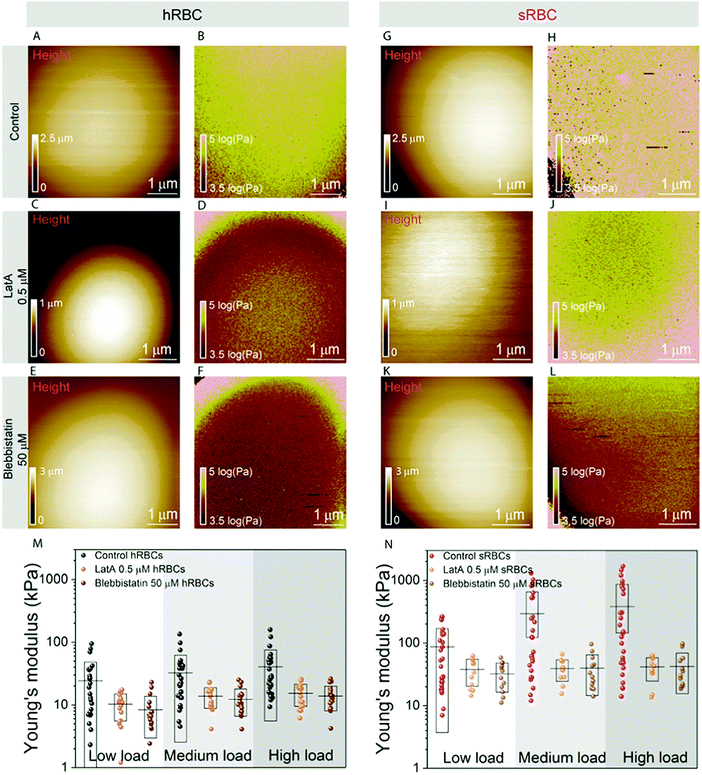
To further assess the influence of the cytoskeleton on the global cell mechanics, we used latrunculin A (LatA) and blebbistatin as inhibitors of actin polymerization and myosin respectively (Fig. 3). Although non-muscle myosin has been identified in mature erythrocytes, its importance in cytoskeleton remodeling was underestimated due to its low cellular abundance (actin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) myosin ratio of 80
myosin ratio of 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).48 While cells treated with either LatA (500 nM during 30 minutes) (Fig. 3C, D, I and J) or blebbistatin (50 μM during 30 minutes) (Fig. 3E, F, K and L) maintain their spherical shape, their height was slightly reduced in comparison with control cells, probably due to the collapse of their cytoskeleton (Fig. 3A, C and E). The effect of LatA and blebbistatin on the mechanical properties of hRBCs and sRBCs is shown in Fig. 3B, D, F and Fig. 3H, J, L respectively. The apparent Young's modulus was displayed as maps and extracted from the high load fits, since we previously observed a dominant effect of the cytoskeleton at this load. Both the depolymerization of actin filaments with LatA and the inhibition of myosin with blebbistatin led to a significant decrease of the Young's modulus for both hRBCs and sRBCs, as compared with the untreated cells. The Young's modulus drop is more pronounced in the case of sRBCs, with a ten-fold factor decrease. Nevertheless, Young's modulus of sRBCs remains higher by a three-fold factor that the one of hRBCs for the three loads used (Fig. 3M, N and Table S1, ESI†), suggesting that mechanical differences could also originate from differences in the vertical cytoskeleton anchorage to integral membrane proteins or from non-cytoskeletal structures such as the glycocalix. Besides the decrease in Young's modulus, we also observed less heterogeneity in the mechanical properties dataset of LatA- and blebbistatin-treated RBCs, as compared to control cells. We hypothesize that these differences could originate from a heterogeneously aging population of RBCs. It has been already established that during their 120 days lifespan, RBCs release microvesicles, which alters their high surface area to volume ratio, thus increasing the cytoskeleton density and reducing deformability.49 This is intrinsic to both hRBCs and sRBCs populations and it leads to a broad distribution of the measured Young's modulus values. Moreover, during our measurements, the AFM tip having a radius of 65 nm is probing the cell surface and recording a pixel every 20–40 nm, depending on the size of the image. Due to the meshwork architecture of the RBC cytoskeleton displaying 200 nm long spectrin filaments, the tip could probe either the freely spanning PM lying above a hole in the cytoskeleton mesh, or the actin–spectrin filaments. When measuring cells with a higher cytoskeleton density, such as upon aging, there is a higher probability of the AFM tip probing the less compliant actin–spectrin filaments, thus rendering higher Young's modulus values. On the other hand, on younger cells, the tip will probe an average distribution of PM and the underlying cytoskeleton. When disrupting the cell cytoskeleton, we mostly probe the PM, which produces more homogenous results. Interestingly, while other groups have shown that LatA affects the cytoskeleton organization of RBCs, the effect of blebbistatin has been neglected so far.11,50 We showed here for the first time that myosin contributes similarly to actin to the mature RBC mechanics. This could be important for future studies on cytoskeleton remodeling during enucleation of erythroblasts and to explore the role of the residual cytoskeletal and cytosolic myosin in the mature RBC deformation.
1).48 While cells treated with either LatA (500 nM during 30 minutes) (Fig. 3C, D, I and J) or blebbistatin (50 μM during 30 minutes) (Fig. 3E, F, K and L) maintain their spherical shape, their height was slightly reduced in comparison with control cells, probably due to the collapse of their cytoskeleton (Fig. 3A, C and E). The effect of LatA and blebbistatin on the mechanical properties of hRBCs and sRBCs is shown in Fig. 3B, D, F and Fig. 3H, J, L respectively. The apparent Young's modulus was displayed as maps and extracted from the high load fits, since we previously observed a dominant effect of the cytoskeleton at this load. Both the depolymerization of actin filaments with LatA and the inhibition of myosin with blebbistatin led to a significant decrease of the Young's modulus for both hRBCs and sRBCs, as compared with the untreated cells. The Young's modulus drop is more pronounced in the case of sRBCs, with a ten-fold factor decrease. Nevertheless, Young's modulus of sRBCs remains higher by a three-fold factor that the one of hRBCs for the three loads used (Fig. 3M, N and Table S1, ESI†), suggesting that mechanical differences could also originate from differences in the vertical cytoskeleton anchorage to integral membrane proteins or from non-cytoskeletal structures such as the glycocalix. Besides the decrease in Young's modulus, we also observed less heterogeneity in the mechanical properties dataset of LatA- and blebbistatin-treated RBCs, as compared to control cells. We hypothesize that these differences could originate from a heterogeneously aging population of RBCs. It has been already established that during their 120 days lifespan, RBCs release microvesicles, which alters their high surface area to volume ratio, thus increasing the cytoskeleton density and reducing deformability.49 This is intrinsic to both hRBCs and sRBCs populations and it leads to a broad distribution of the measured Young's modulus values. Moreover, during our measurements, the AFM tip having a radius of 65 nm is probing the cell surface and recording a pixel every 20–40 nm, depending on the size of the image. Due to the meshwork architecture of the RBC cytoskeleton displaying 200 nm long spectrin filaments, the tip could probe either the freely spanning PM lying above a hole in the cytoskeleton mesh, or the actin–spectrin filaments. When measuring cells with a higher cytoskeleton density, such as upon aging, there is a higher probability of the AFM tip probing the less compliant actin–spectrin filaments, thus rendering higher Young's modulus values. On the other hand, on younger cells, the tip will probe an average distribution of PM and the underlying cytoskeleton. When disrupting the cell cytoskeleton, we mostly probe the PM, which produces more homogenous results. Interestingly, while other groups have shown that LatA affects the cytoskeleton organization of RBCs, the effect of blebbistatin has been neglected so far.11,50 We showed here for the first time that myosin contributes similarly to actin to the mature RBC mechanics. This could be important for future studies on cytoskeleton remodeling during enucleation of erythroblasts and to explore the role of the residual cytoskeletal and cytosolic myosin in the mature RBC deformation.
Following our vertical segmentation approach, we are able to discriminate cytoskeleton contribution to the overall cellular mechanics. The use of drugs supports our hypothesis that at low indentation loads we actually probe mainly the bending of the tensed PM. The three-fold difference observed between the Young's modulus values at low loads for healthy and spherocytosis cells (∼10 kPa for hRBCs vs. ∼30 kPa for sRBCs; see Fig. 3M and N) could originate from structural differences between the PMs of healthy and pathological cells.47,51 The increased line tension of the lipid bilayer in spherocytes could lead to the formation of dimples of lipid domains to liberate tension.52
AFM multiparametric imaging reveals nanoscale structural and mechanical heterogeneities at the RBC surface
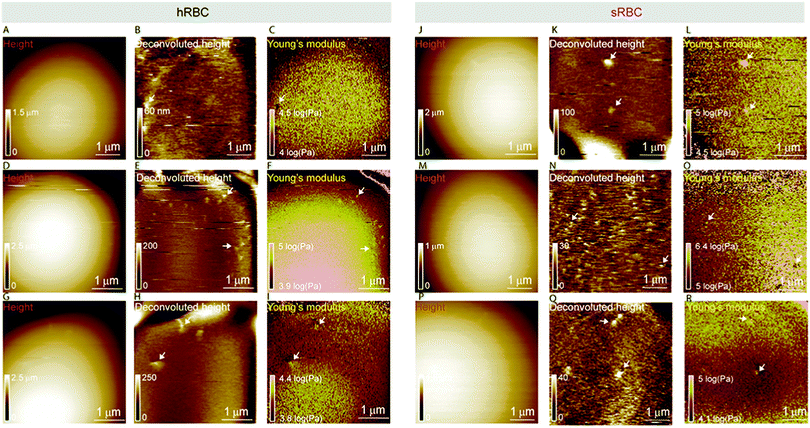
Multiparametric imaging was used with the goal to reveal nanoscale topographical and mechanical heterogeneities on the surface of hRBCs and sRBCs. As can be observed in Fig. 4 and 5, while raw height images showed featureless surfaces, deconvoluted height images revealed numerous submicrometric (∼200 nm) and sub-100 nm protrusions with heights ranging between 5–100 nm for both hRBCS and sRBCs. These heterogeneities were correlated with local minima (Fig. 4B, C, N and O) or local maxima (Fig. 4E, F, H, I, K, L, Q and R) areas in Young's modulus maps. We found these protrusions to have similar morphological and nanomechanical properties on hRBCs and sRBCs. To gain further insights into the intrinsic mechanical properties of RBC membrane domains, we selected two representative images displaying sub-100 nm domains (Fig. 5A and D) and we quantitatively analyzed their domain sizes (Fig. 5B and E) and Young's moduli (Fig. 5C and F) (for further details see Fig. S3 (ESI†) and Methods). Our analysis revealed that these protrusions have heights between 5–20 nm (Fig. 5G) and appear as either local maxima or minima, independently from the region of the cell where they are found. Height and Young's modulus values for each of these domains were plotted and revealed a homogeneous distribution around cell average Young's modulus (Fig. 5H). We hypothesize that such heterogeneities are lipid-enriched domains, with or without recruited membrane proteins. Their mechanical properties will depend both on the specific composition of lipids, as well as on the global organization of the PM and the underlying cytoskeleton and could serve as reservoir for the generation of positive or negative curvature to navigate through human microcirculation vessels and through the pores during spleen filtration.Correlated AFM and fluorescence imaging reveals sphingomyelin submicrometric domains of variable stiffness
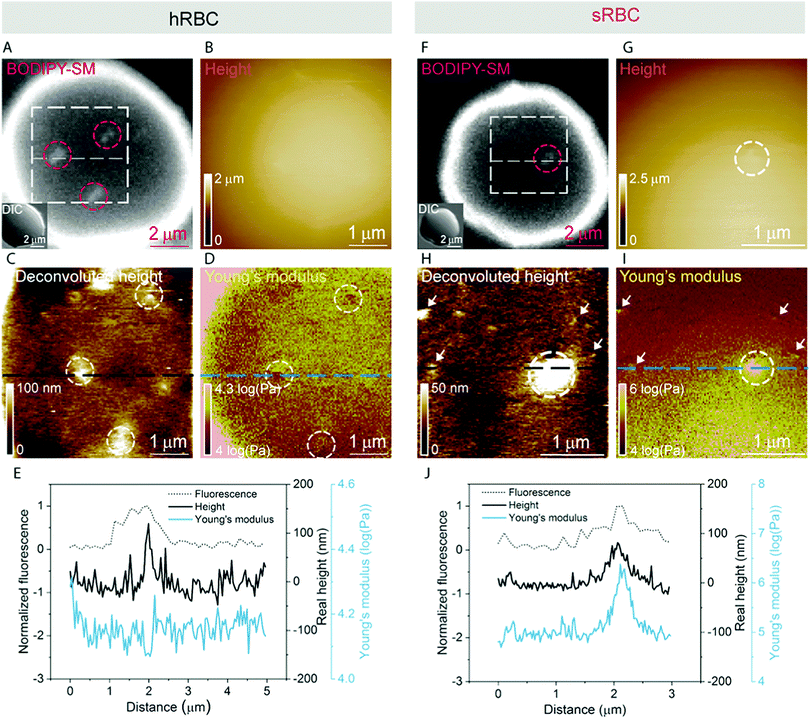
A pertinent question is whether these heterogeneities could arise from submicrometric aggregation of lipids in specific areas of the PM in order to liberate line tension. We therefore tested this hypothesis by combining FD-based AFM experiments with simultaneous fluorescence microscopy of SM-enriched submicrometric domains, revealed by PM insertion of the fluorescent lipid analog BODIPY-SM at low concentration (0.6 μM) to minimize modifications of the intrinsic PM properties, as tested previously.51 Representative images correlating fluorescence microscopy and AFM on hRBCs and spherocytes are shown in Fig. 6A–D and Fig. 6F–I, respectively. The fluorescence was uniform in intensity on the RBC PM, except in the rim of the cells, in vesicles and in lipid domains, where the intensity is more strongly marked (Fig. 6A and F). Differential interference contrast (DIC) microscopy images were recorded simultaneously to exclude the presence of membrane vesicles (Fig. 6A and F, inset). Lipid domains enriched in SM are clearly observed on both hRBCs and sRBCs (Fig. 6A and F, encircled).While height images of the hRBC showed only faint protrusions due to the cell curvature (Fig. 6B), the deconvoluted image allowed several topographical heterogeneities to be observed (Fig. 6C, highlighted by circles). The size of these heterogeneities varies from 50 to 500 nm in diameter and from 10 to 100 nm in height. Correlation with Young's modulus map analyzed at high load revealed that these three domains also present a mechanical behavior that differs from the surrounding environment. This clearly highlights that heterogeneities in the local PM composition are linked to local particular nanomechanical properties. On hRBCs, topographical protrusions coincide with a softer domain (Fig. 6E). To further put in evidence these observations, we performed simultaneous cross-sections of the fluorescence microscopy, height and Young's modulus images on the observed domains. The height measured by the AFM tip on soft surfaces will usually be lower than the true height of the sample, due to the deformation applied while scanning. We circumvented this issue through a pixel-by-pixel reconstruction of the real height of the sample, by adding the deformation and the measured height channels. The corrected real height cross-section along the SM domain shows that it is ∼120 nm high and that its position is well correlated with a peak in the intensity of the fluorescence signal and a valley in the Young's modulus cross-section (Fig. 6E). We exclude the possibility of the identified SM domains being membrane vesicles, based on DIC images. We also performed control experiments on 4% PFA fixed hRBCs and observed the presence of SM-enriched domains with corresponding local nanomechanical heterogeneities similar to the ones found on living hRBCs (Fig. S5, ESI†).
Similarly, we investigated sRBCs (Fig. 6F–J) and observed that a fluorescent SM domain (Fig. 6F) coincides to a protrusion in the height image (Fig. 6G and H) as well as an heterogeneity in the Young's modulus map (Fig. 6I). Simultaneous cross-sections along the SM domain show that the peak in the fluorescence image is again well correlated with the height protrusion and a maximum in the Young's modulus observed by AFM (Fig. 6J). However, in contrast to hRBCs, cross-sections of height and Young's modulus revealed that the protrusion is stiffer that the surrounding PM. Real height of the SM domain is ∼86 nm. Other smaller protrusions are observed in the height images of both hRBCs and sRBCs (indicated by white arrows) and similarly to the SM domain, they appear as local maxima or minima in the Young's modulus maps. We believe that these heterogeneities could also represent lipid domains, however with smaller size below the diffraction limit and not resolved by our optical microscope, or other domains enriched with different lipids (see additional images in Fig. S4, ESI†). Accordingly, combined AFM and fluorescence microscopy imaging identified submicrometric SM-enriched domains on the surface of RBC that were either softer or stiffer than the surrounding areas. These domains could have a direct relevance for cell deformation while navigating through microcirculation vessels and during spleen filtration.
Conclusion
Here, we introduced an AFM-based approach to investigate the 3D architecture and mechanics of RBCs at the nanoscale and to determine how this architecture is altered in pathological states/conditions. We first confirmed that spherocytic RBCs are stiffer than healthy RBCs, a finding that can be directly related to the disease in which RBCs are less deformable, and therefore less compliant to the deformation during the splenic filtration across small pores ∼3 μm of diameter.53,54 Through a novel vertical segmentation approach on RBCs, we were able to isolate the contributions of the cytoskeleton and the PM from overall cell mechanics. We discovered that the cytoskeleton plays a major role in RBC mechanics. For patients with hereditary spherocytosis, the contribution of the cytoskeleton was strongly enhanced and can be related to their β-spectrin gene mutations leading to a reduced spectrin density and a stiffer cytoskeleton.47,55,56 Correlated AFM and fluorescence imaging also revealed that the membrane lateral heterogeneity could influence the local mechanical properties of the RBCs. Therefore, our approach has demonstrated a novel correlation between the local lipid composition and RBC surface nanomechanics. We suggest that these lipid domains contribute to the membrane homeostasis and to their mechanobiological function by their direct implication in local cellular deformations. Our nanoscale observation provides direct links with membrane RBC pathologies (e.g. spherocytosis, elliptocytosis,…) and opens up new avenues for understanding of these RBC diseases.Methods
RBC isolation and immobilization
This study was approved by the Medical Ethics Institutional Committee of the Université catholique de Louvain; each donor gave written informed consent. All methods were performed in accordance with the relevant guidelines and regulations. RBCs were freshly isolated from three healthy volunteers and three splenectomized patients with hereditary spherocytosis with β-spectrin gene mutations. Blood was collected by venipuncture into dry ethylenediaminetetraacetic acid (EDTA) (K+ salt)-coated tubes, then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (v/v) in Dulbecco's Modified Eagle Medium (DMEM containing 4.5 g L−1 of glucose, L-glutamine, and 25 mM HEPES, Gibco) and washed twice by centrifugation at 133g for 2 min. The working solution was obtained by resuspending washed RBCs in DMEM 1
4 (v/v) in Dulbecco's Modified Eagle Medium (DMEM containing 4.5 g L−1 of glucose, L-glutamine, and 25 mM HEPES, Gibco) and washed twice by centrifugation at 133g for 2 min. The working solution was obtained by resuspending washed RBCs in DMEM 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio (v/v). For RBC immobilization, a 1 mg mL−1 poly-L-lysine (PLL, Mw ∼ 70–150 kDa, Sigma-Aldrich) solution in cell culture grade phosphate buffered saline (PBS, Sigma Aldrich) was used. To this aim, glass-bottomed Petri dishes (WillCo) were covered with PLL
10 ratio (v/v). For RBC immobilization, a 1 mg mL−1 poly-L-lysine (PLL, Mw ∼ 70–150 kDa, Sigma-Aldrich) solution in cell culture grade phosphate buffered saline (PBS, Sigma Aldrich) was used. To this aim, glass-bottomed Petri dishes (WillCo) were covered with PLL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMEM (1
DMEM (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and incubated at 37 °C for 30 min. After a washing step with DMEM at room temperature for 3 min, Petri dishes were dried at room temperature in a laminar flow hood for 2 hours. The RBC working solution was plated onto the PLL-coated Petri dishes at room temperature for 4 min. The solution was then replaced by fresh DMEM and attached RBCs were further allowed to spread for another 4 min. After 2 or 3 rapid washings with DMEM, the sample was used for AFM experiments performed in DMEM. For experiments on fixed cells, hRBCs were fixed with a 4% paraformaldehyde (PFA) in PBS solution for 10 min at room temperature. Fixed hRBCs were then immobilized on PLL-coated Petri dishes in the same manner as living cells.
1, v/v) and incubated at 37 °C for 30 min. After a washing step with DMEM at room temperature for 3 min, Petri dishes were dried at room temperature in a laminar flow hood for 2 hours. The RBC working solution was plated onto the PLL-coated Petri dishes at room temperature for 4 min. The solution was then replaced by fresh DMEM and attached RBCs were further allowed to spread for another 4 min. After 2 or 3 rapid washings with DMEM, the sample was used for AFM experiments performed in DMEM. For experiments on fixed cells, hRBCs were fixed with a 4% paraformaldehyde (PFA) in PBS solution for 10 min at room temperature. Fixed hRBCs were then immobilized on PLL-coated Petri dishes in the same manner as living cells.
RBC treatment with latrunculin A and blebbistatin
Stock solutions of 50 μM latrunculin A (Sigma-Aldrich L5163) and 100 mM blebbistatin (Sigma-Aldrich B0560) were obtained by dissolving the drugs in dimethylsulfoxide (DMSO). Washed RBCs were incubated with latrunculin A to a final concentration of 575 nM and with blebbistatin to a final concentration of 50 μM for 30 min. Previous studies have shown that these DMSO concentrations are not toxic for RBCs.50,57 RBCs were then washed two times at 133g for 2 min and suspended in DMEM. RBCs were subsequently spread on PLL-coated Petri dishes as previously described.FD-based AFM on living RBCs
AFM experiments were performed with a Bioscope Catalyst AFM (Bruker) operated in PeakForce QNM mode at ∼25–30 °C in DMEM. PeakForce QNM Live Cell probes (Bruker) with spring constants ranging from 0.08 N m−1 to 0.1 N m−1, a tip radius of curvature of 65 nm, and a tip half-angle of 15° were used. We chose these probes specifically due to their cantilever geometry and 17 μm long tip designed to reduce hydrodynamic forces. The long tip also enables imaging soft surfaces with large differences in height, by reducing the squeeze layer effect. The spring constant of the cantilevers was calibrated with a vibrometer (OFV-551, Polytec, Waldbronn) by the manufacturer. The pre-calibrated spring constant was used to determine the deflection sensitivity using the thermal noise method58 before each experiment. FD-based multiparametric maps were acquired using a force setpoint of 300 pN. The AFM cantilever was oscillated vertically at 0.25 kHz with peak-to-peak oscillation amplitudes ranging from 600 to 1.2 μm. Images were recorded using a scan rate of 0.1 or 0.125 Hz and 128 pixels per line. For experiments at low indentation rates, individual FD curves were recorded in contact mode on the RBC surface. A force setpoint of 300 pN, approach and retraction velocities of 1 μm s−1 and 5 μm ramp size were used. Measurements were performed in the center of the cell, which is the highest part, to avoid substrate effects. Indentations were ranged between 300 nm and 1 μm depending on cell type. At least 60 FD curves were recorded per cell.Combined FD-based AFM and fluorescence microscopy on living RBCs
RBCs spread on PLL-coated Petri dishes were labeled with 0.6 μM BODIPY FL C5-SM (BODIPY, Thermo Fisher) in DMEM supplemented with 1 mg mL−1 bovine serum albumin (BSA fatty acid free, Sigma Aldrich) at room temperature for 15 min. The sample was then rinsed with DMEM and used for AFM experiments. Epifluorescence images of BODIPY-SM-labeled RBCs were acquired with an Axio Observer Z1 (Zeiss) inverted optical microscope coupled to a BioScope Catalyst (Bruker) AFM, using a 40×/100× oil immersion objective. This allowed the localization of SM domains on RBCs prior to AFM imaging. The BODIPY fluorophore was excited at a wavelength of 498 nm and the emission was recorded at 516 nm.Data analysis
Raw FD curves were processed offline using the NanoScope Analysis 1.80 Software (Bruker). We analyzed the retraction part of the FD curves to avoid plastic deformation contributions. For FD curves extracted from PeakForce QNM maps, the best quality of the fit was obtained by fitting the contact part of the curve with the Hertz model:59,60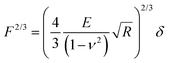 | (1) |
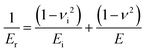 | (2) |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The research of the authors is financially supported by the Fonds National de la Recherche Scientifique (F.R.S.-FNRS grant numbers: PDR T.0090.15 to D. A.), the Research Department of the Communauté française de Belgique (Concerted Research Action), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the ‘MOVE-IN Louvain’ Incoming post-doc Fellowship programme and the Salus Sanguinis Foundation, the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement no. [693630]), and the FNRS-WELBIO (Grant no. WELBIO-CR-2015A-05). Y. F. D. and D. A. are Research Director and Research Associate at the FNRS. We thank Professor Christiane Vermylen (Saint-Luc Hospital & Université catholique de Louvain) for providing blood from spherocytotic patients.References
- N. Mohandas and P. G. Gallagher, Blood, 2008, 112, 3939–3948 CrossRef CAS PubMed.
- D. Discher, N. Mohandas and E. Evans, Science, 1994, 266, 1032–1035 CAS.
- N. Mohandas and E. Evans, Annu. Rev. Biophys. Biomol. Struct., 1994, 23, 787–818 CrossRef CAS PubMed.
- V. M. Fowler, in Current Topics in Membranes, ed. V. Bennett, Academic Press, 2013, vol. 72, pp. 39–88 Search PubMed.
- M. Carquin, L. D’Auria, H. Pollet, E. R. Bongarzone and D. Tyteca, Prog. Lipid Res., 2016, 62, 1–24 CrossRef CAS PubMed.
- C. Leonard, L. Conrard, M. Guthmann, H. Pollet, M. Carquin, C. Vermylen, P. Gailly, P. Van Der Smissen, M. P. Mingeot-Leclercq and D. Tyteca, Sci. Rep., 2017, 7, 4264 CrossRef CAS PubMed.
- H. S. Jacob, A. Ruby, E. S. Overland and D. Mazia, J. Clin. Invest., 1971, 50, 1800–1805 CrossRef CAS PubMed.
- C. F. Reed and S. N. Swisher, J. Clin. Invest., 1966, 45, 777–781 CrossRef CAS PubMed.
- H. S. Jacob, Am. J. Med., 1966, 41, 734–743 CrossRef CAS PubMed.
- G. Bao and S. Suresh, Nat. Mater., 2003, 2, 715–725 CrossRef CAS PubMed.
- R. B. Nowak, J. Papoin, D. S. Gokhin, C. Casu, S. Rivella, J. M. Lipton, L. Blanc and V. M. Fowler, Blood, 2017, 130, 1144–1155 CrossRef CAS PubMed.
- L. Picas, F. Rico, M. Deforet and S. Scheuring, ACS Nano, 2013, 7, 1054–1063 CrossRef CAS PubMed.
- M. Girasole, S. Dinarelli and G. Boumis, Micron, 2012, 43, 1273–1286 CrossRef CAS PubMed.
- M. O’Reilly, L. McDonnell and J. O’Mullane, Ultramicroscopy, 2001, 86, 107–112 CrossRef.
- R. Nowakowski and P. Luckham, Surf. Interface Anal., 2002, 33, 118–121 CrossRef CAS.
- G. Ciasca, M. Papi, S. Di Claudio, M. Chiarpotto, V. Palmieri, G. Maulucci, G. Nocca, C. Rossi and M. De Spirito, Nanoscale, 2015, 7, 17030–17037 RSC.
- I. Dulińska, M. Targosz, W. Strojny, M. Lekka, P. Czuba, W. Balwierz and M. Szymoński, J. Biochem. Biophys. Methods, 2006, 66, 1–11 CrossRef PubMed.
- M. Lekka, M. Fornal, G. Pyka-Fosciak, K. Lebed, B. Wizner, T. Grodzicki and J. Styczen, Biorheology, 2005, 42, 307–317 Search PubMed.
- Y. Li, L. Lu and J. Li, Cell Biochem. Biophys., 2016, 74, 365–371 CrossRef CAS PubMed.
- J. L. Maciaszek and G. Lykotrafitis, J. Biomech., 2011, 44, 657–661 CrossRef PubMed.
- T. Betz, U. Bakowsky, M. R. Müller, C.-M. Lehr and I. Bernhardt, Bioelectrochemistry, 2007, 70, 122–126 CrossRef CAS PubMed.
- Y. F. Dufrene, D. Martinez-Martin, I. Medalsy, D. Alsteens and D. J. Muller, Nat. Methods, 2013, 10, 847–854 CrossRef CAS PubMed.
- Y. F. Dufrêne, T. Ando, R. Garcia, D. Alsteens, D. Martinez-Martin, A. Engel, C. Gerber and D. J. Müller, Nat. Nanotechnol., 2017, 12, 295 CrossRef PubMed.
- D. Alsteens, M. Pfreundschuh, C. Zhang, P. M. Spoerri, S. R. Coughlin, B. K. Kobilka and D. J. Muller, Nat. Methods, 2015, 12, 845–851 CrossRef CAS PubMed.
- D. Alsteens, R. Newton, R. Schubert, D. Martinez-Martin, M. Delguste, B. Roska and D. J. Müller, Nat. Nanotechnol., 2016, 12, 177 CrossRef PubMed.
- A. Calzado-Martín, M. Encinar, J. Tamayo, M. Calleja and A. San Paulo, ACS Nano, 2016, 10, 3365–3374 CrossRef PubMed.
- D. Xia, S. Zhang, J. O. Hjortdal, Q. Li, K. Thomsen, J. Chevallier, F. Besenbacher and M. Dong, ACS Nano, 2014, 8, 6873 CrossRef CAS PubMed.
- D. Alsteens, V. Dupres, S. Yunus, J. P. Latge, J. J. Heinisch and Y. F. Dufrene, Langmuir, 2012, 28, 16738 CrossRef CAS PubMed.
- F. Eghiaian, A. Rigato and S. Scheuring, Biophys. J., 2015, 108, 1330 CrossRef CAS PubMed.
- N. Gavara and R. S. Chadwick, Nat. Nanotechnol., 2012, 7, 733 CrossRef CAS PubMed.
- Y. Ohta, H. Okamoto, M. Kanno and T. Okuda, Artif. Organs, 2002, 26, 10–17 CrossRef PubMed.
- M. Girasole, G. Pompeo, A. Cricenti, G. Longo, G. Boumis, A. Bellelli and S. Amiconi, Nanomedicine, 2010, 6, 760–768 CrossRef CAS PubMed.
- M. Radmacher, Methods in Cell Biology, Academic Press, 2007, vol. 83, pp. 347–372 Search PubMed.
- L. M. Rebelo, J. S. de Sousa, J. Mendes Filho and M. Radmacher, Nanotechnology, 2013, 24, 055102 CrossRef CAS PubMed.
- B. Fabry, G. N. Maksym, J. P. Butler, M. Glogauer, D. Navajas and J. J. Fredberg, Phys. Rev. Lett., 2001, 87, 148102 CrossRef CAS PubMed.
- A. Cartagena and A. Raman, Biophys. J., 2014, 106, 1033–1043 CrossRef CAS PubMed.
- A. Rigato, F. Rico, F. Eghiaian, M. Piel and S. Scheuring, ACS Nano, 2015, 9, 5846–5856 CrossRef CAS PubMed.
- A. Rigato, A. Miyagi, S. Scheuring and F. Rico, Nat. Phys., 2017, 13, 771–775 CrossRef CAS PubMed.
- A. X. Cartagena-Rivera, W. H. Wang, R. L. Geahlen and A. Raman, Sci. Rep., 2015, 5, 11692 CrossRef CAS PubMed.
- A. Raman, S. Trigueros, A. Cartagena, A. P. Z. Stevenson, M. Susilo, E. Nauman and S. A. Contera, Nat. Nanotechnol., 2011, 6, 809 CrossRef CAS PubMed.
- C. Roduit, S. Sekatski, G. Dietler, S. Catsicas, F. Lafont and S. Kasas, Biophys. J., 2009, 97, 674–677 CrossRef CAS PubMed.
- J. R. Staunton, B. L. Doss, S. Lindsay and R. Ros, Sci. Rep., 2016, 6, 19686 CrossRef CAS PubMed.
- A. Wang, K. Vijayraghavan, O. Solgaard and M. J. Butte, ACS Nano, 2016, 10, 257–264 CrossRef CAS PubMed.
- A. Nans, N. Mohandas and D. L. Stokes, Biophys. J., 2011, 101, 2341–2350 CrossRef CAS PubMed.
- N. S. Gov, New J. Phys., 2007, 9, 429 CrossRef.
- L. Lai, X. Xu, C. T. Lim and J. Cao, Biophys. J., 2015, 109, 2287–2294 CrossRef CAS PubMed.
- Y. Park, C. A. Best, K. Badizadegan, R. R. Dasari, M. S. Feld, T. Kuriabova, M. L. Henle, A. J. Levine and G. Popescu, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 6731–6736 CrossRef CAS PubMed.
- V. M. Fowler, J. Q. Davis and V. Bennett, J. Cell Biol., 1985, 100, 47–55 CrossRef CAS PubMed.
- R. Waugh, N. Mohandas, C. Jackson, T. Mueller, T. Suzuki and G. Dale, Blood, 1992, 79, 1351–1358 CAS.
- D. S. Gokhin, R. B. Nowak, J. A. Khoory, A. d. l. Piedra, I. C. Ghiran and V. M. Fowler, Mol. Biol. Cell, 2015, 26, 1699–1710 CrossRef CAS PubMed.
- L. D'Auria, M. Fenaux, P. Aleksandrowicz, P. Van Der Smissen, C. Chantrain, C. Vermylen, M. Vikkula, P. J. Courtoy and D. Tyteca, J. Lipid Res., 2013, 54, 1066–1076 CrossRef PubMed.
- T. S. Ursell, W. S. Klug and R. Phillips, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13301–13306 CrossRef CAS PubMed.
- R. E. Mebius and G. Kraal, Nat. Rev. Immunol., 2005, 5, 606–616 CrossRef CAS PubMed.
- I. V. Pivkin, Z. Peng, G. E. Karniadakis, P. A. Buffet, M. Dao and S. Suresh, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 7804–7809 CrossRef CAS PubMed.
- R. E. Waugh and P. Agre, J. Clin. Invest., 1988, 81, 133–141 CrossRef CAS PubMed.
- H. Li and G. Lykotrafitis, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2015, 92, 012715 CrossRef PubMed.
- K. Ubukawa, Y.-M. Guo, M. Takahashi, M. Hirokawa, Y. Michishita, M. Nara, H. Tagawa, N. Takahashi, A. Komatsuda, W. Nunomura, Y. Takakuwa and K. Sawada, Blood, 2012, 119, 1036–1044 CrossRef CAS PubMed.
- J. L. Hutter and J. Bechhoefer, Rev. Sci. Instrum., 1993, 64, 1868–1873 CrossRef CAS.
- H. Schillers, C. Rianna, J. Schäpe, T. Luque, H. Doschke, M. Wälte, J. J. Uriarte, N. Campillo, G. P. A. Michanetzis, J. Bobrowska, A. Dumitru, E. T. Herruzo, S. Bovio, P. Parot, M. Galluzzi, A. Podestà, L. Puricelli, S. Scheuring, Y. Missirlis, R. Garcia, M. Odorico, J.-M. Teulon, F. Lafont, M. Lekka, F. Rico, A. Rigato, J.-L. Pellequer, H. Oberleithner, D. Navajas and M. Radmacher, Sci. Rep., 2017, 7, 5117 CrossRef PubMed.
- H. Hertz, J. Reine Angew. Math., 1882, 92, 156–171 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7nh00187h |
| ‡ A. C. D. and M. A. P. equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2018 |