MyD88 in antigen-presenting cells is not required for CD4+ T-cell responses during peptide nanofiber vaccination†
Youhui
Si‡
a,
Yi
Wen‡
ab,
Jianjun
Chen
a,
Rebecca R.
Pompano
ac,
Huifang
Han
a,
Joel H.
Collier
 *ab and
Anita S.
Chong
*ab and
Anita S.
Chong
 *a
*a
aDepartment of Surgery, The University of Chicago, 5841 S. Maryland Ave, Chicago, IL 60637, USA. E-mail: joel.collier@duke.edu; achong@surgery.bsd.uchicago.edu
bDepartment of Biomedical Engineering, Duke University, 101 Science Drive, CIEMAS 1393, Campus Box 90281, Durham, NC 27708, USA
cDepartment of Chemistry, The University of Virginia, Charlottesville, VA 22904, USA
First published on 29th November 2017
Abstract
Self-assembled peptide nanofibers raise significant antibody and T cell responses without adjuvants, but the mechanism by which they achieve this has not been fully elucidated. Myeloid differentiation primary response gene 88 (MyD88) has previously been shown to be critical for the antibody response to antigens presented by peptide nanofibers. The present study sought to determine the cell subset in which MyD88 is essential for T cell responses. Mice deficient in MyD88 or CD11c+ cells had severely attenuated T cell responses. However, mice lacking MyD88 in only CD11c+ cells remained capable of internalizing, processing, and presenting nanofiber-derived epitopes to stimulate T cell responses. The necessity of the inflammasome pathway was ruled out. Using adoptive transfer models where MyD88 was eliminated in CD4+ T cells or in the host, we observed that deficiency only in T cells or only in the host had no impact on the T cell response to nanofiber vaccines. Therefore, knocking out MyD88 in either antigen-presenting cells (APCs) or CD4 T cells could not compromise the CD4 T cell responses, suggesting that self-assembled peptide nanofibers trigger redundant MyD88-dependent and MyD88-independent signaling pathways in APCs and T cells. Similar redundancy has been observed for other adjuvants, and this is discussed.
Introduction
Vaccines comprising attenuated viruses, such as measles, mumps, rubella, and varicella, elicit strong immunity that best replicates the immunity elicited by the non-attenuated live virus. However, the possibility that attenuated viruses could revert to a form capable of causing disease or cause disease in individuals with weakened immunity has prompted a shift towards inactivated or subunit vaccines that contain only limited components of the target pathogen.1 While such vaccines are in principle considerably safer, they are also less effective at eliciting protective immune responses, and adjuvants must be incorporated into the vaccine to enhance the strength and durability of the elicited immune responses.2 The most common adjuvants in licensed vaccines include aluminum salts, which are incorporated into several vaccines, monophosphoryl lipid A, which is included in the human papillomavirus (HPV) and hepatitis B vaccines, oil-in-water emulsions (AS03, MF59) for pandemic and seasonal influenza, and virosomes for hepatitis and influenza.2–4 These adjuvants contribute to the initiation of the innate immune response essential for eliciting the adaptive response by causing inflammatory cues to be released at the injection site. These in turn recruit antigen-presenting cells (APCs) and stimulate their activation and migration into the draining lymph nodes where they then activate antigen-specific T cells.4,5We previously reported that supramolecular peptide nanofibers carrying antigenic epitopes raise strong T-dependent antibody responses and T effector responses (TH1 and TH2) without requiring the incorporation of exogenous adjuvants.6–10 For instance, OVAQ11 is obtained by synthesizing the OVA323-339 epitope in tandem with a flexible linker and the self-assembling peptide Q11. OVAQ11 and Q11 co-assemble into supramolecular nanofibers, which raise strong OVA-specific antibody and CD4 T cell responses. These peptide nanofibers are remarkably non-inflammatory,10 but their ability to elicit antibody responses was nevertheless dependent on myeloid differentiation primary response gene 88 (MyD88), the universal adaptor protein used by almost all toll-like receptors (TLRs) and the IL-1R family.11–14 TLRs respond to microbial products as well as endogenous ligands to induce the activation of the antigen-presenting cells and also of T and B cells in some cases.15 The IL-1R family responds to 13 cytokines including IL-1, IL-18, IL-33, and IL-36. IL-1 promotes the proliferation and survival of naive T cells and is crucial for the development of the TH17 cell subset, while IL-18 and IL-33 reinforce differentiation into TH1 cell and TH2 cell subsets, respectively.16 In this study, we focused on defining the mechanisms by which peptide nanofiber vaccines elicit T cell responses by testing the necessity of MyD88 in antigen-presenting cells or in T cells using OVAQ11 nanofibers.
Results
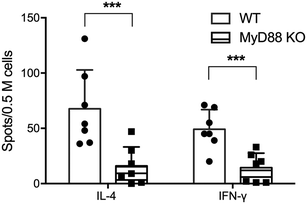
CD4+ T cell responses were significantly ablated in total MyD88 KO mice
CD4+ T cell responses to peptide nanofiber vaccines were significantly compromised in total MyD88 KO mice (Fig. 1). This result corresponds to our previous observation that these materials likewise failed to raise antibody responses in total MyD88 KO mice.7,8 To control possible variations in the microbiota that might then affect the immune response to OVAQ11, MyD88 KO and wild type (WT) C57BL/6 mice were co-housed for at least 8 weeks before immunization. This step was undertaken because it has previously been demonstrated that MyD88 deficiency results in reduced colonic expression of Reg3β and Reg3g antimicrobial peptides and a shift in bacterial diversity that could influence immune responses to vaccines.17 CD4+ T cells from draining lymph nodes were analyzed by ELISPOT 1 week after boost (day 35), which indicated that both IFN-γ and IL-4 OVA-specific CD4+ T cell responses were significantly compromised in total MyD88 KO mice (Fig. 1). This observation indicates that the ability of OVAQ11 nanofibers to raise CD4+ T cell responses is dependent on MyD88.Critical role of DCs in CD4+ T cell responses
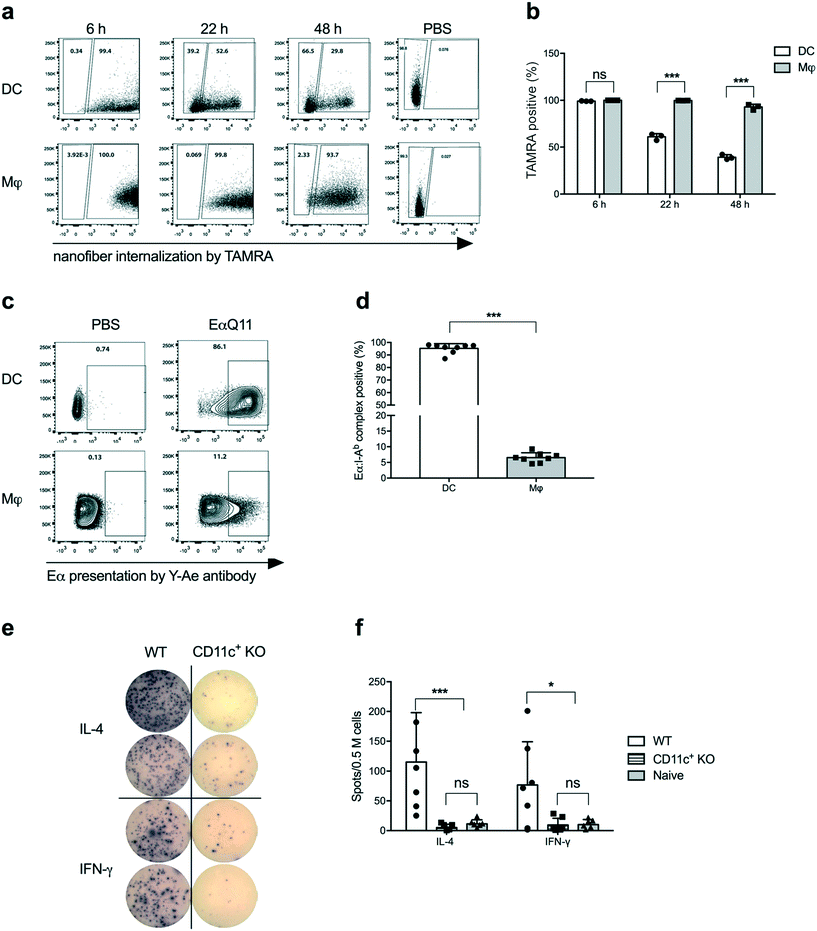
Dendritic cells (DCs) are the most potent APCs and play pivotal roles in innate and adaptive immunity,18–21 by presenting antigenic epitopes in the context of MHC Class II molecules along with co-stimulatory molecules to activate CD4+ T cells.22 We have previously reported that DCs actively acquire nanofibers and upregulate CD80 and CD86 upon OVAQ11 internalization.10 To track the OVAQ11 nanofibers, 0.02 mM OVAQ11 in the nanofibers was labelled with a fluorescent dye, 5-(and-6)-carboxytetramethylrhodamine (TAMRA). Using OVAQ11–TAMRA nanofibers, a time course study of nanofiber uptake by cells in the intraperitoneal cavity was performed. The intraperitoneal route was used as a surrogate for subcutaneous administration because we currently are unable to robustly track nanofiber uptake in the draining lymph nodes. Consistent with our previous report, DCs and macrophages efficiently internalized the nanofibers, detectable as early as 6 h post-immunization. Interestingly, a gradual decrease from 99.2% to 39.2% in the percentage of TAMRA+ cells was observed for DCs over the next 48 h, while the percentage of TAMRA+ macrophages remained persistently high at 92.9% over the same period (Fig. 2a and b). The gradual decline of TAMRA+ DCs over 48 h implied that DCs actively processed nanofibers. The macrophages may have also processed the nanofibers as the MFI of TAMRA-positive macrophages decreased over time, but much less efficiently compared to DCs. Another possibility is that the nanofibers were intact but the fluorochrome was detached and degraded in the DCs; we consider this to be unlikely as the macrophages retained high TAMRA signals over the same period of time.To investigate whether internalized nanofibers were processed and the antigen was presented by MHC Class II molecules, a Q11 nanofiber containing the Eα peptide was employed. Eα is a short peptide from the MHC Class II-I-Eα-chain (not expressed by host C57BL/6 mice) that is presented by host MHC Class II I-Ab, and the Eα:I-Ab complex can be detected with the Y-Ae monoclonal antibody.23 When synthesized in tandem with the self-assembling peptide Q11, EαQ11 was able to form nanofibers with Q11. Experiments reported here utilized a molar ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 EαQ11
1 EαQ11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Q11 co-assembled into mixed nanofibers (Fig. S1a†). In addition, EαQ11 nanofibers were internalized by DCs and macrophages comparably to their OVAQ11 counterparts at 24 h post-administration (Fig. S1b and c†). In contrast to the equally efficient internalization, Eα was presented by significantly higher percentages of DCs than macrophages at 24 h post-injection (Fig. 2c and d). These data demonstrated the ability of DCs to avidly take up the peptide nanofibers, process the Eα peptide, and present it within I-Ab MHC-II on the cell surface, whereas macrophages were much less efficient at processing and presenting the Eα peptide on I-Ab. We therefore hypothesized that DCs are the major antigen-presenting cells for antigens incorporated into nanofiber vaccines.
Q11 co-assembled into mixed nanofibers (Fig. S1a†). In addition, EαQ11 nanofibers were internalized by DCs and macrophages comparably to their OVAQ11 counterparts at 24 h post-administration (Fig. S1b and c†). In contrast to the equally efficient internalization, Eα was presented by significantly higher percentages of DCs than macrophages at 24 h post-injection (Fig. 2c and d). These data demonstrated the ability of DCs to avidly take up the peptide nanofibers, process the Eα peptide, and present it within I-Ab MHC-II on the cell surface, whereas macrophages were much less efficient at processing and presenting the Eα peptide on I-Ab. We therefore hypothesized that DCs are the major antigen-presenting cells for antigens incorporated into nanofiber vaccines.
To test whether DCs were necessary for activating CD4+ T cells following peptide nanofiber vaccination, we used CD11c-DTR mice that allowed for the depletion of CD11c+ cells (which includes DCs as well as macrophage and monocyte subsets) via the administration of diphtheria toxin. Following CD11c+ cell depletion, we adoptively transferred OVA-specific CD4+ OTII cells and then immunized them with OVAQ11. Activation of OTII cells by OVAQ11 nanofibers was analyzed by ELISPOT to show that the depletion of CD11c+ cells significantly compromised IFN-γ and IL-4 responses by OTII cells (Fig. 2e and f). These data confirmed the pivotal role of CD11c+ cells in internalizing nanofibers, processing them, and eliciting CD4+ T cell responses in vivo.
Unaffected DC functions in CD11c-MyD88 KO mice
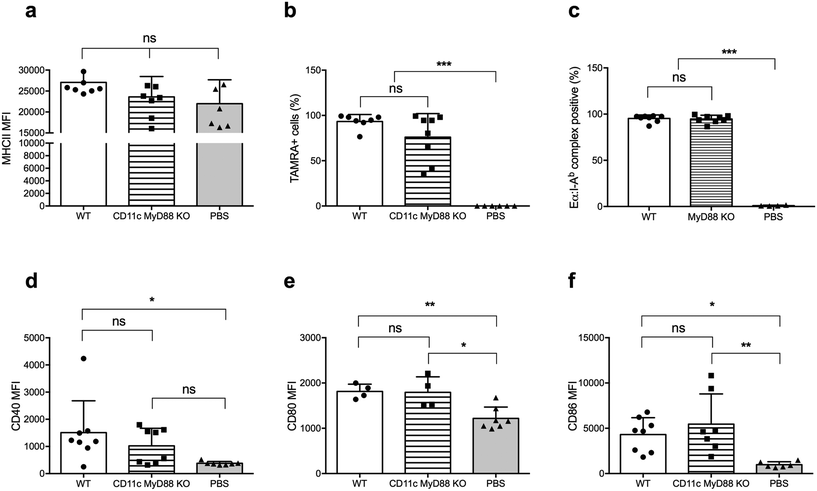
There is an extensive body of literature supporting the notion that MyD88 in DCs is critical for its activation and its ability to promote T cell responses, and vaccines incorporating adjuvants that stimulate the MyD88 pathway downstream of TLRs are more immunogenic than vaccines without these adjuvants.24,25 Therefore, we next investigated whether MyD88 is required for DC uptake, processing, and presentation of peptide nanofibers, and for upregulating CD80 and CD86 expression. We used CD11c-MyD88 KO mice (CD11cCreMyD88flox/flox)26 and first confirmed that the MyD88fl allele in DCs was deleted, by sorting DCs based on CD11c expression and establishing MyD88 deletion by quantitative real-time (qRT) PCR on genomic DNA (Fig. S2†). We also confirmed that MyD88-deficient DCs had comparable levels of MHCII expression to WT DCs (Fig. 3a).Following immunization with OVAQ11–TAMRA, MyD88-deficient and -sufficient DCs were comparably capable of internalizing the nanofibers (Fig. 3b). Using EαQ11 nanofibers, the presentation of Eα peptides by I-Ab was demonstrated to be unaltered by the lack of MyD88 in DCs (Fig. 3c). Finally, DCs were activated upon internalization of peptide nanofibers, as evidenced by the elevated expression of CD40 (Fig. 3d), CD80 (Fig. 3e), and CD86 (Fig. 3f). The absence of MyD88 in DCs did not significantly reduce these responses. Thus, these results collectively demonstrated that MyD88 expression in DCs was not required for nanofiber internalization, presentation, and DC activation.
MyD88 in DCs is not required for CD4+ T cell activation
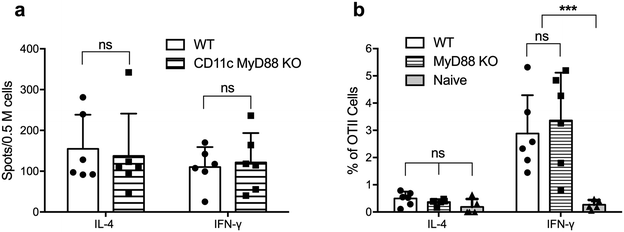
We next tested whether MyD88-deficient DCs were able to activate CD4+ T cells in vivo following immunization with OVAQ11 nanofibers. ELISPOT of endogenous CD4+ T cells from draining lymph nodes of CD11cCreMyD88flox/flox (CD11c-MyD88 KO) mice showed that these CD4+ T cells were strongly activated, when examined on day 7 after the boost (Fig. 4a). To address the possibility that low-level expression of MyD88 could be sufficient to promote CD4+ T cell responses or that the expression of MyD88 is critical in other non-DC subsets that may function as APCs, we tested the role of MyD88 in DCs in a second experimental setting. T cells were purified from OTII mice (5 × 105 per mouse; 95% purity of OTII cells; Fig. S3†) and adoptively transferred to MyD88 KO mice, which were then immunized with OVAQ11 the next day. Five days after immunization, the activation of OTII cells in the draining lymph nodes was quantified by flow cytometry. Consistent with the observations on CD11c-MyD88 KO mice, the OTII cells were activated strongly in total MyD88 KO recipients (Fig. 4b) and differentiated into IFN-γ-producing TH1 cells. In this experiment, MyD88 was deficient in all APCs, not just DCs, yet transferred OT-II cells were normally stimulated to produce IFN-γ. This indicated that there is not a compensating non-classical APC that stimulates T cells when MyD88 is knocked out of DCs. It is also noteworthy that OTII cells are not stimulated in WT mice by the OVA peptide (without the Q11 domain) as previously reported.27 These observations suggested that MyD88 is dispensable in DCs and also in other potential antigen-presenting cells for peptide nanofiber vaccines to stimulate T cells.Inflammasome pathway-independent CD4+ T cell responses
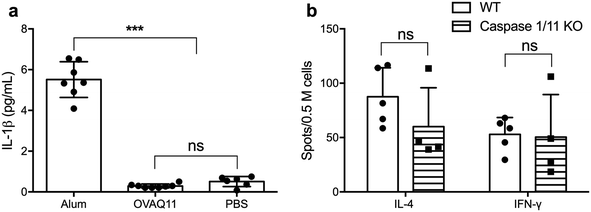
The potent adjuvancy of alum is dependent in part on the activation of the inflammasome pathway, which culminates in activation of caspase-1 and secretion of inflammatory IL-1β,28 a potent stimulator of T cell activation, promoting their proliferation and survival.29 To test whether IL-1β is necessary for the ability of peptide nanofibers to stimulate T cell responses, we first determined whether Q11 nanofibers stimulated the production of IL-1β. In contrast to the high IL-1β concentration in the lavage fluid after alum injection, no IL-1β was detected for OVAQ11 nanofibers (Fig. 5a), consistent with our previous findings that peptide nanofiber immunization elicits minimal local inflammation.10 We also investigated whether activation of the inflammasome pathway is required for CD4+ T cell responses using caspase 1/11 double KO mice.30 Caspase 1/11 double KO mice were immunized twice with peptide nanofiber vaccines, and endogenous CD4+ T cell responses in caspase 1/11 KO mice were quantified by ELISPOT 1 week after the boost immunization. CD4+ T cell responses in caspase 1/11 KO mice were comparable to those in wild type mice (Fig. 5b). Collectively, these observations indicate that nanofiber vaccination is not dependent on the inflammasome–IL-1β pathway for its ability to elicit CD4+ T cell responses.MyD88 expression in CD4+ T cells is not required for nanofiber vaccination
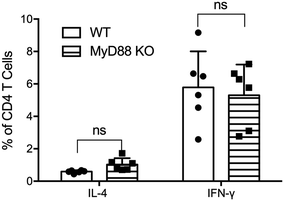
MyD88 in T cells has been shown to be necessary for CD4+ T cell responses following viral infection31 and for their ability to respond to IL-1 (ref. 32) and to differentiate into TFH cells.33 To test whether CD4+ T cells require MyD88 to respond to nanofiber vaccinations, T cells from total MyD88 KO mice were adoptively transferred to TCR KO mice (≥98% T cells; Fig. S4†) to generate a model where all immune cells express MyD88 except for the responding T cells. Following adoptive transfer, recipient mice were immunized twice with OVAQ11 nanofiber vaccines, and CD4+ T cells in draining lymph nodes were analyzed by flow cytometry 1 week after the boost immunization. MyD88-deficient CD4+ T cells responded to peptide nanofiber vaccines comparably to WT CD4+ T cells (Fig. 6). Our observations indicated that while MyD88 was globally required for optimal Q11 nanofiber vaccine responses, knocking out MyD88 in only the DC or T cell populations did not undermine CD4+ T cell responses.Discussion
Early observations of subunit vaccines indicated that foreign antigens alone were insufficient to elicit the adaptive immune response, and that in order to elicit T and B cell responses, these antigens must be delivered along with crude extracts such as a mineral oil containing mycobacteria or aluminum hydroxide, collectively termed as adjuvants. This led to the guiding paradigm that innate immune responses control adaptive immunity,34 and that adaptive immunity requires multiple signals provided by the antigen-presenting cells: antigen-specific signals that engage the T cell receptor, and co-stimulatory signals and cytokines that are upregulated in antigen-presenting cells upon the engagement of pattern recognition receptors (PRRs).22,24,35,36 These PRRs, of which the TLRs are an important subset, recognize a broad class of highly conserved pathogen-associated molecular patterns. Subsequently, Medzhitov et al. reported that MyD88 was an important adaptor protein in the TLR/IL-1 receptor pathway that signaled innate immune activation.37 Since those seminal observations, considerable evidence has accumulated on the necessity of MyD88 expression by antigen-presenting cells for adaptive immunity,38–40 and on DCs being the pivotal antigen-presenting cell that connects innate immunity to adaptive immunity in a naïve animal.18,20 While nanofiber vaccines required global MyD88 and DCs for their immunogenicity, our observations that deficiency of MyD88 in only the DC or T cell populations did not undermine CD4+ T cell responses suggest a more nuanced mechanism of immune activation.Understanding the mechanisms of a vaccine's immunogenicity is key to rationally improving its design. Our previous investigations revealed that self-assembled peptide nanofibers were relatively non-cytotoxic in vitro, and surprisingly, they elicited no measurable inflammation at the injection site and no accumulation of inflammatory cells (neutrophils, inflammatory monocytes or eosinophils), cytokines (IL-1β, IL-6 and IL-5), or chemokines (MCP-1, KC and G-CSF).10 Here, we extend those observations to show that while both dendritic cells and macrophages acquired the nanofiber vaccines, only DCs were capable of presenting antigen acquired from the nanofibers and upregulating their expression of CD80 and CD86. Notably, this occurred despite the absence of inflammatory cues. We also previously reported that B cell/antibody responses to peptide nanofiber vaccines were abrogated in MyD88-deficient mice,7 and here we confirmed that T cell responses were also diminished. However, contrary to expectation, MyD88 in DCs was not required, as the ability of MyD88-deficient DCs to take up the nanofibers, process them, present their epitopes within MHC-II, upregulate the co-stimulatory molecules CD80 and CD86, and stimulate CD4+ T cells was not significantly different from that of MyD88-sufficient DCs. These findings, along with the observation that Q11 nanofibers do not elicit cytokines such as IL-12 or IL-6 (Fig. S6†), are consistent with the nanofiber vaccines harboring no exogenous TLR ligands. Indeed, a survey of individual classical pathways necessary for DC activation, including TLRs 2/4/5, Nalp3, caspase 1/11, Trif and Type I interferon receptor (IFNαR) (Fig. S5† and Table 1), is consistent with the notion that Q11 nanofibers elicit immune responses in ways that are distinct from classical pro-inflammatory adjuvants. The lack of production of IL-6 or IL-12 by peptide nanofiber-stimulated APCs (Fig. S6†) further suggests that NF-κB activation is not a primary pathway involved. We have not examined the TRIF pathway in CD4+ T cell expansion, however the antibodies elicited by Q11 nanofibers are T cell-dependent,27 so it is likely that TRIF is not required for at least the generation of TFH responses. Further knowledge of the more detailed mechanisms by which these nanofibers drive APC activation and T cell responses will require continued investigation.
| Cells or signaling pathways | Function in nanofiber vaccination |
|---|---|
| CD11c+ DCs | Required for cytokine production by T cells |
| CD4+ T cells | Required for antibody response7,8 |
| MyD88 | |
| MyD88 (global expression) | Required for T cell and antibody responses7,8 |
| MyD88 in CD11c+ cells only | Not required for T cell responses |
| MyD88 in T cells only | Not required for T cell responses |
| TLRs and inflammasome | |
| TLR 2/4/5 | Not required for T cell-dependent antibody responses7,10 |
| Caspase 1/11 | Not required for T cell responses |
| IFNαR | Not required for T cell-dependent antibody responses |
| Nalp3 | Not required for T cell-dependent antibody responses |
In addition to TLR ligands, the IL-1R family also recruits MyD88, which is necessary for downstream signaling.13,14 The IL-1–inflammasome pathway has been implicated as a mechanism by which aluminum salts and saponins enhance the immunogenicity of vaccines.28 IL-1 and IL-18 are synthesized as inactive precursors that are then cleaved into active cytokines by caspase 1, a part of the inflammasome complex. Assembly of the inflammasome complex occurs after the sensing of pathogen-associated molecular patterns (PAMPs),41 and caspase 11 is important for caspase 1 activation.42 However, we observed that IL-1β production was not detected following nanofiber vaccination, and caspase 1/11 was not essential for the immunogenicity of peptide nanofiber vaccines, thus confirming that this pathway is also not essential for T cell responses against these materials.
The expression of MyD88 in T cells has been reported to be critical in a number of scenarios. Recently, Schenten et al. reported that MyD88 in CD4 cells was necessary for the ability of T cells to respond to IL-1 and to overcome suppression by regulatory T cells.32 CD4+ T cell responses to lymphocytic choriomeningitis virus (LCMV) infections were reported to be dependent on the expression of MyD88 in T cells, but not DCs, and independent of IL-1 or IL-18 responses.31 TLRs and MyD88 in T cells have also been reported by Gelman et al.43,44 to promote T cell survival by stimulating phosphoinositide 3-kinase (PI3K) and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), while the engagement of TLRs on regulatory T cells inhibits their function, resulting in the overall enhancement of immune responses.45 Likewise, Bartholdy et al.46 reported that T-cell intrinsic expression of MyD88 is required for sustained expansion and TFH differentiation of virus-specific CD8+ T cells in LCMV-infected mice.33 Thus, there is a substantial body of evidence indicating that under certain circumstances T cells can directly engage ligands that stimulate the MyD88 pathway downstream of IL-1R/18R or TLRs, and that MyD88 in DCs are not always necessary. Accordingly, we tested the necessity of MyD88 in T cells for responding to OVAQ11 nanofiber vaccinations, but similar to the result with DCs, we found that MyD88 deficiency in T cells alone did not abrogate the immunogenicity of the self-assembled peptide nanofibers.
The observation that the global deficiency of MyD88 resulted in significantly reduced T cell responses, while the absence of MyD88 in APCs or T cells alone preserved the immunogenicity of the peptide nanofiber vaccines, suggests that Q11 nanofibers trigger redundant MyD88-dependent and independent signaling pathways in APCs and T cells. Comparably, Hou et al.26 reported that the total IgG response to immunization to protein OVA antigen mixed with aggregated TLR ligands in cationic lipid required global MyD88 but not MyD88 in B cells or DC cells alone. One caveat to our conclusion is that complete MyD88 deficiency may have downstream effects that indirectly alter the immune response to Q11 nanofiber vaccination. For instance, gut microbial ecology is altered and shifts towards a greater proportion of segmented filamentous bacteria when gut epithelial cells are deficient in MyD88,17 and segmented filamentous bacteria preferentially drives mucosal TH17 responses.47,48 While co-housing of the MyD88-deficient mice with WT mice that was performed minimized the impact of altered microbiota, it remains possible that other indirect effects of a global MyD88 deficiency may persist and account for the reduced response to Q11 nanofiber vaccines that was not recapitulated when MyD88 was deficient only in DCs or T cells.
We speculate that Q11 nanofiber vaccines may be grouped into a new class of vaccines that are immunogenic without the need for exogenous adjuvants and are relatively non-inflammatory. Included in this class are the self-assembled peptide amphiphile micelles that promote cytotoxic T cell responses49,50 and the self-assembling polypeptide nanoparticles that repetitively display antigenic epitopes and elicit strong antibody and CD8 T cell responses.51 It remains to be determined if such nanoparticles differing in composition, shape, and size have immunological characteristics similar to the Q11 nanofiber system, where inflammation is not required at the injection site and there is a paradoxical requirement for global MyD88 that cannot be attributable necessity in DCs or T cells alone.
Despite much research, available adjuvants are still far from being mechanistically understood or rationally tunable. Indeed, there is still an incomplete understanding of how the widely used aluminum salts function in clinical vaccines. Some common themes that have emerged are that alum acts as a repository, promotes phagocytic activity, and induces inflammatory signals that lead to the recruitment of various types of immune cells.52 There is, however, some dispute regarding the specifics of how inflammation is induced. Kool et al.53 proposed that alum induces cell necrosis and uric acid production, which activates the NACHT, LRR and PYD domain-containing protein 3 (Nlrp3) inflammasome, and Eisenbarth et al.28 confirmed the necessity of the inflammasome. In contrast, Franchi et al.54 found that the antibody responses elicited by alum-adjuvanted vaccines were not inhibited in Nlrp3 inflammasome-deficient mice, while Marichal et al.55 proposed that DNA released from necrotic cells exposed to alum was the basis for the adjuvanting activity. Flach et al.56 reported that aluminum salts induced cell membrane lipid reordering, while Wang et al.57 suggested that the heat shock protein 70 in combination with inflammasomes is critical. Likewise, there is controversy regarding the role of TLR/MyD88 in other classic adjuvants. Early observations by Pasare and Medzhitov emphasized the necessity of the TLR–MyD88 pathway in DCs and B cells for the immunogenicity of OVA–lipopolysaccharide in Freund's incomplete adjuvant or alum.24,25 In contrast, Gavin et al.58 reported that TLR–MyD88–Trif pathways were not necessary for the activity of three classical adjuvants: alum, Freund's complete adjuvant, and monophosphoryl lipid A/trehalose dicorynomycolate (Ribi) adjuvant. An explanation proposed to explain these discordant observations52 is the lack of uniformity in the reagents, so each formulation elicits different effects, which are likely to be further amplified when used in large human populations. Additionally, a redundancy in mechanisms of action may indeed be a feature of a universally potent adjuvant.
Vaccines that can be formulated with precision and elicit immune responses in a mechanistically reproducible manner would be most amenable to rationally designed improvement. Towards that goal, the peptide nanofiber platform is advantageous compared to traditional inflammatory adjuvants, as immunogenicity can be uncoupled from toxicity and inflammation. We propose that self-assembling nanofibers fulfil these criteria in that they have the potential to present antigens in precise quantities to shape the quality and strength of the immune response,59 and that they are nominally inflammatory but have the potential to incorporate TLR ligands to improve immunogenicity.10 Finally, we demonstrate here that Q11 nanofibers are highly capable of eliciting T cell and antibody responses via mechanisms that are MyD88-dependent, but where MyD88 expression in either APCs or T cells is sufficient for optimum T cell responses.
Materials and methods
Peptides and formulations
The peptides OVAQ11 (also named OVA323-339-SGSG-Q11, H2N-ISQAVHAAHAEINEAGR-SGSG-QQKFQFQFEQQ-Am), Q11 (Ac-QQKFQFQFEQQ-Am), and EαQ11 (or Eα52-68-SGSG-Q11, H2N-ASFEAQGALANIAVDKA-SGSG-QQKFQFQFEQQ-Am) were synthesized using standard Fmoc solid-phase synthesis as previously reported.60 5-(and-6)-Carboxytetramethylrhodamine (TAMRA)-conjugated OVAQ11 (OVAQ11–TAMRA) was synthesized via N-terminal on-resin conjugation by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) coupling. All peptides were confirmed using matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) on a Bruker Ultraflextreme MALDI-TOF mass spectrometer. The peptides were purified by reverse-phase high performance liquid chromatography (RP-HPLC), lyophilized, and stored at −20 °C before use.60A typical formulation contains 1.33 mM OVAQ11 (or EαQ11) and 0.67 mM Q11. To prepare the nanofibers, lyophilized peptides were weighed and intermixed as dry powders by vortexing for 30 min. Ultrapure water was added to form 8 mM total peptide solutions. After overnight incubation at 4 °C, the peptide solution was diluted to 2 mM in 1× PBS by adding ultrapure water and sterile 10× PBS (Cat# BP399-500, Fisher, Pittsburgh, PA, USA), and incubated at room temperature for 3 h. Endotoxin in the formulation was measured using the Limulus amebocyte lysate chromogenic endpoint assay (Lonza, Allendale, NJ, USA), which indicated that endotoxin in all vaccine formulations was <0.5 EU mL−1 (<0.05 EU per 100 μL dose). TAMRA-labelled nanofibers comprising 1% (or 0.02 mM) OVAQ11–TAMRA were prepared by intermixing OVAQ11 and Q11 peptides in 0.08 mM OVAQ11–TAMRA, as previously reported in ref. 10.
Mice and immunization
All mice were purchased from Harlan Sprague Dawley and housed in a centralized animal facility at the University of Chicago unless otherwise noted. MyD88 knockout (KO) mice (B6.129P2(SJL)-MyD88tm1.1defr/J) and wild type controls were obtained from Jackson Laboratory and co-housed for at least 8 weeks before use. CD11c MyD88 KO (CD11cCre/CreMyD88flox/flox) mice were bred in-house from CD11c-Cre (B6.Cg-Tg(Itgax-cre)1-1Reiz/J) and MyD88flox/flox (B6.129P2(SJL)-Myd88tm1Defr/J) transgenic mice. All procedures were approved by the University of Chicago Institutional Animal Care and Use Committee and complied with the NIH Guide for the Care and Use of Laboratory Animals. Primary and boost immunizations were performed under anesthesia via subcutaneous injection (50 μL at each location) in the left and right flanks for studying T cell responses. Intraperitoneal injection was performed at the left and right lower abdomen (50 μL each at each location) to study cell uptake and presentation. In some cases, 0.5 million CD4+ T cells in 200 μL PBS were adoptively transferred via tail vein injection 1 day before immunization, and the draining lymph nodes were collected for further analysis.In vivo cell uptake, presentation, and activation
To investigate the efficiency of nanofiber uptake, mice were euthanized at designated time points after intraperitoneal immunization with 100 μL 2 mM OVAQ11–TAMRA nanofibers. Two mL of Hank's balanced salt solution (HBSS) was injected into the intraperitoneal space, and about 1.2–1.5 mL was subsequently withdrawn. Cells were collected by centrifugation and washed once with flow buffer (PBS containing 2% fetal bovine serum). After blocking the Fc receptor with 2.4G2 antibody, cells were stained for F4/80, MHCII, CD11c, CD11b, CD40, CD80, and CD86. Cells were washed and resuspended in flow buffer with 1 μg mL−1 DAPI. Flow cytometry was performed on a BD LSRII and analyzed using FlowJo software. The percentage of TAMRA-positive cells was calculated for both macrophages (gated as F4/80+CD11b+) and dendritic cells (DCs, gated as F4/80−CD11c+MHCIIhigh). To investigate the presentation of the Eα peptide, mice were immunized with 100 μL 2 mM EαQ11 nanofibers intraperitoneally. IP lavage cells were collected and stained as described above. The Eα:I-Ab complexes were stained using Y-Ae antibodies (eBioscience, San Diego, CA, USA). Cells were washed and re-suspended in flow buffer containing 1 μg mL−1 DAPI. Flow cytometry was performed on a BD LSRII and analyzed using FlowJo software.Quantification of CD4+ T cell responses by ELISPOT
ELISPOT was used to analyze T cell responses. Mice were euthanized 7 days after the final boost. Inguinal, brachial, and axillary draining lymph nodes were collected, and single cell suspensions were prepared as previously reported.9 Briefly, 96-well ELISPOT plates (Millipore, Billerica, MA, USA) were washed and coated overnight at 4 °C with capture antibodies (anti-IFN-γ or anti-IL-4, BD, San Jose, CA, USA). Wells were blocked with complete RPMI medium for 2 h at 37 °C; 200 μL 2.5 million per mL cells were seeded and stimulated with 5 μM pOVA for 48 h in a CO2 incubator at 37 °C. After stimulation, plates were washed and incubated with detection antibodies (biotinylated anti-IFN-γ or biotinylated anti-IL-4, BD, San Jose, CA, USA), then with streptavidin–alkaline phosphatase (Mabtech, Cincinnati, OH, USA), and finally with the substrate Sigmafast BCIP/NBT (Sigma, Saint Louis, MO, USA). Plates were dried overnight, and spots were counted using an ELISPOT reader (Cellular Technology Ltd, Cleveland, OH, USA).Analyzing CD4+ T cell responses by flow cytometry
To track the activation of adoptively transferred OTII cells in MyD88 KO mice, spleen and lymph nodes (inguinal, brachial, and axillary) were removed from sacrificed OTII transgenic mice and pressed through a sterile 40 μm nylon strainer (BD, San Jose, CA, USA) to form single-cell suspensions. OTII cells were then isolated by using a CD4+ T cell isolation kit (Miltenyi Biotec, San Diego, CA, USA) according to the manufacturer's protocol. The purity of the CD4+ cells in the enriched fraction was determined by flow cytometry. Five hundred thousand OTII cells were injected into the tail vein of each recipient mouse (MyD88 KO or wild type control mice) 1 day before immunization, and 5 days later single-cell suspensions were obtained from the draining lymph nodes. These were then stimulated with 50 ng mL−1 phorbol 12-myristate 13-acetate (PMA) plus 750 ng mL−1 ionomycin (both from Sigma, Saint Louis, MO, USA) for 6 h at 37 °C. Brefeldin A (10 μg mL−1, Sigma) was added during the last 3 h of incubation. After surface staining with the corresponding cocktail of antibodies, intracellular cytokine staining was sequentially performed for detection of IFN-γ and IL-4.To study the activation of adoptively transferred MyD88 KO T cells in T cell receptor (TCR) KO mice (B6.129S2-Tcratm1Mom/J), single-cell suspensions of spleen and lymph nodes were prepared from MyD88 KO and wild type control mice, respectively. T cells were isolated by using the Pan T Cell Isolation Kit II (Miltenyi Biotec, San Diego, CA, USA), and the purity of the T cell population in the enriched fraction was determined by flow cytometry analysis. Five hundred thousand T cells were adoptively transferred via the tail vein to each recipient TCR KO mouse. Following adoptive transfer, recipient mice were immunized twice with OVAQ11 nanofiber vaccines, and the draining lymph nodes were collected for the detection of IFN-γ and IL-4 by flow cytometry 1 week after the boost immunization as described above.
Statistical analysis
Statistical analysis was performed in Graphpad Prism using Student's t-tests, 1-way, or 2-way ANOVA with Tukey's multiple comparison test as indicated in the figure legends. Means and standard deviations are presented unless otherwise noted.Author contributions
A. S. C. and J. H. C. conceived and designed the framework of this study. Y. S., Y. W., J. C., R. R. P., and H. H. carried out the experiments. All authors analyzed the data, discussed the results, and edited the manuscript. Y. W., A. S. C., J. H. C., and Y. S. wrote the manuscript.Conflicts of interest
None of the authors have financial conflicts of interest to disclose.Acknowledgements
The research described in this article was supported by the National Institutes of Health: The National Institute of Biomedical Imaging and Bioengineering (NIBIB) under grant number 1R01EB009701 and the National Institute of Allergy and Infectious Diseases (NIAID) under grant number 1R01AI118182. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of these agencies.References
- R. Rappuoli, F1000 Med. Rep., 2011, 3, 16 CrossRef PubMed.
- R. Rappuoli, C. W. Mandl, S. Black and E. De Gregorio, Nat. Rev. Immunol., 2011, 11, 865–872 CAS.
- A. Di Pasquale, S. Preiss, F. Tavares Da Silva and N. Garcon, Vaccines, 2015, 3, 320–343 CrossRef PubMed.
- E. De Gregorio, E. Caproni and J. B. Ulmer, Front. Immunol., 2013, 4, 214 Search PubMed.
- S. Awate, L. A. Babiuk and G. Mutwiri, Front. Immunol., 2013, 4, 114 Search PubMed.
- J. S. Rudra, Y. F. Tian, J. P. Jung and J. H. Collier, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 622–627 CrossRef CAS PubMed.
- J. S. Rudra, S. Mishra, A. S. Chong, R. A. Mitchell, E. H. Nardin, V. Nussenzweig and J. H. Collier, Biomaterials, 2012, 33, 6476–6484 CrossRef CAS PubMed.
- J. S. Rudra, T. Sun, K. C. Bird, M. D. Daniels, J. Z. Gasiorowski, A. S. Chong and J. H. Collier, ACS Nano, 2012, 6, 1557–1564 CrossRef CAS PubMed.
- R. R. Pompano, J. Chen, E. A. Verbus, H. Han, A. Fridman, T. McNeely, J. H. Collier and A. S. Chong, Adv. Healthcare Mater., 2014, 3, 1898–1908 CrossRef CAS PubMed.
- J. Chen, R. R. Pompano, F. W. Santiago, L. Maillat, R. Sciammas, T. Sun, H. Han, D. J. Topham, A. S. Chong and J. H. Collier, Biomaterials, 2013, 34, 8776–8785 CrossRef CAS PubMed.
- C. A. Dinarello, Blood, 2011, 117, 3720–3732 CrossRef CAS PubMed.
- N. Warner and G. Nunez, J. Immunol., 2013, 190, 3–4 CrossRef CAS PubMed.
- R. Medzhitov, P. Preston-Hurlburt, E. Kopp, A. Stadlen, C. Chen, S. Ghosh and C. A. Janeway Jr., Mol. Cell, 1998, 2, 253–258 CrossRef CAS PubMed.
- O. Adachi, T. Kawai, K. Takeda, M. Matsumoto, H. Tsutsui, M. Sakagami, K. Nakanishi and S. Akira, Immunity, 1998, 9, 143–150 CrossRef CAS PubMed.
- L. A. O'Neill, D. Golenbock and A. G. Bowie, Nat. Rev. Immunol., 2013, 13, 453–460 CrossRef PubMed.
- J. E. Sims and D. E. Smith, Nat. Rev. Immunol., 2010, 10, 89–102 CrossRef CAS PubMed.
- E. Larsson, V. Tremaroli, Y. S. Lee, O. Koren, I. Nookaew, A. Fricker, J. Nielsen, R. E. Ley and F. Backhed, Gut, 2012, 61, 1124–1131 CrossRef CAS PubMed.
- R. M. Steinman and M. C. Nussenzweig, Immunol. Rev., 1980, 53, 127–147 CrossRef CAS PubMed.
- R. M. Steinman and M. D. Witmer, Proc. Natl. Acad. Sci. U. S. A., 1978, 75, 5132–5136 CrossRef CAS.
- M. C. Nussenzweig, R. M. Steinman, B. Gutchinov and Z. A. Cohn, J. Exp. Med., 1980, 152, 1070–1084 CrossRef CAS PubMed.
- A. Mildner and S. Jung, Immunity, 2014, 40, 642–656 CrossRef CAS PubMed.
- R. H. Schwartz, Cell, 1992, 71, 1065–1068 CrossRef CAS PubMed.
- A. Farr, P. C. DeRoos, S. Eastman and A. Y. Rudensky, Eur. J. Immunol., 1996, 26, 3185–3193 CrossRef CAS PubMed.
- C. Pasare and R. Medzhitov, Immunity, 2004, 21, 733–741 CrossRef CAS PubMed.
- C. Pasare and R. Medzhitov, Nature, 2005, 438, 364–368 CrossRef CAS PubMed.
- B. Hou, B. Reizis and A. L. DeFranco, Immunity, 2008, 29, 272–282 CrossRef CAS PubMed.
- J. Chen, R. R. Pompano, F. W. Santiago, L. Maillat, R. Sciammas, T. Sun, H. Han, D. J. Topham, A. S. Chong and J. H. Collier, Biomaterials, 2013, 34, 8776–8785 CrossRef CAS PubMed.
- S. C. Eisenbarth, O. R. Colegio, W. O'Connor, F. S. Sutterwala and R. A. Flavell, Nature, 2008, 453, 1122–1126 CrossRef CAS PubMed.
- S. Z. Ben-Sasson, J. Hu-Li, J. Quiel, S. Cauchetaux, M. Ratner, I. Shapira, C. A. Dinarello and W. E. Paul, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 7119–7124 CrossRef CAS PubMed.
- N. Kayagaki, S. Warming, M. Lamkanfi, L. Vande Walle, S. Louie, J. Dong, K. Newton, Y. Qu, J. Liu, S. Heldens, J. Zhang, W. P. Lee, M. Roose-Girma and V. M. Dixit, Nature, 2011, 479, 117–121 CrossRef CAS PubMed.
- S. Zhou, E. A. Kurt-Jones, A. M. Cerny, M. Chan, R. T. Bronson and R. W. Finberg, J. Virol., 2009, 83, 1625–1634 CrossRef CAS PubMed.
- D. Schenten, S. A. Nish, S. Yu, X. Yan, H. K. Lee, I. Brodsky, L. Pasman, B. Yordy, F. T. Wunderlich, J. C. Bruning, H. Zhao and R. Medzhitov, Immunity, 2014, 40, 78–90 CrossRef CAS PubMed.
- J. L. Kubinak, C. Petersen, W. Z. Stephens, R. Soto, E. Bake, R. M. O'Connell and J. L. Round, Cell Host Microbe, 2015, 17, 153–163 CAS.
- C. A. Janeway Jr., Cold Spring Harbor Symp. Quant. Biol., 1989, 54(Pt 1), 1–13 CrossRef.
- C. Pasare and R. Medzhitov, Adv. Exp. Med. Biol., 2005, 560, 11–18 CrossRef CAS PubMed.
- D. L. Mueller, M. K. Jenkins and R. H. Schwartz, Annu. Rev. Immunol., 1989, 7, 445–480 CrossRef CAS PubMed.
- T. Horng and R. Medzhitov, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 12654–12658 CrossRef CAS PubMed.
- M. Schnare, G. M. Barton, A. C. Holt, K. Takeda, S. Akira and R. Medzhitov, Nat. Immunol., 2001, 2, 947–950 CrossRef CAS PubMed.
- H. Hemmi, O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda and S. Akira, Nature, 2000, 408, 740–745 CrossRef CAS PubMed.
- T. Kaisho, O. Takeuchi, T. Kawai, K. Hoshino and S. Akira, J. Immunol., 2001, 166, 5688–5694 CrossRef CAS.
- H. Guo, J. B. Callaway and J. P. Ting, Nat. Med., 2015, 21, 677–687 CrossRef CAS PubMed.
- J. M. Blander, Nat. Rev. Immunol., 2014, 14, 601–618 CrossRef CAS PubMed.
- A. E. Gelman, D. F. LaRosa, J. Zhang, P. T. Walsh, Y. Choi, J. O. Sunyer and L. A. Turka, Immunity, 2006, 25, 783–793 CrossRef CAS PubMed.
- A. E. Gelman, J. Zhang, Y. Choi and L. A. Turka, J. Immunol., 2004, 172, 6065–6073 CrossRef CAS.
- D. F. LaRosa, A. E. Gelman, A. H. Rahman, J. Zhang, L. A. Turka and P. T. Walsh, Immunol. Lett., 2007, 108, 183–188 CrossRef CAS PubMed.
- C. Bartholdy, J. E. Christensen, M. Grujic, J. P. Christensen and A. R. Thomsen, J. Gen. Virol., 2009, 90, 423–431 CrossRef CAS PubMed.
- I. I. Ivanov, K. Atarashi, N. Manel, E. L. Brodie, T. Shima, U. Karaoz, D. Wei, K. C. Goldfarb, C. A. Santee, S. V. Lynch, T. Tanoue, A. Imaoka, K. Itoh, K. Takeda, Y. Umesaki, K. Honda and D. R. Littman, Cell, 2009, 139, 485–498 CrossRef CAS PubMed.
- V. Gaboriau-Routhiau, S. Rakotobe, E. Lecuyer, I. Mulder, A. Lan, C. Bridonneau, V. Rochet, A. Pisi, M. De Paepe, G. Brandi, G. Eberl, J. Snel, D. Kelly and N. Cerf-Bensussan, Immunity, 2009, 31, 677–689 CrossRef CAS PubMed.
- M. Black, A. Trent, Y. Kostenko, J. S. Lee, C. Olive and M. Tirrell, Adv. Mater., 2012, 24, 3845–3849 CrossRef CAS PubMed.
- A. Trent, B. D. Ulery, M. J. Black, J. C. Barrett, S. Liang, Y. Kostenko, N. A. David and M. V. Tirrell, AAPS J., 2015, 17, 380–388 CrossRef CAS PubMed.
- S. A. Kaba, M. E. McCoy, T. A. Doll, C. Brando, Q. Guo, D. Dasgupta, Y. Yang, C. Mittelholzer, R. Spaccapelo, A. Crisanti, P. Burkhard and D. E. Lanar, PLoS One, 2012, 7, e48304 CAS.
- S. G. Reed, M. T. Orr and C. B. Fox, Nat. Med., 2013, 19, 1597–1608 CrossRef CAS PubMed.
- M. Kool, T. Soullie, M. van Nimwegen, M. A. Willart, F. Muskens, S. Jung, H. C. Hoogsteden, H. Hammad and B. N. Lambrecht, J. Exp. Med., 2008, 205, 869–882 CrossRef CAS PubMed.
- L. Franchi and G. Nunez, Eur. J. Immunol., 2008, 38, 2085–2089 CrossRef CAS PubMed.
- T. Marichal, K. Ohata, D. Bedoret, C. Mesnil, C. Sabatel, K. Kobiyama, P. Lekeux, C. Coban, S. Akira, K. J. Ishii, F. Bureau and C. J. Desmet, Nat. Med., 2011, 17, 996–1002 CrossRef CAS PubMed.
- T. L. Flach, G. Ng, A. Hari, M. D. Desrosiers, P. Zhang, S. M. Ward, M. E. Seamone, A. Vilaysane, A. D. Mucsi, Y. Fong, E. Prenner, C. C. Ling, J. Tschopp, D. A. Muruve, M. W. Amrein and Y. Shi, Nat. Med., 2011, 17, 479–487 CrossRef CAS PubMed.
- Y. Wang, D. Rahman and T. Lehner, J. Biol. Chem., 2012, 287, 17152–17160 CrossRef CAS PubMed.
- A. L. Gavin, K. Hoebe, B. Duong, T. Ota, C. Martin, B. Beutler and D. Nemazee, Science, 2006, 314, 1936–1938 CrossRef CAS PubMed.
- G. A. Hudalla, J. A. Modica, Y. F. Tian, J. S. Rudra, A. S. Chong, T. Sun, M. Mrksich and J. H. Collier, Adv. Healthcare Mater., 2013, 2, 1114–1119 CrossRef CAS PubMed.
- J. P. Jung, A. K. Nagaraj, E. K. Fox, J. S. Rudra, J. M. Devgun and J. H. Collier, Biomaterials, 2009, 30, 2400–2410 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7md00367f |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2018 |