Defense contracts: molecular protection in insect-microbe symbioses
Ethan B.
Van Arnam
 a,
Cameron R.
Currie
b and
Jon
Clardy
a,
Cameron R.
Currie
b and
Jon
Clardy
 *c
*c
aKeck Science Department, Claremont McKenna, Pitzer, and Scripps Colleges, 925 N. Mills Ave., Claremont, CA 91711, USA
bDepartment of Bacteriology, University of Wisconsin-Madison, Madison, Wisconsin 53706, USA
cDepartment of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 240 Longwood Avenue, Boston, Massachusetts 02115, USA. E-mail: jon_clardy@hms.harvard.edu
First published on 26th July 2017
Abstract
Insects cope with environmental threats using a broad array of strategies. A key strategy, widespread among insects but unappreciated until recently, is the use of molecular defenses from symbiotic microbes. Insect-microbe defensive symbioses span the diversity of insect lineages and microbial partners and use molecules ranging from reactive oxygen species to small molecules to protein toxins to defend against predators, parasites, and microbial pathogens. These systems have a strong initial track record as sources of novel biologically active compounds with therapeutic potential. This review surveys the molecular basis for insect-microbe defensive symbioses with a focus on the ecological contexts for defense and on emerging lessons about molecular diversity from bacterial genomes.
1. Introduction
Insects have evolved diverse strategies to evade predators, parasites, and microbial pathogens, including an impressive array of chemical defenses. The bombardier beetle provides a dramatic example when it mixes hydroquinone and hydrogen peroxide in a specialized abdominal chamber to produce a boiling hot noxious mixture that is squirted at would-be predators.1,2 In a seemingly similar defense strategy the Paederus beetle deters predators with pederin (1, Fig. 1), a potent irritant that can cause blistering in humans.3 These seemingly similar strategies have dramatically different origins. While the bombardier beetle produces its own hydroquinone and hydrogen peroxide, pederin is synthesized by a bacterial symbiont living within its beetle host – a “defensive symbiosis” between an insect and a microbe.4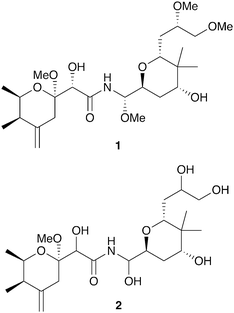 | ||
| Fig. 1 Polyketide toxins produced by insect endosymbionts: pederin (1), diaphorin (2). (Compounds in bold were first described in insect systems.) | ||
Mutualisms with bacteria are ubiquitous in animals ranging from humans to insects. In insects, nutritional symbioses such as the bacteria in a termite's digestive tract are particularly well documented; numerous examples of bacteria living in the gut or within specialized cells of the insect host providing nutrients and/or helping digest food have been described.5–7 Defensive symbioses, like the one between Paederus beetles and its pederin-producing symbiont, are increasingly recognized as important for insect survival as well.8–14
Defensive symbioses documented to date between insects and microbes are strikingly diverse: different threats to be defended against, multiple types of producing microbes, and a rich mixture of chemical scaffolds and biosynthetic pathways. Threats countered by defensive symbioses include predation, parasitism, and microbial pathogens (termed entomopathogens when they infect an insect). Known defensive symbionts span the bacterial kingdom from Proteobacteria to Firmicutes to Actinobacteria. These bacteria reside in diverse insect niches: within specialized insect cells, within the gut, or within specialized anatomical structures on the insect exoskeleton. From their diversity it is clear that defensive symbioses have arisen independently many times and some insect-microbe defensive symbioses are certainly ancient – dating back 60 million years or more.15,16 The transmission mechanism by which symbionts are established on each insect generation can also vary dramatically, from strict ‘vertical’ inheritance from the mother through the germline, to ‘horizontal’ recruitment from the environment in each generation.17,18 As a result, diverse mechanisms reinforce the stability of symbiotic associations, ranging from strict nutritional dependencies that ensure partner fidelity, to selective symbiont choice by the host.19–21
An intriguing feature of the defensive symbiosis paradigm is its parallel to human medicine as both deploy antagonistic molecules (antibiotics) to suppress pathogens. Insect defensive symbioses offer probably the clearest window currently available into antibiotic use in nature; in several cases the identities of the host, pathogen, antibiotic-producing symbiont, and active molecule are all known. These examples undoubtedly hold lessons for optimizing and prolonging antibiotic effectiveness that could inform current efforts to combat the threat of antibiotic resistance. A molecular level understanding of these defensive symbioses provides an opportunity for ecologically-guided drug discovery with a largely unexplored source of compounds with ‘privileged’ scaffolds that were selected for compatibility with an animal host and pharmaceutical potential.
This review considers the molecular basis for defensive symbioses between insects and microbes: the types of molecules involved, their ecological functions, and their genetic origins. Defensive symbioses need not involve secretion of a toxic molecule by the symbiont; effective defense could come from resource competition with a pathogen, or indirectly through stimulation of the host immune system.22,23 This review focuses on insect-microbe symbioses featuring production of antagonistic molecules by a microbial symbiont. This article contributes to an already strong collection of reviews on this topic.24–28
This review begins by describing the best-characterized defensive symbioses in insects, first considering molecular defenses from specific symbiont niches – intracellular compartments or insect guts – across diverse insects. We next consider defenses that support specialized insect ecologies – protection of brood chambers and protection of fungus cultivation, and review molecular discovery efforts from insect-associated microbes. After this examination of defensive molecules and their functions, we move to examples of what can be learned from a genetic and genomic analysis of the biosynthetic gene clusters of defensive molecules. While these studies are still in a relatively early stage, some interesting insights into the evolution of molecular diversity are emerging.
2. Endosymbiont protections against predation, parasitoids, and entomopathogens
Endosymbiotic bacteria – residing within insect cells or within specialized host organs – are widespread in insects and many are known to provide essential nutrients to their hosts. Often a single insect contains multiple endosymbionts.6,29 As a result of their extreme adaptation to this niche, these bacteria tend to have dramatically reduced genome sizes and cannot be cultured outside their insect hosts.30 In addition to nutritional roles, a growing number are now known to provide chemical defenses to their hosts. While molecular mechanisms of these defenses remain poorly understood, small molecule and protein toxins have been implicated in some cases.As noted in the introduction, certain rove beetles in the genus Paederus carry the polyketide toxin pederin (1, Fig. 1), which appears to protect them from predation by wolf spiders.31 While pederin has been known since the 1950s, when an astonishing 25 million beetles were harvested for elucidation of its structure,32 its biosynthetic origins were elusive until recently. The toxin is only observed in beetles when a bacterial endosymbiont is present: a Gammaproteobacterium in the genus Pseudomonas.33,34 Partial sequencing of this uncultured endosymbiont's genome revealed a cluster of genes consistent with pederin biosynthesis, the first compelling evidence of an endosymbiont producing a toxin beneficial to its host.35,36
A distantly related insect, the Asian citrus psyllid Diaphorina citri, contains a polyketide toxin named diaphorin (2) that shares high structural homology with pederin and is also produced by an endosymbiont.37 The endosymbiont producer in this case is a Betaproteobacterium, “Candidatus Profftella armatura,” and the biosynthetic genes for diaphorin account for an impressive 15% of its tiny 465 kb genome. The ecological role for the toxin has not been established, but it is toxic to mammalian cells. Interestingly, the genome of the host insect suggests a simplified innate immune system (lacking common insect antimicrobial peptides); small molecule defenses from this endosymbiont may compensate.38
Endosymbionts can also provide defenses against insect parasites. The fruit fly Drosophila neotestacea is susceptible to infection by the parasitic nematode Howardula aoronymphium, which leaves female flies sterile. These infections, and the ensuing sterility, are attenuated when the flies harbor endosymbiotic bacteria in the genus Spiroplasma.39,40 The protection offered by Spiroplasma was traced to a secreted ribosome inactivating peptide (RIP) that shares homology with Shiga-like toxins from E. coli. This 403 amino acid protein depurinates and disables Howardula rRNA, but importantly does not alter the Drosophila host's rRNA. Similar RIPs are found in the genomes of other Spiroplasma endosymbionts and could serve similar defensive roles.41,42
Aphids also belong to this diverse roster of insects that host defensive endosymbionts. Individuals of the pea aphid Acyrthosiphon pisum, a common pest of legumes, can host multiple representatives from a roster of at least 7 distinct endosymbionts, several of which appear to have defensive roles.11 The best-documented defense from these endosymbionts is protection from the specialized parasitoid wasp Aphidius ervi, a major threat to these aphids in North America. These wasps attack the aphid and lay an egg within its abdomen. The wasp larva develops within the living aphid and eventually kills it.43 Aphids that host the endosymbiont Hamiltonella defensa are protected even though they are attacked by wasps as frequently as unprotected aphids. Wasp eggs in protected aphids are less likely to develop into larvae and aphid survival is improved.44–46 Further examination of this system revealed that bacteriophage harbored by the Hamiltonella symbiont are actually responsible for suppressing wasp development.47 Several phage variants are known, each of which encodes homologs of known eukaryotic protein toxins such as tyrosine–aspartic acid repeat-containing protein (YD-repeat), cytolethal distending toxin (cdtB), and Shiga-like toxin (stx) (though genetic experiments confirming the role of these toxins are currently lacking).48,49 Endosymbionts may function broadly in the defense of these aphids: several other aphid endosymbionts provide protection from fungal pathogens, but the mechanism of this effect is not known.50–52
3. Gut bacteria defenses against parasites and microbial pathogens
As in other animals, insects typically host a consortium of gut microbes that participate in food digestion. In addition to the digestive consortium, the gut, which is topologically equivalent to an insect's exterior surface, is routinely exposed to a variety of environmental bacteria during feeding.7 Insect gut microbes have been implicated in suppression of unwanted microorganisms using secreted chemical defenses, with examples provided by mosquitoes, bees, and caterpillars.Mosquitoes are vectors of a wide variety of human diseases and there has been intense interest in understanding how their resident microbial communities might interact with disease-causing agents. The malaria-causing parasite Plasmodium falciparum carries out a portion of its lifecycle in the mosquito midgut, a site also populated by bacteria. A strain of Enterobacter isolated from the midgut of some wild mosquitoes is able to inhibit parasite development in the gut and effectively block Plasmodium infection of the mosquito.53 This inhibition is caused by reactive oxygen species from the bacteria and can be recapitulated in vitro. A variety of other bacterial isolates from mosquito guts have also been shown to inhibit Plasmodium, including Chromobacterium, Pantoea, Pseudomonas, Serratia, and Escherichia, but their inhibitory mechanisms are unknown.54–56
The gut bacteria of social bees also appear to play defensive roles. In the bumblebee Bombus terrestris, gut bacteria can protect against infection by the protozoan parasite Crithidia bombi.57 The mechanism of inhibition and potential molecular players are unknown, but complex feedback between the bee's immune system and its microbiota appears to be required for the protective effect.58 A honeybee gut isolate, Frischella perrara, produces an analog of the DNA damaging compound colibactin, a compound known from E. coli in the human gut that is implicated in colon cancer, whose detailed structure remains elusive.59–61E. coli isolates from the mosquito gut also appear to produce colibactin and to damage DNA, though the significance of this compound in insect guts is currently unknown.56
Competitive exclusion of pathogenic gut bacteria appears critical to healthy development of the cotton leaf worm caterpillar Spodoptera littoralis. Their gut bacterial community becomes dominated by Enterococci in older caterpillars.62 The species Enterococcus faecalis can cause disease symptoms and inhibit larval development, but the species Enterococcus mundtii can effectively compete with and suppress this pathogen, both in vivo and in vitro.63 Fractionation of E. mundtii cultures led to identification of the 43-amino acid bacteriocin mundticin KS as the active antibacterial agent, which has the ability to kill other Enterococci and shape the gut microbial community.
4. Insect brood protection by microbes
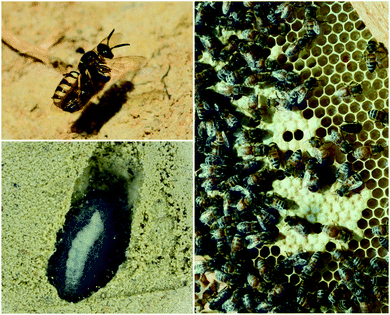
Insect eggs and larvae, which are immobile and nutrient rich, are susceptible to a variety of threats, especially microbial pathogens from the environment. Several insect species are known to benefit from defensive symbioses during these vulnerable but obligatory development periods, and many more undoubtedly adopt similar strategies.The solitary wasps known as beewolves deploy symbiotic Streptomyces bacteria to protect their developing pupae in underground brood chambers – welcoming environments for molds and other microbial pathogens. These wasps paralyze bees or other prey that they carry into underground burrows as food for their larvae (Fig. 2). Before laying their eggs, they apply bacteria to these underground brood chambers. The larvae eventually assimilate these bacteria and distribute them on their cocoons. The presence of these bacteria has been documented to improve larval survival by warding off fungi.64 The Streptomyces produce a suite of antimicrobial compounds that collectively provide broad spectrum antibacterial and antifungal activity including streptochlorin (3, Fig. 3) and a variety of piericidin analogs including piericidin A1 (4). Due to the complementary activities of these antibiotics, this may represent an example of “combination prophylaxis” in an ecological context.65 The bacteria appear to be highly specific to the wasps and are transmitted from one generation to the next by maternal inoculation of the brood cells. Taxonomic characterization of bacteria from three different genera of these wasps supports a vertical mode of transmission and suggests that this symbiosis dates back at least 68 million years.21
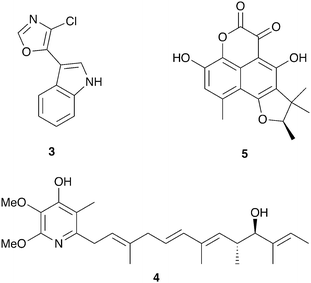 | ||
| Fig. 3 Molecules implicated in insect brood protection: streptochlorin (3), piericidin A1 (4), scleroderolide (5). | ||
Honeybees harbor a highly specific bacterial community in their guts and in the brood cells where their larvae develop (Fig. 2).66,67In vitro, members of this bacterial community, especially lactic acid bacteria, have been demonstrated to inhibit bee pathogens such as the bacteria Paenibacillus larvae (causes American foulbrood disease), Melissococcus plutonius (causes European foulbrood disease), and others.68–70 Lactic acid bacteria appear to have a protective effect against European foulbrood disease in developing larva.71 Some of these bacterial isolates produce bacteriocins, which may be responsible for this inhibitory activity.72,73
A final example of microbial brood protection comes from the solitary leaf-rolling weevil Euops chinensis, which rolls up leaf pieces inside which they lay their eggs. They inoculate these leaf rolls with a specific fungus (Penicillium herquei),74 the presence of which promotes larval survival.75 This symbiotic fungus does not appear to provide a nutritional benefit to the larvae but rather has protective antifungal activity. Activity guided fractionation of cultures of this Penicillium revealed the antibiotic scleroderolide (5) as the active antimicrobial agent, which is active against a variety of fungi, both ascomycetes and basidiomycetes, isolated from leaf rolls.76
5. Defense against microbial pathogens in fungus-growing insects
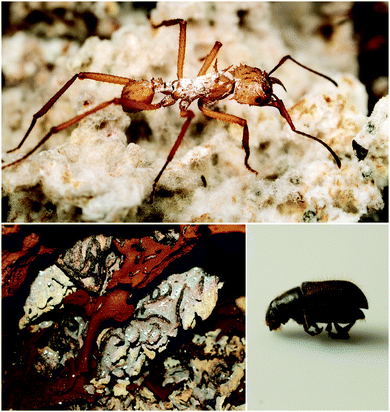
Fungus-growing insects are the original practitioners of agriculture in the animal kingdom. The major insect lineages of fungus-growers – the fungus-growing ants, termites, and beetles (Fig. 4) – share common nutritional strategies, even though the evolution of fungal agriculture occurred independently in each case.77–79 While defensive symbioses have many potential roles in these systems, protection against these ascomycete pathogens has emerged as a general trend, and a fruitful area for discovery of novel antifungal compounds. Antibacterials have been discovered as well, and they could play additional defensive roles in these systems.5.1. Fungus-growing ants
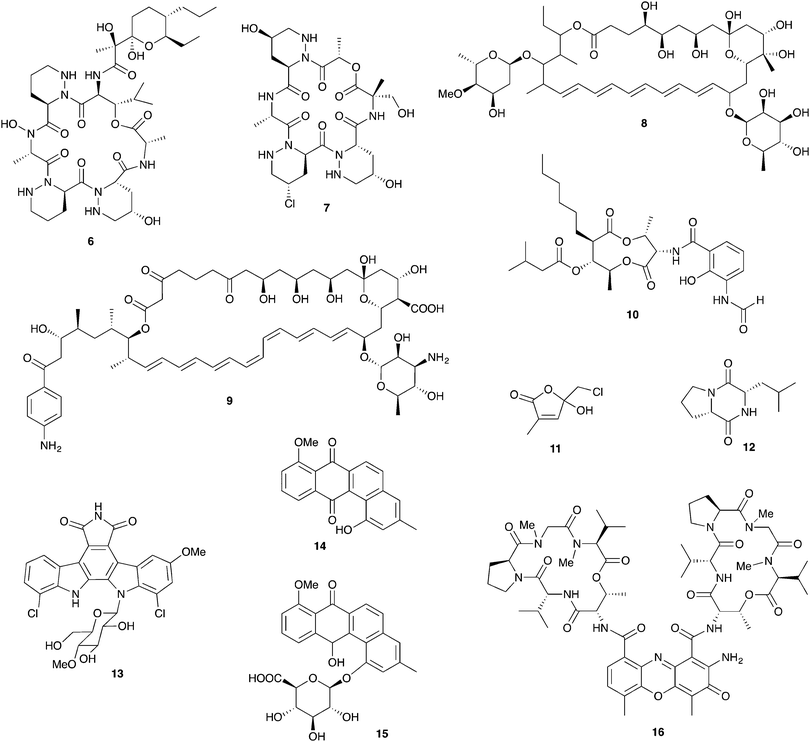
Fungus-growing ants, a large group that includes the charismatic leafcutters, gather plant matter and feed it to their basidiomycete cultivar fungus that is housed in underground nest chambers. These ants participate in a complex, multipartite symbiosis involving this obligate cultivar fungus in addition to an array of other microbial players.80 The best studied bacterial symbionts are the antibiotic-producing Actinobacteria in the genus Pseudonocardia. These symbiotic bacteria, whose association with this family of ants is thought to date back tens of millions of years, live off of secretions from specialized anatomical features on the ant exoskeleton.16,81 Antibiotics produced by these bacteria have now been characterized for a variety of different host ant species from different sites in Central and South America. These studies reveal a diverse suite of molecules including antifungals and antibacterials that can defend against microbial competitors.Ascomycete fungal pathogens represent a major threat to these ants and their fungal gardens, especially the specialized pathogen Escovopsis, which attacks and feeds directly on the cultivar fungus and can wipe out a colony.82–84 The presence of Pseudonocardia on the ants improves their efficacy at suppressing Escovopsis infections in their fungal gardens.85 Further, ant-derived Pseudonocardia show antifungal activity against Escovopsis in in vitro plate-based assays.86,87 The first insight into the molecular basis for this anti-Escovopsis activity was the discovery of dentigerumycin A (6, Fig. 5), a novel antifungal depsipeptide with a mixed polyketide/nonribosomal peptide architecture.88 This molecule, produced by Pseudonocardia isolated off the basal fungus-growing ant species Apterostigma dentigerum in Panama, shows selective antifungal activity. It is active against Escovopsis as well as the human fungal pathogen Candida albicans, but not the basidiomycete fungal cultivar of this ant.88,89 A different antifungal compound was isolated from two strains of Pseudonocardia isolated from neighboring A. dentigerum nests in Costa Rica. These strains produce the antifungal agent selvamicin (8), a new member of the well-known polyene macrolide family of polyketide-derived antifungals, which includes indispensable drugs such as amphotericin. Selvamicin bears several unusual structural features, including a second glycosylation and a lack of charged groups. Selvamicin appears to have a distinct mechanism of action from other polyene antifungals and is currently under evaluation for clinical use.90 Another novel polyene natural product, with presumed antifungal activity, is produced by a Pseudonocardia strain isolated off of the ant Acromyrmex octospinosus. While the complete structure of this molecule has not yet been elucidated, it appears to be a close analog of the antifungal polyene nystatin.91 In addition to protection from Escovopsis, antifungals produced by bacterial symbionts could play other defensive roles. In one study, removal of Actinobacteria from the cuticle of Acromyrmex ants increased the susceptibility of young ants to the entomopathogen Metarhizium anisopliae.92
Antifungal production has also been documented for other genera of Actinobacteria, such as Streptomyces, recovered from fungus-growing ant exoskeletons and fungal gardens.93 The role of these bacteria in this symbiosis has not been well established, and they may represent transient microbes from the environment rather than true symbionts. Several Streptomyces isolates from Acromyrmex ants or their colonies produce yet another polyene antifungal: candicidin (9).91,94 Other Streptomyces isolates from Acromyrmex have been shown to produce antimycins including antimycin A1 (10), toxic mitochondrial electron transport chain inhibitors with antifungal activity.95,96
In addition to Actinobacteria housed on the cuticle of the ants, defenses derived from the fungal garden itself have been documented in several fungus-growing ants. For example, the cultivar fungus of the ant Cyphomyrmex costatus produces the antibacterial lepiochlorin (11),97 while a different cultivar fungus from the ant Cyphomyrmex minutus produces a variety of antifungal diketopiperazines including 12. These diketopiperazines have activity against ascomycete yeasts such as Candida and may play a role in suppression of unwanted pathogenic fungi in the garden.98 A variety of other microbes isolated from ant fungal gardens also have demonstrated antifungal activity, though ecological roles and underlying chemistry are currently unknown. These include both ascomycete and basidiomycete yeasts from the nests of the leafcutter Atta texana as well as Proteobacteria in the genus Burkholderia from Atta sexdens nests.99,100
In addition to antimicrobial activity that suppresses host pathogens, defensive symbionts associated with fungus-growing ants (or indeed any other host) may produce antimicrobials that serve as niche defense against their own microbial competitors. One example are Pseudonocardia living on the ant exoskeleton; these bacteria are expected to face niche competition from other closely related strains of ant-associated Pseudonocardia. Agar plate-based competition assays show that closely related strains of ant-associated Pseudonocardia frequently have antagonistic activity against one another.101 In one clear demonstration of the chemical basis for this antagonism, a novel analog (13) of the bis-indole anticancer agent rebeccamycin is produced by one strain of Pseudonocardia from the ant A. dentigerum in Panama. This strain has nanomolar antagonistic activity against other ant associated Pseudonocardia from the same region.102
Other antibacterials from ant-associated Actinobacteria include 14 and other analogs of the angucycline antibiotic rabelomycin, including the glycosylated pseudonocardone 15, from another Apterostigma dentigerum-derived Pseudonocardia strain from Panama.103Pseudonocardia isolated from Acromyrmex ants in Panama show inhibitory activity against both Gram-negative and Gram-positive bacteria, though the molecules responsible for this inhibition are currently unknown.104 Some Streptomyces bacteria isolated off of Acromyrmex ants produce a series of antibacterial actinomycin compounds such as actinomycin D (16), which interestingly have activity against both Streptomyces and Pseudonocardia.95
5.2. Fungus-growing termites
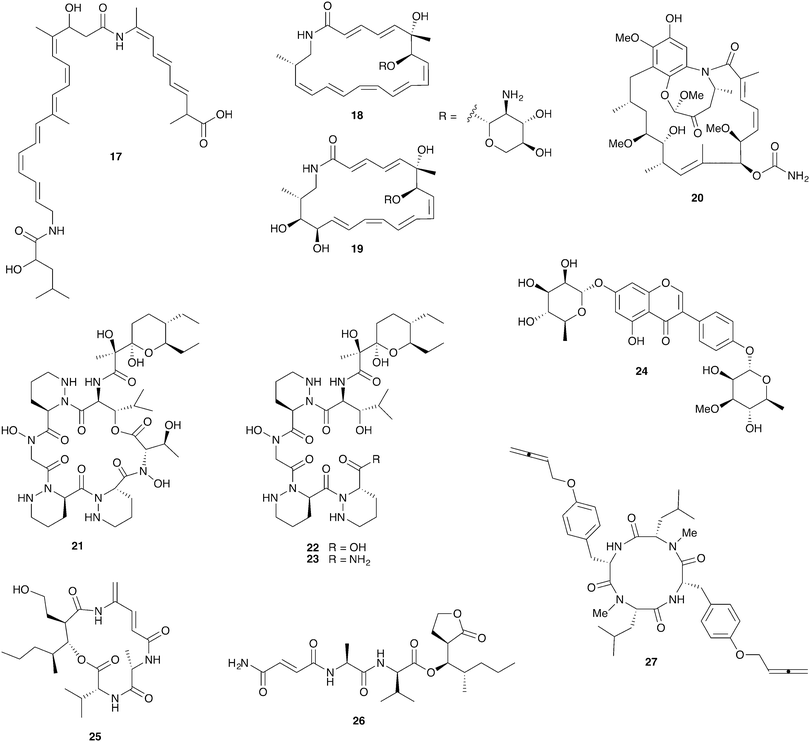
Fungus-growing termites are found throughout Africa and Asia and have been engaged in a symbiotic relationship with their basidiomycete fungal cultivar Termitomyces for approximately 30 million years.105 Ascomycete fungi such as Pseudoxylaria and Trichoderma are common in these termite nests where they are thought to be competitors with the basidiomycete cultivar; they can rapidly overgrow unoccupied termite nests.106,107 The fungal comb structures within these nests host rich bacterial communities that are at least partially derived from the termite gut.108–111Strains of the bacterial genus Bacillus with antifungal activity have been isolated from termites and from fungal comb material from both the Asian fungus-growing termite Odontotermes formosanus and the African fungus-growing termite Macrotermes natalensis.107,112 Intriguingly, the Bacillus isolates from both termite species selectively inhibit the ascomycete putative pathogens Pseudoxylaria and Trichoderma but not the basidiomycete cultivar fungus. This activity was traced to the antifungal polyketide bacillaene A (17, Fig. 6) in the M. natalensis-derived Bacillus strains.112
Actinobacteria are routinely isolated from fungus-growing termite nests and while their ecological role is unclear they have been a highly productive source for chemical discovery. These bacteria do not appear to be termite-specific and while they frequently display antifungal activity, a large-scale sampling of these bacteria revealed cultivar inhibition to be more frequent than Pseudoxylaria inhibition.108 However, one recently reported isolate, the Amycolatopsis strain M39 from a M. natalensis colony, does selectively inhibit Pseudoxylaria but not Termitomyces. This activity was traced to the novel glycosylated polyketide macrolactam antifungals macrotermycins A (18) and C (19).113
Streptomyces isolates from colonies of the South African termite M. natalensis have yielded a number of interesting novel compounds. Close investigation of one Streptomyces strain with especially strong antifungal activity led to the discovery of natalamycin A (20), a highly unusual analog of the ansamycin polyketide antifungal geldanamycin (although natalamycin A lacks apparent antimicrobial activity).114 Another Streptomyces isolate unexpectedly produces a novel cyclic analog 21 of dentigerumycin, the antifungal from the fungus-growing ant system, and several linear analogs (22, 23) that may represent premature termination of this molecule's biosynthesis.115 Finally, a series of glycosylated isoflavonoid compounds, including termisoflavone A (24), were recovered from another Streptomyces isolate.116
A screen of 30 Streptomyces strains from a different South African fungus-growing termite, Microtermes sp., led to the identification of the novel mixed polyketide/nonribosomal peptides microtermolide A (25) and microtermolide B (26). Neither of these compounds have antibacterial or antifungal activity.117 Interestingly, an isolate of the fungal pathogen Pseudoxylaria from a South African Microtermes sp. colony strongly antagonizes the Termitomyces cultivar in in vitro assays. While the compounds responsible for this antifungal activity have not yet been identified, this fungus also produces a series of novel antibacterial peptides with highly unusual allene groups, including pseudoxyallemycin B (27).118
5.3. Pine beetle-fungus symbioses
Bark beetles in the genus Dendroctonus (meaning “tree killer”) have gained notoriety in recent years for the decimation of conifer forests.119 Certain species within this genus engage in obligate nutritional symbioses with fungi transmitted by specialized anatomical structures on their exoskeleton called mycangia. These fungi grow in galleries bored under the tree bark, where beetle larvae develop.79,120 One such species is the Southern pine beetle, Dendroctonus frontalis, which cultivates the basidiomycete fungus Entomocorticium within its galleries. The ascomycete fungus Ophiostoma minus is an antagonist in this system; it can outcompete the cultivar fungus, negatively impacting larval survival.121,122 Two strains of Streptomyces bacteria, one white and one red, were repeatedly isolated from D. frontalis beetles sampled from a site in Mississippi. The red Streptomyces inhibits the antagonistic Ophiostoma fungus, but not the mutualistic fungus Entomocorticium, suggesting a defensive role that supports the beetle-fungus mutualism. The molecule responsible for this selective antifungal activity is mycangimycin (28, Fig. 7), a novel polyene polyketide with a highly unusual peroxide group.123,124 While the white Streptomyces strain showed no antifungal activity in initial in vitro bioassays, laboratory cultivation in different rich growth media revealed that this strain can also produce antifungal agents with activity against Ophiostoma, the novel polycyclic tetramate macrolactams frontalamides A (29) and B (30).125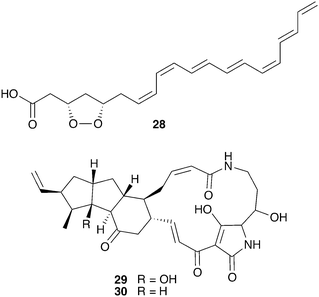 | ||
| Fig. 7 Molecules from microbes associated with Dendroctonus pine beetles: mycangimycin (28), frontalamide A (29), frontalamide B (30). | ||
While the above data suggest a protective role for Streptomyces among one beetle population, it is currently unclear whether Streptomyces play a general role as defensive symbionts of Dendroctonus beetles. A survey of various North American Dendroctonus species repeatedly yielded Streptomyces isolates, but revealed no pattern of a specific association.126 Other bacterial isolates from Dendroctonus beetles have shown antifungal activity against putative pathogens such as Trichoderma and Aspergillus, but the specificity of these associations and their chemical bases are currently unknown.127,128
6. Chemistry from other insect-associated bacteria
The productivity of insect-associated microbes as a source of novel bioactive chemistry has inspired efforts to discover new defensive symbioses and to screen insect-associated microbes for novel molecules. The ecological role for the microbes isolated is not firmly established, in many cases, but even absent this proof these studies support the notion that insect-associated microbes are an untapped source of novel biologically active molecules. A particular emphasis has been placed on actinomycetes associated with insects and three examples of these efforts are summarized below: mud dauber wasps, dung beetles, and Allomerus ants.Solitary mud dauber wasps create mud nests with chambers containing insect prey on which they lay their eggs. Two of these wasp species native to Wisconsin, Sceliphron caementarium and Chalybion californicum, were sampled for associated Actinobacteria, yielding a large and diverse collection of Streptomyces, of which 15 were analyzed in depth for antimicrobial chemistry. Many of these strains showed robust antibacterial and antifungal activity, attributable to a variety of previously known molecules, including the polyene antifungal mycangimycin (28), which had been seen in Streptomyces from Dendroctonus pine beetles.129 Two Streptomyces strains from this collection isolated from the wasp S. caementarium produced a novel antifungal compound, an unusual polyene macrolactam named sceliphrolactam (31, Fig. 8), which has antifungal activity against amphotericin-resistant C. albicans.130
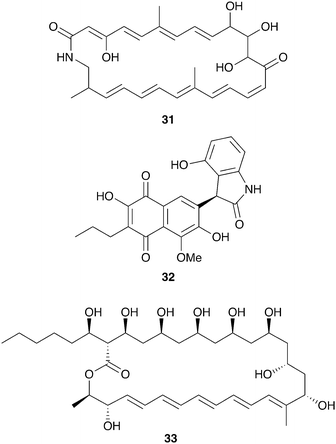 | ||
| Fig. 8 Molecules described from other insect-associated bacteria: sceliphrolactam (31), coprisidin A (32), filipin III (33). | ||
Dung beetles are a large group of insects with global distribution that collect fecal matter, typically from herbivores, as a source of nutrition. The dung beetle Copris tripartitus, native to Korea, forms brood balls out of feces in which it lays its eggs and which serve as food for larval development.131Streptomyces isolated from these beetles, their larva, and their dung balls have recently served as a prolific source of novel bioactive compounds.132–136 An ecological role for these bacteria has not been established, although in other dung beetles maternal transmission of bacteria to the brood ball has been documented, and could play a role in suppression of pathogenic microbes.137,138 Among the Streptomyces isolates from Copris tripartitus, one strain was repeatedly isolated from the gut of adult beetles over the course of three years. This strain produces the antifungal macrolactam sceliphrolactam (31), and also produces novel naphthoquinone-oxindole alkaloids including coprisidin A (32).136
Another group of insects recently examined for defensive chemistry from bacterial symbionts are the Allomerus ants of South America. These ants, only recently described, practice ‘ambush hunting’ from fungal galleries built on plant stems.139 These fungal galleries are built from propagated ascomycete fungi in the order Chaetothyriales, and serve as hiding places from which the ants wait for insect prey. It has been hypothesized that selective antifungals from bacterial symbionts could help maintain the specific fungal composition of these galleries.140 While culture-independent methods indicate that Proteobacteria dominate the microbial community on the cuticle of Allomerus, Actinobacteria with antifungal activity have been isolated off of two species of these ants.141 This activity was observed against two non-symbiotic fungi isolated from the galleries, although the beneficial fungus itself was not evaluated.142 Using activity-guided fractionation, the antifungal activity from one of these strains, a Streptomyces isolate, was traced to the antifungal polyene filipin III (33).143
7. Insights from bacterial genomes: molecular diversity and evolution
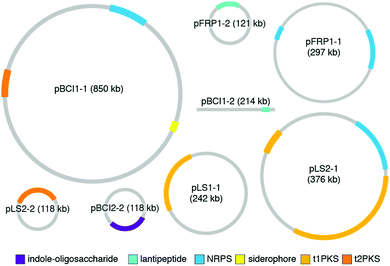
While the studies reviewed in the earlier sections provide a wonderful collection of molecules and functions for defensive symbioses, the rapidly growing catalog of microbial genomes provides an alternative and much larger window into symbiont adaptation and the evolutionary histories of host-microbe relationships.29,144–146 A number of genomes for defensive microbial symbionts of insects, in which the chemistry of defense has been established, are now available, and these genomes provide an authoritative description of the biosynthesis of specialized metabolites. In bacteria, genes for a given molecule's biosynthetic enzymes, transport, resistance, and regulation are typically adjacent to one another. The genomic context of these biosynthetic gene clusters (BGCs), as well as their gene content, can inform the ways in which these molecules came to be adapted for defensive symbiosis. The major strategy for molecular diversification and dissemination is horizontal gene transfer, but the tactics used to implement this strategy vary.In a surprising number of recently studied insect defensive symbiont genomes, plasmids and other mobile genetic elements host the defense molecule's BGC. Especially in the case of fungus-growing ant Pseudonocardia symbionts, these plasmids drive population-level diversity of defensive chemistry among otherwise identical bacterial isolates (Fig. 9). This scenario of variable plasmids encoding defensive molecules means that conventional measures for assessing bacterial phylogeny, such as 16S rRNA gene sequencing, do not capture the relevant differences in BGCs that underlie functionally important defenses. In one example, the antibacterial molecule 9-methoxyrebeccamycin (13), produced by a Pseudonocardia symbiont of the ant Apterostigma dentigerum, is encoded on a 119 kb circular plasmid. The Pseudonocardia symbiont isolated from a nearby colony of the same ant species, on the same island in Panama, does not produce 9-methoxyrebeccamycin, and lacks this plasmid encoding its biosynthesis. With the exception of plasmid content, these two Pseudonocardia are virtually indistinguishable at a whole genome level.102 Other plasmid-encoded molecules produced by Pseudonocardia from the fungus-growing ant system include the antifungal polyene selvamicin (8) and the depsipeptide gerumycin C (7).89 In the case of selvamicin, the molecule's BGC is plasmid-encoded in the Pseudonocardia from one ant colony, yet is found on the chromosome of the Pseudonocardia from a neighboring ant colony.90 In all of these cases, the Pseudonocardia genomes are rich in mobile genetic element proteins, including putative transposases and integrases, and these mobile element proteins often lie at the boundaries of the BGCs.89,90,102,147 An analysis of 10 Pseudonocardia genomes from Acromyrmex colonies collected in Panama confirm the existence of two major “phylotypes” of Pseudonocardia distinguishable at the level of 16S rRNA sequence. This analysis shows a core of BGCs conserved across all strains, a subset of BGCs conserved within each phylotype, as well as five BGCs unique to individual Pseudonocardia genomes. While plasmids were not resolved in these sequences, this BGC variability underscores the mobility of gene clusters in this symbiotic niche.104
In another example of plasmid-encoded defense, the antimicrobial bacteriocin mundticin from an enterococcus gut symbiont of the caterpillar Spodoptera littoralis is encoded on a plasmid, and curing this strain of that plasmid results in a loss of antibacterial activity.63
Another situation in which functionally relevant molecular defenses are encoded by variable mobile genetic elements are the protein toxins defending against parasitoid wasps in Hamiltonella bacterial symbionts of aphids. This suite of protein toxins is phage-encoded, and variations in phage infection are reflected in varying defense against parasitoids.47–49 Once again, defensive symbionts need to be considered not just at the level of bacterial strain identity, but also at the level of mobile genetic elements harbored. Mobile element-encoded defenses certainly provide an evolutionary shortcut for responding to changing threats from the environment.
Available genomes of insect defensive symbionts provide clues for how horizontal gene transfer has allowed recruitment of useful molecules to symbiotic niches. For example, the antifungal polyene selvamicin (8) is produced by two different strains of ant-associated Pseudonocardia from nearby colonies. These two strains are notably different from one another: the average nucleotide identity across their chromosomes is only 83%. The nucleotide identity between the two selvamicin BGCs, however, is greater than 98%, suggesting that one or both strains have recently acquired this cluster by horizontal gene transfer.90
Pederin (1) and diaphorin (2), two structurally related polyketide toxins produced by endosymbionts of insects, also appear to have arrived at their current symbiotic contexts by horizontal gene transfer. The entire genome of the pederin-producing Pseudomonas endosymbiont has not been sequenced, but the available sequence for a large portion including the polyketide BGC for pederin reveals that this BGC resides on a ‘symbiosis island,’ a region with low homology to closely related free-living Pseudomonas bacteria. This genomic island is bounded by degraded mobile genetic element proteins: hallmarks of acquisition by horizontal gene transfer.36 The diaphorin BGC is highly similar to the cluster for pederin, yet it resides in a very different bacterial endosymbiont, a Betaproteobacterium in the genus Profftella, within a hemipteran bug of a completely different insect order than the Paederus beetles that produce pederin. It is unclear whether horizontal gene transfer of the diaphorin cluster into Profftella predated this bacteria's association with its insect host.37 Interestingly, similar polyketide toxins in the same structural class are produced by bacterial symbionts of marine sponges and by cyanobacteria from lichens, but have yet to be identified in any free-living.24,148 These molecules appear to have special utility in defensive symbioses as indicated by their spread to multiple symbiotic associations by horizontal gene transfer.
Further genome sequencing of the bacteria participating in defensive symbioses with insects will surely contribute not only to our understanding of these symbioses and the role of specialized metabolites but also more general features of bacterial secondary metabolism. A ubiquitous feature of bacterial genomes is ‘orphan’ BGCs that encode ‘cryptic’ metabolites – gene clusters that appear to presumably encode the instructions for making metabolites that have never been isolated. Insect symbiont genomes are no exception; ant Pseudonocardia genomes contain 10 or more putative BGCs predicted by the cluster-detection algorithm antiSMASH, yet only a handful of molecules have been characterized from these strains.89,90,102,104 Access to these cryptic molecules, and evaluation of their activity, could improve our understanding of the molecular basis for symbiosis. Some molecules may be coaxed into production by expanding bacterial culture conditions, especially exploring ecologically relevant co-cultures with other microorganisms or their metabolites. In addition, synthetic biology is creating a number of approaches to coax the expression of these molecules through heterologous expression or genome engineering such as pathway refactoring.149–151
8. Conclusion
The study of insect-microbe defensive symbioses became a robust research area only recently, but it has already provided a large number of examples of what is clearly a widespread strategy with highly variable tactics. Insect-microbe defensive symbioses are highly diverse in terms of the types of insects and bacteria represented, the threats guarded against, and the types of defensive molecules deployed by microbes range from reactive oxygen species to small molecules to proteins. Given the vast diversity of insects on the planet, most of which have never been described and many of those that have been described have never been studied in any depth, and the apparent ubiquity of microbial symbioses, we clearly have only begun to mine the chemical and biological lessons of these molecular defenses.Most discoveries to date on insect-microbe defensive symbioses have been spurred by careful studies on the natural history and ecology of insects: studies that identified the environmental threats and defensive symbionts. Chemical analysis has filled in the molecular underpinnings. However, the study of microbial chemistry from these systems has the potential to offer more general insights. For example, these studies repeatedly discover antifungal molecules suggesting that fungi constitute an even more substantial existential threat to insects than currently recognized.
These studies will also contribute in related areas like human health. Today new antibiotics are sorely needed and while conventional microbial natural product discovery efforts suffer from high rediscovery rates, underexplored niches such as the defensive microbes of insects hold considerable promise. The relatively short track record for discovery efforts in this area has been promising, with over a dozen novel and potentially useful compounds discussed in this review. Most intriguing, focusing discovery efforts on symbiotic microbes aligns ecology with the desired therapeutic activity as these molecules are already compatible with an animal host.
Conflict of interest
There are no conflicts of interest to declare.References
- E. M. Arndt, W. Moore, W.-K. Lee and C. Ortiz, Science, 2015, 348, 563–567 CrossRef CAS PubMed.
- T. Eisner and D. J. Aneshansley, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 9705–9709 CrossRef CAS.
- J. H. Frank and K. Kanamitsu, J. Med. Entomol., 1987, 24, 155–191 CrossRef CAS PubMed.
- R. Kellner, Entomol. Exp. Appl., 2003, 107, 115–124 CrossRef.
- A. E. Douglas, Annu. Rev. Entomol., 1998, 43, 17–37 CrossRef CAS PubMed.
- A. E. Douglas, Annu. Rev. Entomol., 2015, 60, 17–34 CrossRef CAS PubMed.
- P. Engel and N. A. Moran, FEMS Microbiol. Rev., 2013, 37, 699–735 CrossRef CAS PubMed.
- E. R. Haine, Proc. R. Soc. B, 2008, 275, 353–361 CrossRef PubMed.
- R. F. Seipke, M. Kaltenpoth and M. I. Hutchings, FEMS Microbiol. Rev., 2012, 36, 862–876 CrossRef CAS PubMed.
- M. Kaltenpoth and T. Engl, Funct. Ecol., 2013, 28, 315–327 CrossRef.
- K. M. Oliver, A. H. Smith and J. A. Russell, Funct. Ecol., 2013, 28, 341–355 CrossRef.
- K. Clay, Funct. Ecol., 2014, 28, 293–298 CrossRef.
- N. M. Gerardo and B. J. Parker, Curr. Opin. Insect. Sci., 2014, 4, 1–8 CrossRef PubMed.
- S. A. Ford and K. C. King, PLoS Pathog., 2016, 12, e1005465 Search PubMed.
- M. Kaltenpoth, K. Roeser-Mueller, S. Koehler, A. Peterson, T. Y. Nechitaylo, J. W. Stubblefield, G. Herzner, J. Seger and E. Strohm, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6359–6364 CrossRef CAS PubMed.
- C. R. Currie, M. Poulsen, J. Mendenhall, J. J. Boomsma and J. Billen, Science, 2006, 311, 81–83 CrossRef CAS PubMed.
- M. Bright and S. Bulgheresi, Nat. Rev. Drug Discovery, 2010, 8, 218–230 CAS.
- H. Salem, L. Florez, N. Gerardo and M. Kaltenpoth, Proc. R. Soc. B, 2015, 282, 20142957 CrossRef PubMed.
- J. L. Sachs, U. G. Mueller, T. P. Wilcox and J. J. Bull, Q. Rev. Biol., 2004, 79, 135–160 CrossRef PubMed.
- S. E. Marsh, M. Poulsen, A. Pinto-Tomás and C. R. Currie, PLoS One, 2014, 9, e103269 Search PubMed.
- M. Kaltenpoth, K. Roeser-Mueller, S. Koehler, A. Peterson, T. Y. Nechitaylo, J. W. Stubblefield, G. Herzner, J. Seger and E. Strohm, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6359–6364 CrossRef CAS PubMed.
- N. M. Gerardo and B. J. Parker, Curr. Opin. Insect. Sci., 2014, 4, 8–14 CrossRef PubMed.
- I. Eleftherianos, J. Atri, J. Accetta and J. C. Castillo, Front. Physiol., 2013, 4, 46 Search PubMed.
- J. Piel, Nat. Prod. Rep., 2009, 26, 338–362 RSC.
- A. T. Dossey, Nat. Prod. Rep., 2010, 27, 1737–1757 RSC.
- T. R. Ramadhar, C. Beemelmanns, C. R. Currie and J. Clardy, J. Antibiot., 2013, 67, 53–58 CrossRef PubMed.
- L. V. Flórez, P. H. W. Biedermann, T. Engl and M. Kaltenpoth, Nat. Prod. Rep., 2015, 32, 904–936 RSC.
- C. Beemelmanns, H. Guo, M. Rischer and M. Poulsen, Beilstein J. Org. Chem., 2016, 12, 314–327 CrossRef CAS PubMed.
- N. A. Moran, J. P. McCutcheon and A. Nakabachi, Annu. Rev. Genet., 2008, 42, 165–190 CrossRef CAS PubMed.
- Y. Kikuchi, Microbes Environ., 2009, 24, 195–204 CrossRef PubMed.
- R. L. L. Kellner and K. Dettner, Oecologia, 1996, 107, 293–300 CrossRef PubMed.
- M. Pavan and G. Bo, Physiol. Comp. Oecol., 1953, 3, 307–312 CAS.
- R. L. L. Kellner, Insect Biochem. Mol. Biol., 2002, 32, 389–395 CrossRef CAS PubMed.
- M. Kador, M. A. Horn and K. Dettner, FEMS Microbiol. Lett., 2011, 319, 73–81 CrossRef CAS PubMed.
- J. Piel, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 14002–14007 CrossRef CAS PubMed.
- J. Piel, I. Hofer and D. Hui, J. Bacteriol., 2004, 186, 1280–1286 CrossRef CAS PubMed.
- A. Nakabachi, R. Ueoka, K. Oshima, R. Teta, A. Mangoni, M. Gurgui, N. J. Oldham, G. van Echten-Deckert, K. Okamura, K. Yamamoto, H. Inoue, M. Ohkuma, Y. Hongoh, S.-Y. Miyagishima, M. Hattori, J. Piel and T. Fukatsu, Curr. Biol., 2013, 23, 1478–1484 CrossRef CAS PubMed.
- A. P. Arp, W. B. Hunter and K. S. Pelz-Stelinski, Front. Physiol., 2016, 7, 417–418 Search PubMed.
- J. Jaenike, R. Unckless, S. N. Cockburn, L. M. Boelio and S. J. Perlman, Science, 2010, 329, 212–215 CrossRef CAS PubMed.
- P. T. Hamilton and S. J. Perlman, PLoS Pathog., 2013, 9, e1003808 Search PubMed.
- P. T. Hamilton, F. Peng, M. J. Boulanger and S. J. Perlman, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 350–355 CrossRef CAS PubMed.
- P. T. Hamilton, J. S. Leong, B. F. Koop and S. J. Perlman, Mol. Ecol., 2014, 23, 1558–1570 CrossRef CAS PubMed.
- R. Sequeira and M. Mackauer, Ecology, 1992, 73, 183–189 CrossRef.
- K. M. Oliver, J. A. Russell, N. A. Moran and M. S. Hunter, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 1803–1807 CrossRef CAS PubMed.
- K. M. Oliver, N. A. Moran and M. S. Hunter, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 12795–12800 CrossRef CAS PubMed.
- L. Cayetano and C. Vorburger, Ecol. Entomol., 2015, 40, 85–93 CrossRef.
- K. M. Oliver, P. H. Degnan, M. S. Hunter and N. A. Moran, Science, 2009, 325, 992–994 CrossRef CAS PubMed.
- P. H. Degnan and N. A. Moran, Appl. Environ. Microbiol., 2008, 74, 6782–6791 CrossRef CAS PubMed.
- S. R. Weldon and K. M. Oliver, The Mechanistic Benefits of Microbial Symbionts, 2016 Search PubMed.
- C. L. Scarborough, Science, 2005, 310, 1781 CrossRef CAS PubMed.
- P. Łukasik, M. van Asch, H. Guo, J. Ferrari, H. Charles and J. Godfray, Ecol. Lett., 2012, 16, 214–218 CrossRef PubMed.
- B. J. Parker, C. J. Spragg, B. Altincicek and N. M. Gerardo, Appl. Environ. Microbiol., 2013, 79, 2455–2458 CrossRef CAS PubMed.
- C. M. Cirimotich, Y. Dong, A. M. Clayton, S. L. Sandiford, J. A. Souza-Neto, M. Mulenga and G. Dimopoulos, Science, 2011, 332, 855–858 CrossRef CAS PubMed.
- J. L. Ramirez, S. M. Short, A. C. Bahia, R. G. Saraiva, Y. Dong, S. Kang, A. Tripathi, G. Mlambo and G. Dimopoulos, PLoS Pathog., 2014, 10, e1004398 Search PubMed.
- A. C. Bahia, Y. Dong, B. J. Blumberg, G. Mlambo, A. Tripathi, O. J. BenMarzouk-Hidalgo, R. Chandra and G. Dimopoulos, Environ. Microbiol., 2014, 16, 2980–2994 CrossRef CAS PubMed.
- M. T. Tchioffo, L. Abate, A. Boissière, S. E. Nsango, G. Gimonneau, A. Berry, E. Oswald, D. Dubois and I. Morlais, Infect., Genet. Evol., 2016, 43, 22–30 CrossRef PubMed.
- H. Koch and P. Schmid-Hempel, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 19288–19292 CrossRef CAS PubMed.
- K. Näpflin and P. Schmid-Hempel, Proc. R. Soc. B, 2016, 283, 20160312 CrossRef PubMed.
- E. P. Balskus, Nat. Prod. Rep., 2015, 32, 1534–1540 RSC.
- G. Cuevas-Ramos, C. R. Petit, I. Marcq, M. Boury, E. Oswald and J.-P. Nougayrède, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 11537–11542 CrossRef CAS PubMed.
- P. Engel, M. I. Vizcaino and J. M. Crawford, Appl. Environ. Microbiol., 2015, 81, 1502–1512 CrossRef PubMed.
- B. Chen, B.-S. Teh, C. Sun, S. Hu, X. Lu, W. Boland and Y. Shao, Sci. Rep., 2016, 6, 1–14 CrossRef PubMed.
- Y. Shao, B. Chen, C. Sun, K. Ishida, C. Hertweck and W. Boland, Cell Chem. Biol., 2017, 24, 66–75 CrossRef CAS PubMed.
- M. Kaltenpoth, W. Göttler, G. Herzner and E. Strohm, Curr. Biol., 2005, 15, 475–479 CrossRef CAS PubMed.
- J. Kroiss, M. Kaltenpoth, B. Schneider, M.-G. Schwinger, C. Hertweck, R. K. Maddula, E. Strohm and A. Svatos, Nat. Chem. Biol., 2010, 6, 261–263 CrossRef CAS PubMed.
- W. K. Kwong and N. A. Moran, Nat. Rev. Microbiol., 2016, 14, 374–384 CrossRef CAS PubMed.
- P. Engel, W. K. Kwong, Q. McFrederick, K. E. Anderson, S. M. Barribeau, J. A. Chandler, R. S. Cornman, J. Dainat, J. R. de Miranda, V. Doublet, O. Emery, J. D. Evans, L. Farinelli, M. L. Flenniken, F. Granberg, J. A. Grasis, L. Gauthier, J. Hayer, H. Koch, S. Kocher, V. G. Martinson, N. Moran, M. Munoz-Torres, I. Newton, R. J. Paxton, E. Powell, B. M. Sadd, P. Schmid-Hempel, R. Schmid-Hempel, S. J. Song, R. S. Schwarz, D. vanEngelsdorp and B. Dainat, mBio, 2016, 7, e02164 CAS.
- J. D. Evans and T.-N. Armstrong, BMC Ecol., 2006, 6, 4 CrossRef PubMed.
- E. Forsgren, T. C. Olofsson, A. Vásquez and I. Fries, Apidologie, 2009, 41, 99–108 CrossRef.
- A. Endo and S. Salminen, Syst. Appl. Microbiol., 2013, 36, 444–448 CrossRef PubMed.
- A. Vásquez, E. Forsgren, I. Fries, R. J. Paxton, E. Flaberg, L. Szekely and T. C. Olofsson, PLoS One, 2012, 7, e33188 Search PubMed.
- M. C. Audisio, H. R. Terzolo and M. C. Apella, Appl. Environ. Microbiol., 2005, 71, 3373–3375 CrossRef CAS PubMed.
- I. Butler, M. Alsterfjord, T. C. Olofsson, C. Karlsson, J. Malmström and A. Vásquez, BMC Microbiol., 2013, 13, 1 CrossRef PubMed.
- K. Sakurai, J. Ethol., 1985, 3, 151–156 CrossRef.
- X. Li, W. Guo and J. Ding, J. Insect Physiol., 2012, 58, 867–873 CrossRef CAS PubMed.
- L. Wang, Y. Feng, J. Tian, M. Xiang, J. Sun, J. Ding, W.-B. Yin, M. Stadler, Y. Che and X. Liu, ISME J., 2015, 9, 1793–1801 CrossRef PubMed.
- T. R. Schultz and S. G. Brady, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 5435–5440 CrossRef CAS PubMed.
- D. K. Aanen, P. Eggleton, C. Rouland-Lefevre, T. Guldberg-Froslev, S. Rosendahl and J. J. Boomsma, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 14887–14892 CrossRef CAS PubMed.
- D. L. Six, J. Chem. Ecol., 2013, 39, 989–1002 CrossRef CAS PubMed.
- C. R. Currie, Annu. Rev. Microbiol., 2001, 55, 357–380 CrossRef CAS PubMed.
- M. J. Cafaro, M. Poulsen, A. E. F. Little, S. L. Price, N. M. Gerardo, B. Wong, A. E. Stuart, B. Larget, P. Abbot and C. R. Currie, Proc. R. Soc. B, 2011, 278, 1814–1822 CrossRef PubMed.
- C. R. Currie, U. G. Mueller and D. Malloch, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 7998–8002 CrossRef CAS.
- S. A. Steffan, Y. Chikaraishi, C. R. Currie, H. Horn, H. R. Gaines-Day, J. N. Pauli, J. E. Zalapa and N. Ohkouchi, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 15119–15124 CrossRef CAS PubMed.
- H. T. Reynolds and C. R. Currie, Mycologia, 2004, 96, 955–959 CrossRef PubMed.
- C. R. Currie, A. Bot and J. J. Boomsma, Oikos, 2003, 101, 91–102 CrossRef.
- C. R. Currie, J. A. Scott, R. C. Summerbell and D. Malloch, Nature, 1999, 398, 701–704 CrossRef CAS.
- M. Poulsen, M. J. Cafaro, D. P. Erhardt, A. E. F. Little, N. M. Gerardo, B. Tebbets, B. S. Klein and C. R. Currie, Environ. Microbiol. Rep., 2009, 2, 534–540 CrossRef PubMed.
- D.-C. Oh, M. Poulsen, C. R. Currie and J. Clardy, Nat. Chem. Biol., 2009, 5, 391–393 CrossRef CAS PubMed.
- C. S. Sit, A. C. Ruzzini, E. B. Van Arnam, T. R. Ramadhar, C. R. Currie and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 13150–13154 CrossRef CAS PubMed.
- E. B. Van Arnam, A. C. Ruzzini, C. S. Sit, H. Horn, A. A. Pinto-Tomás, C. R. Currie and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 12940–12945 CrossRef CAS PubMed.
- J. Barke, R. F. Seipke, S. Grüschow, D. Heavens, N. Drou, M. J. Bibb, R. J. Goss, D. W. Yu and M. I. Hutchings, BMC Biol., 2010, 8, 109 CrossRef PubMed.
- T. C. Mattoso, D. D. O. Moreira and R. I. Samuels, Biol. Lett., 2012, 8, 461–464 CrossRef PubMed.
- R. Sen, H. D. Ishak, D. Estrada, S. E. Dowd, E. Hong and U. G. Mueller, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 17805–17810 CrossRef CAS PubMed.
- S. Haeder, R. Wirth, H. Herz and D. Spiteller, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 4742–4746 CrossRef CAS PubMed.
- I. Schoenian, M. Spiteller, M. Ghaste, R. Wirth, H. Herz and D. Spiteller, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1955–1960 CrossRef PubMed.
- R. F. Seipke, J. Barke, C. Brearley, L. Hill, D. W. Yu, R. J. M. Goss and M. I. Hutchings, PLoS One, 2011, 6, e22028 CAS.
- M. Nair and A. Hervey, Phytochemistry, 1979, 18, 326–327 CrossRef CAS.
- Y. Wang, U. G. Mueller and J. Clardy, J. Chem. Ecol., 1999, 25, 935–941 CrossRef CAS.
- A. Rodrigues, R. N. Cable, U. G. Mueller, M. Bacci and F. C. Pagnocca, Antonie van Leeuwenhoek, 2009, 96, 331–342 CrossRef PubMed.
- A. V. Santos, R. J. Dillon, V. M. Dillon, S. E. Reynolds and R. I. Samuels, FEMS Microbiol. Lett., 2004, 239, 319–323 CrossRef CAS PubMed.
- M. Poulsen, D. P. Erhardt, D. J. Molinaro, T.-L. Lin and C. R. Currie, PLoS One, 2007, 2, e960 Search PubMed.
- E. B. Van Arnam, A. C. Ruzzini, C. S. Sit, C. R. Currie and J. Clardy, J. Am. Chem. Soc., 2015, 137, 14272–14274 CrossRef CAS PubMed.
- G. Carr, E. R. Derbyshire, E. Caldera, C. R. Currie and J. Clardy, J. Nat. Prod., 2012, 75, 1806–1809 CrossRef CAS PubMed.
- N. A. Holmes, T. M. Innocent, D. Heine, M. A. Bassam, S. F. Worsley, F. Trottmann, E. H. Patrick, D. W. Yu, J. C. Murrell, M. Schiøtt, B. Wilkinson, J. J. Boomsma and M. I. Hutchings, Front. Microbiol., 2016, 7, 4307–4316 Search PubMed.
- D. K. Aanen and P. Eggleton, Curr. Biol., 2005, 15, 851–855 CrossRef CAS PubMed.
- A. A. Visser, P. W. Kooij, A. J. M. Debets, T. W. Kuyper and D. K. Aanen, Fungal Ecology, 2011, 4, 322–332 CrossRef.
- G. M. Mathew, Y.-M. Ju, C.-Y. Lai, D. C. Mathew and C. C. Huang, FEMS Microbiol. Ecol., 2011, 79, 504–517 CrossRef PubMed.
- A. A. Visser, T. Nobre, C. R. Currie, D. K. Aanen and M. Poulsen, Microb. Ecol., 2011, 63, 975–985 CrossRef PubMed.
- S. Otani, L. H. Hansen, S. J. Sørensen and M. Poulsen, Microb. Ecol., 2016, 71, 207–220 CrossRef CAS PubMed.
- S. Otani, A. Mikaelyan, T. Nobre, L. H. Hansen, N. A. Koné, S. J. Sørensen, D. K. Aanen, J. J. Boomsma, A. Brune and M. Poulsen, Mol. Ecol., 2014, 23, 4631–4644 CrossRef CAS PubMed.
- F. O. Aylward, G. Suen, P. H. W. Biedermann, A. S. Adams, J. J. Scott, S. A. Malfatti, T. Glavina del Rio, S. G. Tringe, M. Poulsen, K. F. Raffa, K. D. Klepzig and C. R. Currie, mBio, 2014, 5, e02077 CrossRef PubMed.
- S. Um, A. Fraimout, P. Sapountzis, D.-C. Oh and M. Poulsen, Sci. Rep., 2013, 3, 1–7 Search PubMed.
- C. Beemelmanns, T. R. Ramadhar, K. H. Kim, J. L. Klassen, S. Cao, T. P. Wyche, Y. Hou, M. Poulsen, T. S. Bugni, C. R. Currie and J. Clardy, Org. Lett., 2017, 19, 1000–1003 CrossRef CAS PubMed.
- K. H. Kim, T. R. Ramadhar, C. Beemelmanns, S. Cao, M. Poulsen, C. R. Currie and J. Clardy, Chem. Sci., 2014, 5, 4333–4338 RSC.
- T. P. Wyche, A. C. Ruzzini, C. Beemelmanns, K. H. Kim, J. L. Klassen, S. Cao, M. Poulsen, T. S. Bugni, C. R. Currie and J. Clardy, Org. Lett., 2017, 19, 1772–1775 CrossRef CAS PubMed.
- H. R. Kang, D. Lee, R. Benndorf, W. H. Jung, C. Beemelmanns, K. S. Kang and K. H. Kim, J. Nat. Prod., 2016, 79, 3072–3078 CrossRef CAS PubMed.
- G. Carr, M. Poulsen, J. L. Klassen, Y. Hou, T. P. Wyche, T. S. Bugni, C. R. Currie and J. Clardy, Org. Lett., 2012, 14, 2822–2825 CrossRef CAS PubMed.
- H. Guo, N. B. Kreuzenbeck, S. Otani, M. Garcia-Altares, H.-M. Dahse, C. Weigel, D. K. Aanen, C. Hertweck, M. Poulsen and C. Beemelmanns, Org. Lett., 2016, 18, 3338–3341 CrossRef CAS PubMed.
- J. F. Negrón and C. J. Fettig, For. Sci., 2014, 60, 409–413 Search PubMed.
- D. L. Six and K. D. Klepzig, Symbiosis, 2004, 37, 189–206 Search PubMed.
- K. D. Klepzig and R. T. Wilkens, Appl. Environ. Microbiol., 1997, 63, 621–627 CAS.
- R. W. Hofstetter, J. T. Cronin, K. D. Klepzig, J. C. Moser and M. P. Ayres, Oecologia, 2006, 147, 679–691 CrossRef PubMed.
- J. J. Scott, D. C. Oh, M. C. Yuceer, K. D. Klepzig, J. Clardy and C. R. Currie, Science, 2008, 322, 63 CrossRef CAS PubMed.
- D.-C. Oh, J. J. Scott, C. R. Currie and J. Clardy, Org. Lett., 2009, 11, 633–636 CrossRef CAS PubMed.
- J. A. V. Blodgett, D.-C. Oh, S. Cao, C. R. Currie, R. Kolter and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 11692–11697 CrossRef CAS PubMed.
- J. Hulcr, A. S. Adams, K. Raffa, R. W. Hofstetter, K. D. Klepzig and C. R. Currie, Microb. Ecol., 2011, 61, 759–768 CrossRef PubMed.
- Y. J. Cardoza, K. D. Klepzig and K. F. Raffa, Ecol. Entomol., 2006, 31, 636–645 CrossRef.
- J. Therrien, C. J. Mason, J. A. Cale, A. Adams, B. H. Aukema, C. R. Currie, K. F. Raffa and N. Erbilgin, Oecologia, 2015, 179, 467–485 CrossRef PubMed.
- M. Poulsen, D.-C. Oh, J. Clardy and C. R. Currie, PLoS One, 2011, 6, e16763 CAS.
- D.-C. Oh, M. Poulsen, C. R. Currie and J. Clardy, Org. Lett., 2011, 13, 752–755 CrossRef CAS PubMed.
- G. T. Fincher, J. Ga. Entomol. Soc., 1981, 16, 316–333 Search PubMed.
- S.-H. Kim, H. Ko, H.-S. Bang, S.-H. Park, D.-G. Kim, H. C. Kwon, S. Y. Kim, J. Shin and D.-C. Oh, Bioorg. Med. Chem. Lett., 2011, 21, 5715–5718 CrossRef CAS PubMed.
- S.-H. Park, K. Moon, H.-S. Bang, S.-H. Kim, D.-G. Kim, K.-B. Oh, J. Shin and D.-C. Oh, Org. Lett., 2012, 14, 1258–1261 CrossRef CAS PubMed.
- S.-H. Kim, S. H. Kwon, S.-H. Park, J. K. Lee, H.-S. Bang, S.-J. Nam, H. C. Kwon, J. Shin and D.-C. Oh, Org. Lett., 2013, 15, 1834–1837 CrossRef CAS PubMed.
- S. Um, S. H. Park, J. Kim, H. J. Park, K. Ko, H.-S. Bang, S. K. Lee, J. Shin and D.-C. Oh, Org. Lett., 2015, 17, 1272–1275 CrossRef CAS PubMed.
- S. Um, D.-H. Bach, B. Shin, C.-H. Ahn, S.-H. Kim, H.-S. Bang, K.-B. Oh, S. K. Lee, J. Shin and D.-C. Oh, Org. Lett., 2016, 18, 5792–5795 CrossRef CAS PubMed.
- S. P. Shukla, J. G. Sanders, M. J. Byrne and N. E. Pierce, Mol. Ecol., 2016, 25, 6092–6106 CrossRef CAS PubMed.
- A. M. Estes, D. J. Hearn, E. C. Snell-Rood, M. Feindler, K. Feeser, T. Abebe, J. C. Dunning Hotopp and A. P. Moczek, PLoS One, 2013, 8, e79061 CAS.
- A. Dejean, P. J. Solano, J. Ayroles, B. Corbara and J. Orivel, Nature, 2005, 434, 973 CrossRef CAS PubMed.
- M. X. Ruiz-Gonzalez, P. J. G. Male, C. Leroy, A. Dejean, H. Gryta, P. Jargeat, A. Quilichini and J. Orivel, Biol. Lett., 2011, 7, 475–479 CrossRef PubMed.
- R. F. Seipke, J. Barke, D. Heavens, D. W. Yu and M. I. Hutchings, MicrobiologyOpen, 2013, 2, 276–283 CrossRef CAS PubMed.
- R. F. Seipke, J. Barke, M. X. Ruiz-Gonzalez, J. Orivel, D. W. Yu and M. I. Hutchings, Antonie van Leeuwenhoek, 2011, 101, 443–447 CrossRef PubMed.
- H. Gao, S. Grüschow, J. Barke, R. F. Seipke, L. M. Hill, J. Orivel, D. W. Yu, M. Hutchings and R. J. M. Goss, RSC Adv., 2014, 4, 57267–57270 RSC.
- A. Moya, J. Peretó, R. Gil and A. Latorre, Nat. Rev. Genet., 2008, 9, 218–229 CrossRef CAS PubMed.
- S. Sudakaran, C. Kost and M. Kaltenpoth, Trends in Microbiology, 2017, 25, 375–390 CrossRef CAS PubMed.
- C. Toft and S. G. E. Andersson, Nat. Rev. Genet., 2010, 11, 465–475 CrossRef CAS PubMed.
- A. C. Ruzzini and J. Clardy, Curr. Biol., 2016, 26, R859–R864 CrossRef CAS PubMed.
- A. Kampa, A. N. Gagunashvili, T. A. M. Gulder, B. I. Morinaka, C. Daolio, M. Godejohann, V. P. W. Miao, J. Piel and Ó. S. Andrésson, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E3129–E3137 CrossRef CAS PubMed.
- M. R. Seyedsayamdost and J. Clardy, ACS Synth. Biol., 2014, 3, 745–747 CrossRef CAS PubMed.
- R. Breitling and E. Takano, Cold Spring Harbor Perspect. Biol., 2016, 8, a023994 CrossRef PubMed.
- C.-J. Guo, F.-Y. Chang, T. P. Wyche, K. M. Backus, T. M. Acker, M. Funabashi, M. Taketani, M. S. Donia, S. Nayfach, K. S. Pollard, C. S. Craik, B. F. Cravatt, J. Clardy, C. A. Voigt and M. A. Fischbach, Cell, 2017, 168, 517–526 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |