An engineered cell-imprinted substrate directs osteogenic differentiation in stem cells†
Khorshid
Kamguyan
ab,
Ali Asghar
Katbab
 *a,
Morteza
Mahmoudi
*a,
Morteza
Mahmoudi
 c,
Esben
Thormann
d,
Saeed
Zajforoushan Moghaddam
c,
Esben
Thormann
d,
Saeed
Zajforoushan Moghaddam
 d,
Lida
Moradi
e and
Shahin
Bonakdar
*b
d,
Lida
Moradi
e and
Shahin
Bonakdar
*b
aDepartment of Polymer Engineering and Colour Technology, Amirkabir University of Technology, Tehran, 1599637111, Iran. E-mail: katbab@aut.ac.ir; Fax: +9821 664 69162; Tel: +98 21 644 18107
bNational Cell Bank of Iran, Pasteur Institute of Iran, 1316943551, Tehran, Iran. E-mail: sh_bonakdar@pasteur.ac.ir; Fax: +98 21 66465132; Tel: +98 21 66953311
cDepartment of Nanotechnology and Nanotechnology Research Center, Faculty of Pharmacy, Tehran University of Medical Sciences, Tehran, Iran
dDepartment of Chemistry, Technical University of Denmark, 2800, Kgs. Lyngby, Denmark
ePediatric Urology and Regenerative Medicine Research Center, Children's Medical Center, Tehran University of Medical sciences, 1417613151, Tehran, Iran
First published on 20th November 2017
Abstract
A cell-imprinted poly(dimethylsiloxane)/hydroxyapatite nanocomposite substrate was fabricated to engage topographical, mechanical, and chemical signals to stimulate and boost stem cell osteogenic differentiation. The physicochemical properties of the fabricated substrates, with nanoscale resolution of osteoblast morphology, were probed using a wide range of techniques including scanning electron microscopy, atomic force microscopy, dynamic mechanical thermal analysis, and water contact angle measurements. The osteogenic differentiation capacity of the cultured stem cells on these substrates was probed by alizarin red staining, ALP activity, osteocalcin measurements, and gene expression analysis. The outcomes revealed that the concurrent roles of the surface patterns and viscoelastic properties of the substrate provide the capability of directing stem cell differentiation toward osteogenic phenotypes. Besides the physical and mechanical effects, we found that the chemical signaling of osteoinductive hydroxyapatite nanoparticles, embedded in the nanocomposite substrates, could further improve and optimize stem cell osteogenic differentiation.
1. Introduction
Stem cell differentiation holds one of the key lines of regenerative medicine and tissue engineering.1–3 The natural extracellular matrix (ECM) guides the tissue regeneration process through various biochemical signals.4–6 Accordingly, a synthetic microenvironment should be similar to the natural ECM in inducing analogous biochemical signals in vitro.7,8 Differentiation is strongly governed by chemical, physical, and mechanical factors/stimuli related to the substrate, which together are recognized as the triangle of control.2 Chemical stimuli have been employed more commonly due to their feasibility and efficiency; however, physical and mechanical factors have been vastly investigated lately, to replace the less safe chemical features such as growth factors.9–11 It is well known that physical features (e.g., surface topography) and mechanical factors (e.g., substrate elastic modulus) have substantial effects on stem cell proliferation, migration, and differentiation through different mechanisms, including by mimicking the natural niche of the target cells.12–15 Accordingly, a substrate mimicking the surface topography and morphology of the natural ECM can be employed as an ideal culture substrate for stimulating stem cell differentiation.8,16–19 In this regard, the topography of a patterned surface plays the most important role in directing cellular behavior,20–22 implying that each specific pattern may have the capacity to lead stem cells to a distinctive lineage.23 For instance, it has been shown that nanopit topographies can direct stem cell fates to osteogenesis,20,24,25 while nanoscale ridge/groove patterns promote neurogenic differentiation of these cells.26–29Besides the pattern structure, the dimensions of the topographical features are another decisive property of the culture substrate.12,30 For instance, micro-sized topographies have been demonstrated to stimulate cytoskeleton formation and differentiation;31–33 on the other hand, nano-sized patterns are known to generate interconnected cell–cell interactive networks.34–36
Several studies have been conducted on stem cell differentiation using a cell-imprinting method, in which the footprint of the target cells has been employed as the substrate surface topography, suggesting that topographical features alone have the capacity to stimulate cell behaviour and lineage specification.37–39 Further research has been carried out on the differentiation of stem cells towards chondrocytes,37 tenocytes,39 and keratinocytes38via the cell-imprinting method, which has offered a new line of research to the field of physical stem cell differentiation.
The mechanical properties of substrates have been proposed as another key factor affecting cellular behaviour, functioning, and phenotype.40–44 Stem cells can sense the physical and mechanical characteristics of their environment, through a so-called “mechanotransduction” process.45,46 The recent literature shows that several mechanical stimuli can affect stem cell differentiation.13,47–50 For instance, Engler and co-workers revealed that the elastic modulus of culturing substrates can direct stem cell differentiation.13 In another study, Gonzalez-Garcia et al. have proposed that the main mechanical property directing stem cell differentiation is the surface mobility, which indeed provided a deeper understanding of the role of the mechanical interaction between the substrate and the stem cells.50
In the present work, we employed a cell-imprinting method to direct adipose-derived stem cells (ADSCs) toward osteogenic differentiation based on the topography-driven approach. In addition, through the fabrication of poly(dimethylsiloxane)/hydroxyapatite (PDMS/HA) nanocomposites, we investigated the effect of the substrate mechanical properties (in particular, the viscoelastic properties) on stem cell osteogenic differentiation. Finally, by increasing the amount of HA nanoparticles in the fabricated nanocomposite substrates, we could enhance the chemical signalling to optimize osteogenic differentiation. Through fabricating a two-phase substrate that consists of an inorganic, rigid, and osteoconductive phase (HA) together with a flexible viscoelastic polymeric phase (PDMS), we aimed at fabricating an artificial microenvironment which mechanically, physically and chemically mimics the natural bone ECM.51 The topography of the cell-imprinted substrates was assessed using scanning electron microscopy and atomic force microscopy. The surface hydrophilicity and viscoelastic properties of the substrates were examined by water contact angle measurements and dynamic mechanical thermal analysis. Next, osteogenic differentiation of stem cells on the substrates was investigated through alizarin red staining, alkaline phosphatase activity, and osteocalcin measurements, as well as gene expression analysis.
2. Experimental
2.1. Isolation and culture of adipose-derived stem cells (ADSCs)
Adipose tissue samples were withdrawn from healthy thirty year old humans with ethical consent, and grown in Dulbecco's modified Eagle's medium (DMEM; Gibco, Scotland, UK), containing 10% (v/v) fetal bovine serum (FBS, Seromed, Germany) and penicillin (100 IU mL−1)–streptomycin (100 μg mL−1, Sigma, the United States) at 25 °C for a maximum of 1 day before use. To extract stem cells, the adipose tissue samples were washed three times with a 3% solution of DMEM/penicillin–streptomycin, then were chopped into 1 × 1 mm pieces and digested in an incubator with 0.02 mg mL−1 collagenase type I (Sigma, the United States) for 2 h. The prepared suspension was centrifuged at 1300 rpm for 5 min, and the obtained cell pellets were separated. The solution containing suspended ADSCs was then transferred to the culture medium consisting of DMEM/Ham's F12 supplemented with 10% FBS, 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin in a humidified incubator (37 °C, 5% CO2). After 24 h, the non-adherent cells and debris were removed, and fresh culture medium was added.2.2. Isolation and culture of human osteoblasts
Bone tissue samples were obtained from healthy thirty year old human bone tissues in a sterilized operating room and then preserved in a DMEM/10% FBS solution containing penicillin–streptomycin to transfer to the laboratory. Samples were washed three times with a washing solution (DMEM containing penicillin–streptomycin) for removal of the probable infection. Tissue samples were chopped into small pieces, washed again, and then transferred to a culture medium as used for the stem cells. Suspended cells were transferred every 48 h to add fresh culture medium.Live subject statement: All experiments concerning live human tissue were performed according to the laws and guidelines of Iran's Ministry of Health and Medical Education and approved by the Ethics Committee at the National Cell Bank of Iran. All experiments concerning a human patient were performed after obtaining informed consent.
2.3. Characterization of osteoblasts
Osteoblast cells were characterized using crystal violet staining, alizarin red staining, and alkaline phosphatase (ALP) activity measurements. The culture medium of cells was drawn for ALP activity measurements; then the cells were fixed using a 4% glutaraldehyde solution for 45 min. Alizarin red and crystal violet were then added to the cells, kept for 45 min, and finally washed with ice-cold phosphate-buffered saline (PBS).2.4. Fabrication of cell-imprinted substrates
Poly(dimethylsiloxane) (PDMS, SYLGARD 184, RTV, Dow Corning, the United States) was used to fabricate the cell-imprinted substrates. The silicone resin and the curing agent were mixed in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. Osteoblasts and ADSCs were cultured for 24 h on polystyrene well plates according to the standard cell culture procedures (both at passage two).52,53 Cells were fixed using a 4% glutaraldehyde solution for 45 min and washed with deionized water. The resin–curing agent mixture was poured onto samples and retained at 37 °C for 48 h. The cured silicone rubber was then peeled off the cell culture plates, followed by multiple washing steps using boiled water and a 1 M NaOH solution to remove the remaining cells/debris and other chemicals from the substrates.
1 ratio. Osteoblasts and ADSCs were cultured for 24 h on polystyrene well plates according to the standard cell culture procedures (both at passage two).52,53 Cells were fixed using a 4% glutaraldehyde solution for 45 min and washed with deionized water. The resin–curing agent mixture was poured onto samples and retained at 37 °C for 48 h. The cured silicone rubber was then peeled off the cell culture plates, followed by multiple washing steps using boiled water and a 1 M NaOH solution to remove the remaining cells/debris and other chemicals from the substrates.
2.5. Synthesis of hydroxyapatite nanoparticles
Hydroxyapatite powders were synthesized at 50 °C using calcium nitrate (Ca (NO3)2·4H2O, 99%, Merck, Germany), and diammonium hydrogen phosphate ((NH4)2HPO4, 99%, Merck, Germany). The raw materials were dissolved in distilled water at pH = 10, which was adjusted using a 25% NH4OH solution (Merck, Germany). Stoichiometric hydroxyapatite powder (HA) was synthesized by the dropwise addition of a calcium nitrate solution into the diammonium hydrogen phosphate solution. All precipitates were aged at 25 °C for 24 h, filtered, washed three times with distilled water, and finally dried in an oven at 80 °C for 24 h.542.6. Fabrication of cell-imprinted HA/PDMS nanocomposite substrates
After the mechanical mixing of HA powder and silicone resin, the mixture was sonicated in an ultrasound bath for 30 min, then poured onto the prefixed osteoblast cells and kept at 37 °C for 48 h to ensure full crosslinking. The cured nanocomposite was then peeled off the plates followed by several washing steps using boiled water and a 1 M NaOH solution to remove the remaining cells, debris and other chemicals.2.7. Stem cell seeding on substrates
ADSCs (3 × 103 cells per cm2 in 100 μL of culture medium) were cultured on the fabricated substrates (at passages three to five), and placed into polystyrene plates. After 24 h, 600 μL cm−2 fresh DMEM/Ham's F12 (with a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) and 10% (v/v) FBS medium were added to each substrate to completely cover the substrate surface.
1 ratio) and 10% (v/v) FBS medium were added to each substrate to completely cover the substrate surface.
2.8. Scanning electron microscopy (SEM)
The topography of the cell-imprinted substrates and the morphology of stem cells cultured on the substrates were examined using SEM (Philips XL30, Germany). ADSCs were fixed in a 4% glutaraldehyde solution for 45 min after 24 h of culture on the substrates. The samples were dehydrated using graded dilutions of ethanol (10–100% v/v). A thin layer (around 0.8 nm) of gold was coated onto the sample surface using a sputter coater, before taking images using 20 keV voltage.2.9. Atomic force microscopy (AFM) imaging
The topography of the cell-imprinted samples was investigated using AFM contact mode imaging (NanoWizard 4a NanoScience, JPK Instruments AG, Berlin, Germany). A rectangular cantilever (HQ:NSC18/Al BS, MikroMasch, Bulgaria) with an approximate spring constant of 2.8 N m−1 and a conic tip of 8 nm radius was used. Surface areas up to 100 × 100 μm were scanned using a set point force of 0.5 nN and a scan rate of 0.05 Hz. The standard software of the instrument (JPKSPM Data Processing) was employed to analyse the images as well as obtain the cross-section height profiles.2.10. Water contact angle measurements
To compare the surface hydrophilicities of the neat and HA-reinforced PDMS nanocomposite substrates, sessile drop measurements (Kruss G40, Germany) were carried out. The experiments were conducted at an ambient temperature of 23.0 ± 0.1 °C using a constant water droplet volume of 1.5 μl. The advancing contact angle value after 10 s was recorded. Five measurements at different spots of the surface were conducted.2.11. Surface modulus measurements by AFM
To assess the mechanical reinforcement of the PDMS/HA samples, AFM force measurements were performed, using a tipless cantilever (HQ: CSC38/Cr-Au, MikroMasch). The accurate normal spring constant of the cantilever was found to be 0.190 N m−1, according to the thermal noise method.55 A spherical silica particle with a diameter of 9.1 ± 0.1 μm was then glued to the end of the cantilever.56 The force measurements were performed with a set point force of 20 nN on 64 points over a 50 × 50 μm surface area and repeated three times on new regions of the surface. A Hertz model for the spherical probe was used to estimate the surface elastic modulus. The initial 10 nm of indentation was considered to fit the model to the force curves. The instrument software (JPKSPM Data Processing) was used to analyse the force curves and data fitting.2.12. Dynamic mechanical thermal analysis (DMTA)
The viscoelastic behaviour of PDMS-based substrates was analysed using DMTA. The measurements were performed using a STARe system DMA1 instrument (Mettler-Toledo, Switzerland) at a frequency of 1 Hz in a tensile dynamic field with a strain amplitude of 20 μm and the temperature range of −150 to 120 °C. Samples were prepared in 5 × 8 × 0.5 mm dimensions. The loss modulus, storage modulus and dissipation factor were measured as a function of temperature.2.13. Characterization of stem cells cultured on PDMS substrates by alizarin red and crystal violet staining
To analyse viable cells and investigate osteogenic differentiation, alizarin red staining and crystal violet staining were performed on ADSCs cultured on PDMS substrates and culture plates for 14 days. After culturing 5 × 103 stem cells in 24-well plates, the cells were fixed in 4% glutaraldehyde solution for 24 h and stained with alizarin red for 45 min, and then washed with PBS solution to remove residue dye. To ascertain the optical density of viable cells cultured on substrates, the maximum absorbance at a wavelength of 260 nm in a spectrophotometer (NanoDrop, Eppendorf, Germany) was measured.2.14. Characterization of stem cells cultured on PDMS substrates by serum osteocalcin and ALP measurements
Osteogenic differentiation was investigated via serum osteocalcin and ALP activity measurements. ADSCs were cultured on different substrates in 24-well plates (3 × 103 cells per well). After 14 days, the medium was removed and analysed using an auto-analyser (Hitachi 902, Germany) and an alkaline phosphatase kit (Pars Azmoon, Iran) employing the DGKC method to measure the ALP activity; the absorbance was read at 1 min intervals at 405 nm. Serum osteocalcin was measured with N-MID Osteocalcin ELIZA kits (Immunodiagnostic Systems, UK). Substrate turnover was determined colourimetrically by measuring the absorbance at 450 nm and a four parameter algorithm was employed to generate the standard curve.2.15. Gene expression profiling of stem cells cultured on PDMS substrates
2−ΔΔ(CT).
| Gene | Forward primer | Reverse primer |
|---|---|---|
| GAPDH | TCAAGGCTGAGAACGGGAA | TGGGTGGCAGTGATGGCA |
| Collagen I | CGATGGCTGCACGAGTCA | GGTTCAGTTTGGGTTGCTTGTC |
| Osteocalcin | CAGCGAGGTAGTGAAGAGACC | TCTGGAGTTTATTTGGGAGCAG |
| RUNX2 | GGTCAGATGCAGGCGGCCC | TACGTGTGGTAGCGCGTGGC |
3. Results
3.1. Characterization of the cell-imprinted substrates
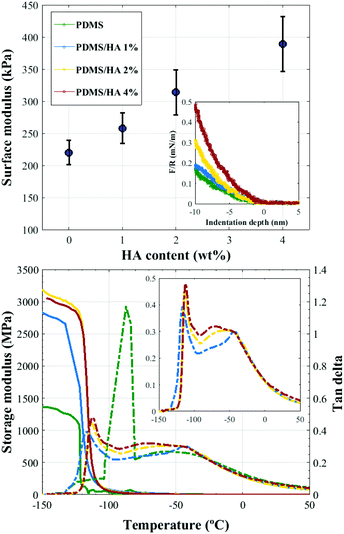
3.1.2.1. Surface elastic modulus. AFM force measurements were employed to assess the surface elastic modulus of the substrates.57,58 Comparison between the surface elastic moduli of neat PDMS and PDMS/HA nanocomposites of different HA contents is demonstrated in Fig. 2 (top). Accordingly, an increment in the surface modulus and stiffness is obtained with an enhanced amount of HA in the polymer matrix, which can also be inferred from the slope of the force-indentation curves (inset, Fig. 2 (top)). It is noteworthy that the spectrum of surface elastic modulus obtained for the PDMS-based substrates is considerably differing from those suggested for stimulating osteogenic differentiation by the earlier reports.13,49
3.1.2.2. Viscoelastic behaviour. DMTA was employed to analyse the viscoelastic behaviour of PDMS-based substrates. Fig. 2 (bottom) illustrates the storage modulus and tan delta (damping) curves for PDMS and 1, 2, and 4 wt% PDMS/HA nanocomposites as a function of temperature. As shown, addition of 1 wt% HA nanoparticles to the PDMS matrix significantly affects the viscoelastic behaviour of the substrates, i.e., a large gain in the storage modulus and a substantial decrement in the tan delta. On the other hand, the effect of increasing the amount of nanoparticles on the storage modulus and damping is practically negligible, which can be interpreted as due to the rather similar viscoelastic behaviours of the PDMS/HA nanocomposites with different HA contents. Moreover, the addition of HA nanoparticles gives rise to a small secondary peak in the tan delta curves, which can be more clearly seen in the inset figure. This observation might be attributed to the aggregation of HA nanoparticles in the nanocomposite, which can give rise to two distinct viscoelastic behaviours occurring in the PDMS matrix. It should be mentioned that due to the weak interaction between PDMS and HA nanoparticles,59–63 we were unable to fabricate PDMS/HA nanocomposites of higher HA contents.
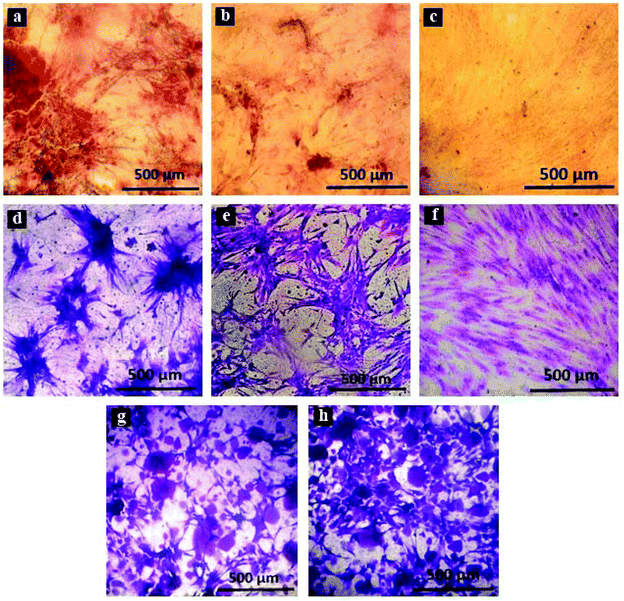
3.1.3.1. Alizarin red staining. Dispersion of HA nanoparticles in the PDMS matrix, as well as the presence of nanoparticles at the surface of PDMS substrates, was probed by alizarin red staining (displayed in Fig. 3). The results demonstrate that HA nanoparticles aggregate throughout the nanocomposite. The aggregated entities grow in size and number as the amount of the nanoparticles is increased. The average size of the aggregates in the 1 wt% nanocomposite is nearly 50 micrometres, while aggregated clusters as large as around 200 micrometres were found in the 4 wt% nanocomposite.
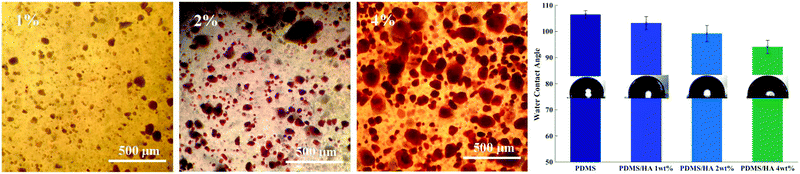 | ||
| Fig. 3 (left) Alizarin red staining of 1, 2 and 4 wt% PDMS/HA nanocomposites. (right) The water contact angle on the PDMS and PDMS/HA nanocomposite substrates. | ||
3.1.3.2. Water contact angle. Water contact angle measurements were carried out to roughly assess the presence of nanoparticles at the surface of substrates, which may change the surface properties of the nanocomposites. Fig. 3 depicts the average water contact angle on PDMS, as well as PDMS/HA nanocomposites. Accordingly, a decline in contact angle is observed as the amount of nanoparticles in the matrix increases, which can indicate an enhanced surface hydrophilicity for the substrates. It should be mentioned that the 1 wt% PDMS/HA nanocomposite has a rather comparable contact angle relative to PDMS, which can suggest the negligible presence of HA nanoparticles at the surface, and therefore an insignificant change in the surface chemistry is expected. For this reason, the 1 wt% nanocomposite was not used as the culture substrate for osteogenic differentiation assessment.
3.2. Osteogenic differentiation
Adipose-derived stem cells were cultured on culture plates, flat and osteoblast-imprinted PDMS, and osteoblast-imprinted PDMS/HA nanocomposite substrates with 2 and 4 wt% nanoparticle content, in order to define the osteogenic differentiation capacity and the mechanism of action of different substrates.Crystal violet staining was conducted to study the viable stem cells cultured on the surface of different substrates. Fig. 5 presents crystal violet staining of the cultured stem cells for 14 days on patterned PDMS (d), flat PDMS (e), the culture plate (f), as well as 2 and 4 wt% PDMS/HA nanocomposites (g, h).
The results confirm considerable differences between the shape and morphology of the cultured stem cells on the culture plate and those cultured on PDMS-based substrates. We also noticed that the cultured stem cells on the surface of the culture plate are more spread out, unlike those cultured on PDMS-based substrates, which are found to be rather large aggregates. In addition, the stem cells cultured on neat PDMS substrates aggregate and form larger clusters, while those cultured on PDMS/HA substrates are found to be relatively smaller entities.
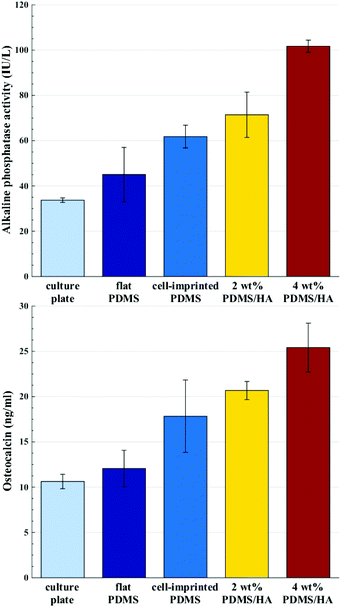 | ||
| Fig. 6 Alkaline phosphatase activity (top) and serum osteocalcin (bottom) of stem cells cultured for 14 days on the culture plate, flat PDMS, cell-imprinted PDMS, 2 and 4 wt% cell-imprinted PDMS/HA nanocomposites. All the data are normalized with respect to the optical densities of the stem cells cultured on each substrate (optical density data are available in the ESI, Section S3†). | ||
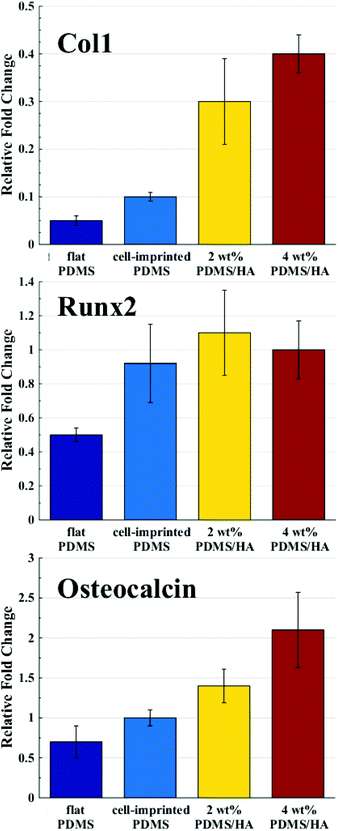
4. Discussion
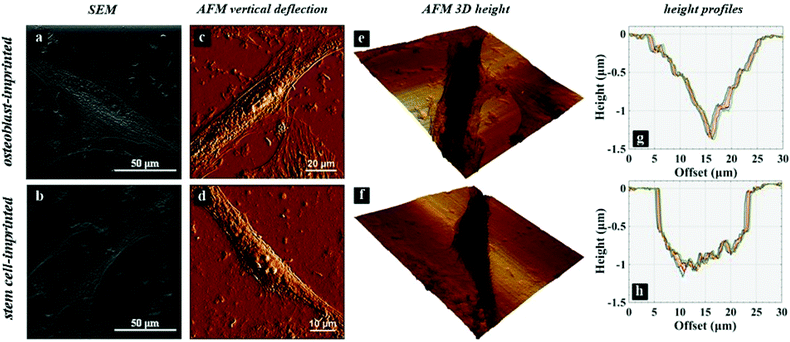
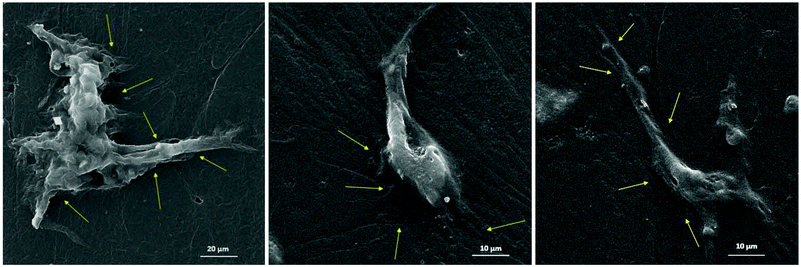
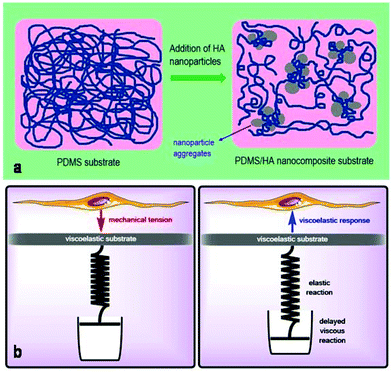
The chemical, physical and mechanical factors of the culture substrate can direct stem cell lineage specification.1–3 As established in previous investigations, patterned substrates that resemble the topographical features of a specific cell can give rise to improved stem cell differentiation.37–39 Based on this idea, we fabricated the exact pattern of osteoblast cells on the PDMS-based substrates using the cell-imprinting method, with the aim of stimulating osteogenesis. Scanning electron microscopy and atomic force microscopy were employed to investigate the surface topography of cell-imprinted PDMS substrates (Fig. 1) and cell-imprinted PDMS/HA nanocomposite substrates (Fig. S5†), which indicated successful imprinting of the topographical features of osteoblast and stem cells, resembling the optical microscopy images of the cultured cells. From the obtained images, it can be inferred that both imprinted osteoblast and stem cells possess a spindle-like shape from the plain view. However, the AFM cross-section height profiles of the imprinted entities reveal that the imprinted osteoblast cells apparently have a pyramidal shape, while the stem cells seem to have a cubical geometry. These differences in the substrates, with the patterns of osteoblasts and stem cells, reveal that both micro- and nano-scale topographical details can effectively direct stem cell differentiation, even in the absence of chemical signalling. This can be concluded from the results of alizarin red and crystal violet staining (Fig. 5), ALP and osteocalcin measurements (Fig. 6), and gene expression analysis (Fig. 7), which confirmed an enhanced osteogenic differentiation in the stem cells cultured on the osteoblast-imprinted PDMS substrate compared to those cultured on the flat substrate.In addition, HA nanoparticles were incorporated into the polymer matrix to modify the mechanical properties and surface hydrophilicity, in order to elevate osteogenesis in stem cells cultured on the patterned substrates. To better understand the role of mechanical interaction between stem cells and the polymeric surface, the viscoelastic behaviour of the nanocomposite substrates was examined using DMTA measurements (Fig. 2). According to the dissipation curves, the addition of HA nanoparticles resulted in a two-phase polymeric system (Fig. 8a), which can originate from the considerable difference between the surface energies of HA nanoparticles and the PDMS matrix.59–63 Accordingly, dispersion of HA nanoparticles in PDMS is thermodynamically unfavourable, which then leads to aggregation of nanoparticles and the formation of rather large clusters. As a result, some polymer chains can be trapped between the HA aggregates, which then produces an entropic loss in the system. To compensate for this entropic loss, the non-trapped polymer chains in the matrix increase the free volume of the system, implying easier segmental motion, an increment in the elastic response and then a shorter response time of the free chains to the received mechanical tension.
Several studies have been conducted for understanding the effect of the mechanical properties of the substrate on cell behaviour and differentiation; however, it has not been clearly addressed which mechanical parameter has the main contribution.2 The range of the surface elastic modulus estimated for the fabricated PDMS/HA substrates exceeds those suggested earlier for osteogenic differentiation.13,43,44,49 Herein, the ALP and osteocalcin measurements (Fig. 6) alongside gene expression analysis (Fig. 7) of stem cells cultured on the cell-imprinted nanocomposite substrates with 2 and 4 wt% nanoparticle demonstrated an elevated osteogenesis compared to those cultured on neat PDMS surfaces. The obtained results require a hard look at the nature of the materials used as the substrate for stem cell culture. Mechanotransduction is based on the applied mechanical tension from the cells toward the surface and conversely the mechanical response received by the cells from the surface.45,46 In this case, the viscoelastic nature of polymers enables them to render a twofold response to the received mechanical tension, i.e., an elastic reaction and a viscous response.64 This dual mechanical behavior can be described according to Maxwell's model for viscoelasticity, also known as the spring-dashpot model, in which the spring represents the immediate elastic reaction, and the dashpot indicates the time-dependent viscous component.65 Based on the ratio of these two reactions, the cells can sense the physical and mechanical nature of the substrate (Fig. 8b). For instance, polystyrene-based culture substrates have a relatively high dissipation factor, which prolongs the substrate's response time to the mechanical tension received from the cultured cells.66,67 In contrast, elastomers such as PDMS have a much lower ratio of viscous to elastic reaction; in other words, they render a higher segmental motion, an enhanced chain mobility, a lower glass transition temperature and a much shorter response time to the received mechanical tension,64,65,68 which consequently results in improved cell–substrate mechanical interaction.
Besides the mechanically induced effect, the osteoinductive nature of HA nanoparticles stimulates osteogenic chemical signals in the stem cells, which may lead to bone regeneration.51,69–71 The elevated hydrophilicity and the increased amount of HA nanoparticles revealed by alizarin red staining (Fig. 3), and the raised level of calcium and phosphate ions on the surface of PDMS/HA nanocomposite substrates (Fig. S6†), all point to higher levels of chemical signalling and protein absorption through increasing the nanoparticle amount in the surface of PDMS-based substrates. Accordingly, osteogenesis was optimized in the stem cells cultured on the cell-imprinted PDMS/HA substrates through increasing the amount of nanoparticles up to 4 wt%, thus showing the synergistic effects of topography, mechanical stimulation and chemical signalling on stem cell differentiation (Fig. 6, 7 and S8†). Moreover, the former studies have demonstrated better attachment and more spreading for various cell types cultured on hydrophilic surfaces, compared to those cultured on hydrophobic surfaces where the cells rather attach to each other and form aggregates.72–74 In this respect, it is evident from crystal violet images of the cells cultured on PDMS substrates and polystyrene plates that stem cells cultured on the more hydrophilic polystyrene surface are finely spread out. On the other hand, stem cells cultured on the relatively hydrophobic PDMS surfaces formed large aggregates. Besides, the HA nanoparticles were employed to alter the surface chemistry of the PDMS substrates. It can be seen from the crystal violet images that with an increase in HA content and thus surface hydrophilicity of the substrates, the cultured stem cells are spotted in relatively smaller clusters and are more outspread on the surface.
5. Conclusions
The substrates produced for osteogenic differentiation effectively simulate the natural ECM of osteoblast cells in multiple aspects. The cell-imprinted substrates mimic the topography and shape of natural osteoblast ECM, which can physically direct stem cells toward osteogenesis. In addition, the surface viscoelastic behaviours of PDMS such as mechanical response time are represented as the effective parameters stimulating stem cell mechanical differentiation. Finally yet importantly, the enhanced amount of HA nanoparticles in the matrix elevates chemical signalling and thus imitates the osteoconductive ECM of osteoblasts. The cell-imprinted PDMS/HA substrates contain a flexible and viscoelastic polymeric phase much like the collagenous matrix of bone, and a stiff phase composed of nanoparticle aggregates that mimic the inorganic rigid matrix of bone rich in HA.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors gratefully acknowledge funding from the Iran National Science Foundation (94017533) and the Iranian Ministry of Health and Medical Education Fund for Young Researchers.References
- A. Dolatshahi-Pirouz, M. Nikkhah, K. Kolind, M. Dokmeci and A. Khademhosseini, J. Funct. Biomater., 2011, 88–106 CrossRef CAS PubMed.
- O. Mashinchain, L. Turner, M. Dalby, S. Laurent, M. Shokrgozar, S. Bonakdar, M. Imani and M. Mahmoudi, Nanomedicine, 2015, 829–847 CrossRef PubMed.
- O. Fisher, A. Khademhosseini, R. Langer and N. Peppas, Acc. Chem. Res., 2010, 419–428 CrossRef CAS PubMed.
- S. Park and G. I. Im, J. Biomed. Mater. Res., Part A, 2014, 1238–1245 Search PubMed.
- R. A. Marklein and J. A. Burdick, Adv. Mater., 2010, 22, 175–189 CrossRef CAS PubMed.
- A. Higuchi, Q.-D. Ling, Y. Chang, S.-T. Hsu and A. Umezawa, Chem. Rev., 2013, 3297–3328 CrossRef CAS PubMed.
- B. Joddar, T. Hoshiba, G. Chen and Y. Ito, Biomater. Sci., 2014, 1595–1603 RSC.
- L. Turner and M. Dalby, Biomater. Sci., 2014, 1574–1594 RSC.
- K. Lee, E. A. Silva and D. Mooney, J. R. Soc., Interface, 2011, 153–170 CrossRef CAS PubMed.
- M. Simons and J. Ware, Nat. Rev. Drug Discovery, 2003, 863–871 CrossRef CAS PubMed.
- M. Simons, B. Annex, R. Laham, N. Kleiman, T. Henry, H. Dauerman, J. Udelson, E. Gervino, M. Pike and M. Whitehouse, et al. , Circulation, 2002, 788–793 CrossRef CAS.
- H. Jeon, C. Simon and G. Kim, J. Biomed. Mater. Res., Part B, 2014, 1580–1594 CrossRef CAS PubMed.
- A. Engler, S. Sen, H. Sweeney and D. Discher, Cell, 2006, 677–689 CrossRef CAS PubMed.
- M. Dalby, N. Gadegaard and R. Oreffo, Nat. Mater., 2014, 558–569 CrossRef CAS PubMed.
- C. Y. Tay, C. G. Koh, N. S. Tan, D. T. Leong and L. P. Tan, Nanomedicine, 2013, 623–638 CrossRef CAS PubMed.
- M. Lord, M. Foss and F. Besenbacher, Nano Today, 2012, 66–78 Search PubMed.
- P. Tsimbouri, R. McMurray and K. Burgess, et al. , ACS Nano, 2012, 10239–10249 CrossRef CAS PubMed.
- E. Yim, E. Darling, K. Kulangara, F. Guilak and K. Leong, Biomaterials, 2010, 1299–1306 CrossRef CAS PubMed.
- M. Scherthaner, B. Reisinger and H. Wolinski, et al. , Acta Biomater., 2012, 2953–2962 CrossRef PubMed.
- M. Dalby, N. Gadegaard, R. Tare, A. Andar, M. Riehle, P. Herzyk, C. Wilkinson and R. Oreffo, Nat. Mater., 2007, 997–1003 CrossRef CAS PubMed.
- M. Frey, I. Ysai, T. Russel, S. Hanks and Y. Wang, Biophys. J., 2006, 3774–3782 CrossRef CAS PubMed.
- J. Recknor, J. Recknor, D. Sakaguchi and S. Mallapragada, Biomaterials, 2004, 2753–2767 CrossRef CAS PubMed.
- S. Li, S. Kuddannaya, Y. J. Chuah, J. Bao, Y. Zhang and D. Wang, Biomater. Sci., 2017, 2056–2067 RSC.
- J. Yang, L. McNamara, N. Gadegaard, E. Alakpa, K. Burgess, R. Dominic Meek and M. Dalby, ACS Nano, 2014, 10239–10249 Search PubMed.
- J. Wang, S. Ahmed, N. Gadegaard, R. Dominic Meek, M. Dalby and S. Yarwood, Growth Horm. IGF Res., 2014, 245–250 CrossRef PubMed.
- M. Lee, K. Kwon, H. Jung, H. Kim, K. Suh, K. Kim and K. S. Kim, Biomaterials, 2010, 4360–4366 CrossRef CAS PubMed.
- A. Beduer, C. Vieu, F. Arnauduc, J. C. Sol, I. Loubinoux and L. Vaysse, Biomaterials, 2012, 504–514 CrossRef CAS PubMed.
- A. M. Rajnicek, S. Britland and C. D. McCaig, J. Cell Sci., 1997, 2905–2913 CAS.
- P. Clark, P. Connolly, A. S. G. Curtis, J. A. T. Dow and C. D. W. Wilkinson, J. Cell Sci., 1991, 73–77 CAS.
- T. Kawano, M. Sato, H. Yabu and M. Shimomura, Biomater. Sci., 2014, 52–56 RSC.
- C. Wilkinson, M. Riehle, M. Wood, J. Gallagher and A. Curtis, Mater. Sci. Eng., 2002, 263–269 CrossRef CAS.
- H. Cha, J. Hong, T. Kang, J. Jung and D. Cho, J. Micromech. Microeng., 2012 Search PubMed , 12.
- J. Charest, A. Garcia and W. King, Biomaterials, 2007, 2202–2210 CrossRef CAS PubMed.
- A. Andersson, F. Backhed, A. Euler, A. Richter-Dahlfors, D. Sutherland and B. Kasemo, Biomaterials, 2003, 3427–3436 CrossRef CAS.
- W. Hu, E. Yim, R. Reano, K. Leong and S. Pang, J. Vac. Sci. Technol., B: Microelectron. Nanometer Struct. – Process., Meas., Phenom., 2005, 2984–2989 CrossRef CAS PubMed.
- E. Yim, R. Reano, S. Pang, A. Yee, C. Chen and K. Leong, Biomaterials, 2005, 5405–5413 CrossRef CAS PubMed.
- M. Mahmoudi, S. Bonakdar, M. Shokrgozar, H. Aghaverdi, R. Hartmann, A. Pick, G. Witte and W. Parak, ACS Nano, 2013, 8379–8384 CrossRef CAS PubMed.
- O. Mashinchian, S. Bonakdar, H. Taghinejad, V. Satarifard, M. Heidari, M. Majidi, S. Sharifi, A. Peirovi, S. Saffar, M. Taghinejad, M. Abdolahad, S. Mohajerzade, M. Shokrgozar, M. Rezayat, M. Ejtehadi, M. Dalby and M. Mahmoudi, ACS Appl. Mater. Interfaces, 2014, 13280–13292 CAS.
- S. Bonakdar, M. Mahmoudi, L. Montazeri, M. Taghipoor and A. Bretsch, et al. , ACS Appl. Mater. Interfaces, 2016, 13777–13784 CAS.
- F. Guilak, D. Cohen, B. Estes, J. Gimble, W. Liedtke and C. Chen, Cell Stem Cell, 2009, 17–26 CrossRef CAS PubMed.
- Y. Han, S. Wang and X. Zhang, et al. , Drug Discovery Today, 2014, 763–773 CrossRef PubMed.
- N. Shah, Y. Morsi and R. Manasseh, Cell Biochem. Funct., 2014, 309–325 CrossRef CAS PubMed.
- K. Chatterjee, S. Lin-Gibson, W. Wallace, S. H. Parekh, Y. J. Lee, M. T. Cicerone, M. F. Young and C. G. J. Simon, Biomaterials, 2010, 5051–5062 CrossRef CAS PubMed.
- X. Hu, S. H. Park, E. S. Gil, X. X. Xia, A. S. Weiss and D. L. Kaplan, Biomaterials, 2011, 8979–8989 CrossRef CAS PubMed.
- D. Discher, P. Janmey and Y. Wang, Science, 2005, 1139–1143 CrossRef CAS PubMed.
- D. Fletcher and R. Mullins, Nature, 2010, 485–492 CrossRef CAS PubMed.
- J. Park, J. Chu, A. Tsou, R. Diop, Z. Tang, A. Wang and S. Li, Biomaterials, 2011, 3921–3930 CrossRef CAS PubMed.
- K. Saha, A. Keung, E. Irwin, Y. Li, L. Little, D. Schaffer and K. Healy, Biophys. J., 2008, 4426–4438 CrossRef CAS PubMed.
- A. Rowlands, P. Goeorge and J. Cooper-white, Am J Physiol Cell Physiol, 2008, 1037–1044 CrossRef PubMed.
- C. Gonzalez-Garcia, D. Mortatal, R. Oreffeo, M. Dalby and M. Salmero n-Sanchez, Integr. Biol., 2012, 531–539 RSC.
- B. Ratner, A. Hoffman, F. Schoen and J. Lemons, Biomaterials science: An introduction to materials in medicine, Academic Press, Oxford, UK, 2012 Search PubMed.
- J. Picot, Human cell culture protocols, Springer, Berlin, Germany, 2nd edn, 2005 Search PubMed.
- R. I. Freshney, G. N. Stacey and J. M. Auerbach, Culture of human stem cells, Wiley, Hoboken, NJ, 5th edn, 2007 Search PubMed.
- M. Mehrjoo, J. Javadpour, M. Shokrgozar, M. Farokhi, S. Javadian and S. Bonakdar, Mater. Express, 2015, 41–48 CrossRef CAS.
- J. E. Sader, J. W. Chon and P. Mulvaney, Rev. Sci. Instrum., 1999, 70, 3967–3969 CrossRef CAS.
- W. A. Ducker, T. J. Senden and R. M. Pashley, Nature, 1991, 353, 239 CrossRef CAS.
- J. Domke and M. Radmacher, Langmuir, 1998, 14, 3320–3325 CrossRef CAS.
- L.-Y. Lin and D.-E. Kim, Polym. Test., 2012, 31, 926–930 CrossRef CAS.
- J. Jovanovic, B. Adnadjevic, M. Kicanovic and D. Uskokovic, Colloids Surf., B, 2004, 181–186 CrossRef CAS PubMed.
- L. Rydén, O. Omar, A. Johansson, R. Jimbo, A. Palmquist and P. Thomsen, J. Mater. Sci.: Mater. Med., 2017, 28, 9 CrossRef PubMed.
- N. Roy, T. Dey, S. C. Kundu and A. K. Bhowmick, J. Mater. Sci., 2013, 48, 5132–5139 CrossRef CAS.
- S. A. Redey, M. Nardin, D. Bernache–Assolant, C. Rey, P. Delannoy, L. Sedel and P. J. Marie, J. Biomed. Mater. Res., Part A, 2000, 50, 353–364 CrossRef CAS.
- O. J. B. Ferreira, K. B. Demétrio and L. A. L. dos Santos, Composites, Part B, 2017, 2056–2067 Search PubMed.
- H. F. Brinson and L. C. Brinson, Polymer Engineering Science and Viscoelasticity: An Introduction, Springer, New York, 2nd edn, 2015 Search PubMed.
- M. T. Shaw and W. J. Macknight, Introduction to Polymer Viscoelasticity, John Wiley and Sons, Inc., New Jersey, 3rd edn, 2005 Search PubMed.
- D. J. Plazek and V. M. O'Rourke, J. Polym. Sci., 1971, 209–243 CAS.
- Y. H. Lin, Polymer Viscoelasticity: Basics, Molecular Theories, and Experiments, World Scientific Publishing, Singapore, 2003 Search PubMed.
- D. Ponnamma and S. Thomas, Non-Linear Viscoelasticity of Rubber Composites and Nanocomposites: Influence of Filler Geometry and Size in Different Length Scales, Springer, Switzerland, 2014 Search PubMed.
- L. Cheng, F. Ye, R. Yang, X. Lu, Y. Shi, L. Li, H. Fan and H. Bu, Acta Biomater., 2009, 1569–1574 Search PubMed.
- P. Habibovic, T. M. Sees, M. A. Van den Doel, C. A. van Blitterswijk and K. de Groot, InterScience, 2006, 747–762 CAS.
- H. Yuan, H. Fernandes, P. Habibovic, J. de Boer, A. M. C. Barradas, A. de Ruiter, W. R. Walsh, C. A. van Blitterswijk and J. D. de Bruijin, Proc. Natl. Acad. Sci. U. S. A., 2012, 13614–13619 Search PubMed.
- K. Webb, V. Hlady and P. Tresco, J. Biomed. Mater. Res., 1998, 422–430 CrossRef CAS.
- S. Kennedy, N. Washburn, C. Simon and E. Amis, Biomaterials, 2006, 3817–3824 CrossRef CAS PubMed.
- K. Anselme, Biomaterials, 2000, 667–681 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7bm00733g |
| This journal is © The Royal Society of Chemistry 2018 |