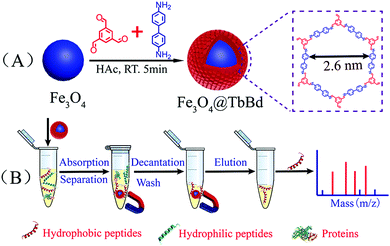
Facile synthesis of core–shell structured magnetic covalent organic framework composite nanospheres for selective enrichment of peptides with simultaneous exclusion of proteins†
Chaohong
Gao‡
a,
Guo
Lin‡
 a,
Zhixian
Lei
a,
Qiong
Zheng
a,
Jiashi
Lin
b and
Zian
Lin
*a
a,
Zhixian
Lei
a,
Qiong
Zheng
a,
Jiashi
Lin
b and
Zian
Lin
*a
aMinistry of Education Key Laboratory of Analytical Science of Food Safety and Biology, Fujian Provincial Key Laboratory of Analysis and Detection Technology for Food Safety, College of Chemistry, Fuzhou University, Fuzhou, Fujian 350116, China. E-mail: zianlin@fzu.edu.cn; Fax: +86-591-22866165
bCollege of Physical Education, Jimei University, Xiamen 361021, China
First published on 21st August 2017
Abstract
Selective enrichment of peptides from complex biosamples is essential for mass spectrometry-based proteomics but still remains a challenge. In this work, a facile approach was developed for rapid room-temperature synthesis of core–shell structured magnetic covalent organic framework composite nanospheres (denoted as Fe3O4@TbBd) by using monodisperse Fe3O4 nanoparticles as the magnetic core and 1,3,5-triformylbenzene (Tb) and benzidine (Bd) as two building blocks in the presence of dimethyl sulfoxide (DMSO). The resultant core–shell structured Fe3O4@TbBd nanospheres exhibited high adsorption capacity (28.5 mg g−1), fast adsorption kinetics (<5 min) and excellent reusability (more than 30 times) for peptides, owing to their specific properties of high surface area (196.21 m2 g−1), large pore volume (0.63 cm3 g−1), narrow pore size distribution (∼2.8 nm), strong magnetic response (41.5 emu g−1), as well as good thermal and chemical stability. Moreover, the Fe3O4@TbBd nanospheres also showed good selectivity towards hydrophobic peptides and a size-exclusion effect against proteins due to the inherent π–π stacking interaction and interconnected mesoporous channels of covalent organic framework (COF) shells. Taking advantage of these composite nanospheres, selective extraction and efficient enrichment of low abundance hydrophobic peptides from human serum in the presence of high abundance proteins were achieved. Based on the HPLC-Q-TOF/MS results, 29 hydrophobic peptides assigned to 12 proteins were clearly identified in 5 ng μL−1 human serum digestion upon treatment with Fe3O4@TbBd nanospheres, much better than those obtained without treatment, confirming the outstanding performance of the Fe3O4@TbBd nanospheres in proteome analysis.
Introduction
Peptide mapping by liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis has become one of the powerful tools for protein identification in proteomic analysis.1,2 Although the MS-based method shows some advantages of high throughput and good sensitivity,3–5 it seems insufficient to directly detect low-abundance peptides owing to the inevitable challenges in peptide isolation and enrichment from complex biological samples, such as coexistence of high-abundance proteins and other contaminants, which may severely interfere with MS signals. Therefore, developing an effective approach for selective enrichment of low-abundance peptides and simultaneous exclusion of high-abundance proteins from complex biological samples is highly desirable.Although great progress has been made in the development of nanomaterial-based approaches for peptide enrichment from complex biosamples, these nanomaterials with different surface functional groups and compositions, including silica nanospheres,6,7 carbon nanotubes,8 polymeric functionalized nanoparticles,9 zeolite nanocrystals,10 ZnO-PMMA nanobeads11 and magnetic nanoparticles,12–14 often co-enriched proteins to the outer surface through hydrophobic- and/or electrostatic interactions, thus resulting in nonspecific protein adsorption. To address this problem, various types of mesoporous materials, such as mesoporous silica microspheres,15 mesoporous carbon,16 metal–organic frameworks (MOFs)17 and magnetic mesoporous microspheres,18–20 have been developed successively. Among them, magnetic mesoporous composites, which combine the advantages of mesoporous structures, high magnetic responsiveness, and ease of manipulation, have attracted considerable attention and have been widely applied to the selective enrichment of peptides from complex biological samples. Furthermore, the superior properties of the magnetic mesoporous composites with large surface area, well-defined pore structure, and narrow pore size distribution endow them with high selectivity for capturing peptides and repelling proteins. Nevertheless, elegant fabrication of magnetic mesoporous composites with desirable functionalities still remains a big challenge due to the difficulty in controlling the synthetic conditions, tuning the pore size and maintaining the structural stability.21–23
Covalent organic frameworks (COFs), a class of novel crystalline porous polymers with well-defined structural regularity, large surface area and tunable pore structure, which could be synthesized from lightweight elements (H, B, C, N, O, Si), have received great attention since their first report in 2005.24 In contrast to MOFs, the fascinating features of COFs like tunable pore size, high chemical stability and outstanding structural stability enable their superior potential in a wide variety of applications, such as catalysis,25–27 sensing,28–30 optoelectronic devices,31–33 and gas storage.34–36 Against such a backdrop, the design and construction of porous architectures that mimic structures found in nature in synthesized materials, down to microscale and nanoscale ranges, have become an important research topic. Recently, Tan and co-workers37 successfully synthesized imine-linked magnetic COFs and reconstructed them under thermodynamic control to reform the polyimine amorphous networks into well-defined core–shell crystals for the first time. Since then, there has been rapid progress in the preparation and applications of magnetic COFs.38,39 More recently, an encouraging breakthrough in the room-temperature synthesis of core–shell structured magnetic COFs was achieved by our group,40 which has been successfully applied to capture peptides with simultaneous exclusion of proteins from complex biological samples. Nevertheless, the potential advantages of the magnetic COFs and the facile synthetic strategy have not been fully demonstrated. Further development is very necessary to address the merits and explore new applications of this class of magnetic COFs.
Herein, we developed a facile approach for rapid room-temperature synthesis of the core–shell structured Fe3O4@TbBd composite nanospheres. The as-prepared Fe3O4@TbBd nanospheres showed high magnetization saturation, large surface area, accessible mesopores, and outstanding thermal and chemical stability. Taking these advantages together, the Fe3O4@TbBd nanospheres could be applied to the selective enrichment of peptides and simultaneous exclusion of proteins prior to HPLC-MS analysis. The adsorption performance and selectivity were evaluated by examining the adsorption capacity, adsorption kinetics and chromatographic selectivity of the Fe3O4@TbBd nanospheres towards peptides. In addition, the practicabilities for real-world applications were further assessed by the enrichment peptides for human serum samples and their tryptic digestion.
Experimental
Materials
All chemicals and reagents were commercially available and used without further purification. Ferric chloride hexahydrate (FeCl3·6H2O), trisodium citrate dihydrate (Na3Cit·2H2O), dimethyl sulfoxide, tetrahydrofuran, ethanol and methanol were supplied by Sinopharm Chemical Reagent, Co., Ltd (Shanghai, China). 1,3,5-Triformylbenzene (Tb) was purchased from J&K Chemical Ltd (Shanghai, China). Trifluoroacetic acid (TFA), formic acid (FA), sodium acetate, ethylene glycol, trypsin (DPCC treated, from bovine pancreas), concanavalin A (Con A), insulin, lysozyme (Lyz), and benzidine (Bd) were obtained from Aladdin Chemistry Co., Ltd (Shanghai, China). Four peptides (Phe-Gly-Phe-Gly-Phe (FGFGF, Mw = 573.66, Grand Average of Hydropathy (GRAVY) = 1.52), Gly-Gly-Phe-Gly-Gly (GGFGG, Mw = 393.35, GRAVY = 0.24), Tyr-Gly-Gly-Phe-Me (YGGFM, Mw = 573.67, GRAVY = 0.52), and Tyr-Gly-Gly-Phe-Met (YGGFL, Mw = 555.64, GRAVY = 0.9)) were purchased from Guotai Bio-technology Co. Ltd (Hefei, China). Healthy human serum was kindly donated by Fujian Provincial Official Hospital (Fuzhou, China). Deionized water (18.2 MΩ cm−1) was produced by using a Milli-Q water purification system (Millipore, USA).Preparation of Fe3O4 magnetic nanoparticles
The magnetic nanoparticles (MNPs) were prepared according to the previous literature with some modification.41,42 Firstly, FeCl3·6H2O (8.1 g, 29.97 mmol) and Na3Cit·2H2O (1.5 g, 5.10 mmol) were first dissolved in ethylene glycol (200 mL) by stirring vigorously, followed by addition of sodium acetate (12.0 g, 146.29 mmol). After that, the obtained homogeneous faint yellow suspension was transferred to a Teflon-lined stainless-steel autoclave (50 mL capacity), and then heated up to 200 °C for 12 h. After cooling to room temperature, the black product was washed with deionized water and ethanol several times. Finally, the product was dried under vacuum at room temperature.Preparation of core–shell structured Fe3O4@TbBd composite nanospheres
The synthesis of core–shell structured Fe3O4@TbBd nanospheres was carried out as follows. In brief, the Fe3O4 MNPs (0.15 g, 0.65 mmol) were suspended in a solution of 50 mL of DMSO containing Tb (0.3 mmol) and Bd (0.45 mmol), and then sonicated for 5 min. After that, acetic acid (17.5 M, 2 mL) was slowly added to the mixture. Then, the reaction was allowed to proceed at room temperature for 5 min and the brown precipitates (Fe3O4@TbBd) were taking shape in the beaker. After incubation for 15 min, the yielded brown precipitate was collected with the help of a magnet. After being washed with tetrahydrofuran and anhydrous methanol three times, the brown precipitate was dried under vacuum at room temperature prior to use.Characterization
Scanning electron microscopy (SEM) images were obtained on an FEI Emission Scanning Electron Microscope (SU8020, Hitachi, Japan). Transmission electron microscopy (TEM) images were observed using a Hitachi HT7700 operating at 100 kV. Fourier-transform infrared (FTIR) spectra were collected with a Nicolet 6700 spectrometer (Thermo Fisher, USA) using KBr pellets in the wavenumber region of 4000–400 cm−1. The Brunauer–Emmett–Teller (BET) surface areas were calculated from N2 adsorption–desorption isotherms at 77 K using an ASAP 2020 (Micromeritics, USA). The samples were degassed in a vacuum at 150 °C for 8 h before measurement. The total pore volume, VT, was recorded using the desorption branch of the N2 isotherm at P/P0 = 0.992. The pore volume and pore size distribution data were calculated based on the Barrett–Joyner–Halenda (BJH) model in the Micromeritics ASAP2020 software package. Thermogravimetric analysis (TGA) was performed on a Shimadzu DTG-60AH in the temperature range of 100 to 800 °C under flowing N2 (100 mL min−1) with a heating rate of 10 °C min−1. Powder X-ray diffraction (XRD) patterns were carried out on an X'Pert Pro MPD (Philips, Holland) using Cu Kα radiation at 40 mA to 40 kV. The magnetization curves were conducted on an MPMS XL-7 superconducting quantum interference device (SQUID) magnetometer (Quantum Design, USA) at 300 K.HPLC analysis was performed on a Prominence LC-20A (Shimadzu, Kyoto, Japan) equipped with an ultraviolet detector (UV-detector). A Venusil XBP C8 column (100 mm × 4.6 mm, 5 μm, 300 Å) (Agela Technologies, Tianjin, China) and an InertSustain C18 column (150 mm × 4.6 mm, 5 μm) (GLSciences Inc., Tokyo, Japan) were adopted unless stated otherwise. The peptide and protein mixture was separated with a binary solvent mixture of acetonitrile–water (ACN/H2O) under gradient mode: mobile phase A (0.1% TFA aqueous solution) and mobile phase B (ACN with 0.08% TFA). The flow rate was set as 1.0 mL min−1 and the column temperature was maintained at 40 °C steadily.
Adsorption performance evaluation of peptides
The adsorption experiments were conducted in a centrifuge tube by using deionized water as the solvent of all samples under ambient conditions. Briefly, 0.3 mg of Fe3O4@TbBd nanospheres were incubated with 0.3 mL of FGFGF (GGFGG, YGGFM or YGGFL) solution at different concentrations. The nanosphere–peptide mixture was first vortex-mixed for 1 min and was subjected to additional shaking for another 20 min. After that, an external magnet was employed to isolate the mixture. Then the supernatants were collected and analyzed using high performance liquid chromatography with ultraviolet/visible spectroscopy (HPLC-UV/Vis). The equilibrium adsorption capacity (Qe, mg g−1) was given as follows:| Qe = (C0 − Ce) V/m | (1) |
In the time-dependent absorption experiments, 0.3 mg of Fe3O4@TbBd nanospheres were incubated with 0.3 mL of FGFGF (GGFGG, YGGFM or YGGFL) aqueous solution (each of peptides, 70 μg mL−1) at different given times in the range of 0–60 min. The nanoparticle–peptide mixture was first vortex-mixed and then was subjected to additional shaking at given times. After that, an external magnet was employed to isolate the mixture. Then the supernatants were collected prior to HPLC-UV/Vis analysis.
In the specific adsorption experiments, 3 mg of Fe3O4@TbBd nanospheres were incubated with 1 mL of the mixture containing FGFGF (25 μg mL−1), Con A (25 μg mL−1), insulin (25 μg mL−1) and Lyz (25 μg mL−1), (or incubated with 1 mL of 100-fold diluted human serum spiked with 0.5 μg mL−1 FGFGF) for 10 min, respectively. After magnetic separation, the supernatants were collected. The peptide-captured Fe3O4@TbBd nanospheres were eluted with an eluent (50% ACN aqueous solution). The supernatants and the eluates were analyzed using HPLC-UV/Vis and electrospray ionisation-quadrupole-time-of-flight mass spectrometry (ESI-Q-TOF/MS).
Reusability of the Fe3O4@TbBd nanospheres
To estimate the reusability of the Fe3O4@TbBd nanospheres, 0.3 mg of Fe3O4@TbBd nanospheres were incubated with 0.3 mL of FGFGF at a concentration of 70 μg mL−1 for 10 min at room temperature. After magnetic separation, the supernatants were collected. The nanosphere–peptide conjugates were then washed with 300 μL of eluent (50% ACN aqueous solution). After washing with water several times, the obtained Fe3O4@TbBd nanospheres were reused for the next cycle adsorption. Both the supernatants and the eluates were analyzed using HPLC-UV/Vis.Real sample preparation and analysis
0.1 mL of human serum was dissolved in 5.9 mL of 50 mM ammonium bicarbonate (NH4HCO3) buffer (pH 8.3) to a concentration of 1000 μg mL−1 and then treated in a water bath at 100 °C for 5 min. After being reduced, the products were alkylated using iodoacetic acid (IAA) (final concentration: 10 mM) in the dark for 1 h at room temperature. Subsequently, the digestion was carried out via adding trypsin into the above sample with a substrate-to-enzyme ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w) for 24 h at 37 °C. Subsequently, 1% FA was used to terminate the reaction process. All the obtained samples were stored at −20 °C and further diluted to the concentration of 5 ng μL−1 and 0.5 ng μL−1 before analysis.
1 (w/w) for 24 h at 37 °C. Subsequently, 1% FA was used to terminate the reaction process. All the obtained samples were stored at −20 °C and further diluted to the concentration of 5 ng μL−1 and 0.5 ng μL−1 before analysis.
In the experiments of real sample analysis, 5 mg of Fe3O4@TbBd was incubated with 1 mL of human serum digestion (∼5 and 0.5 ng μL−1, respectively) for 10 min. After magnetic separation, the supernatants were collected. The captured peptides were eluted with 300 μL of eluent (50% ACN aqueous solution) and collected, followed by HPLC-Q-TOF/MS analysis.
Results and discussion
Preparation and characterization of Fe3O4@TbBd nanospheres
The general scheme for the facile synthesis of core–shell magnetic Fe3O4@TbBd composite nanospheres is illustrated in Fig. 1(A), which mainly involved two steps: (1) firstly, solvothermal synthesis of the citrate-stabilized Fe3O4 nanoparticles, which could significantly enhance the aqueous dispersion stability. (2) Secondly, rapid room-temperature synthesis of the core–shell structured Fe3O4@TbBd nanospheres in a one pot process by using the monodisperse Fe3O4 nanoparticles (0.65 mmol) as the magnetic core and 1,3,5-triformylbenzene (Tb, 0.3 mmol) and benzidine (Bd, 0.45 mmol) as two building blocks in the presence of DMSO. The developed approach in current work was very simple, efficient and time-saving. As displayed in Fig. 1(B), the as-prepared Fe3O4@TbBd nanospheres could be applied to selectively enrich peptides and simultaneously repel macromolecules (e.g. proteins) from complex biological samples with the help of an external magnet.The morphologies of the bare Fe3O4 MNPs and Fe3O4@TbBd nanospheres were verified by means of scanning electron microscopy (SEM) and transmission electron microscopy (TEM). The SEM images (Fig. 2(A)) showed that the bare Fe3O4 MNPs had good monodispersity and were nearly spherical in shape with an average size of 200 nm. Although the Fe3O4@TbBd nanospheres displayed the same morphology as the bare Fe3O4 MNPs, its surface became rough, suggesting the formation of COF shells (Fig. 2(B)). A comparison between Fig. 2(C) and (D) confirmed that the Fe3O4@TbBd nanospheres have a distinct core–shell structure and the thickness of COF shells surrounding the Fe3O4 core was approximately 40 nm.
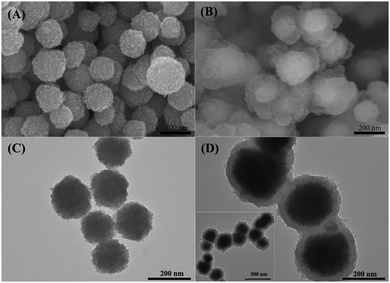 | ||
| Fig. 2 SEM (A and B) and TEM (C and D) images of the Fe3O4 MNPs and Fe3O4@TbBd nanospheres, inset: image of a larger scale of Fe3O4@TbBd nanospheres. | ||
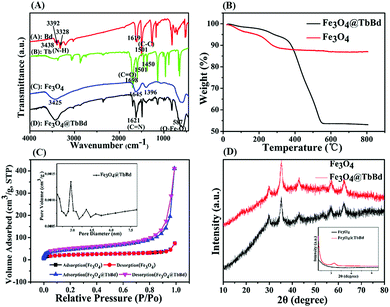
Fourier transform infrared (FT-IR) spectroscopy was further carried out to verify the successful preparation of the imine-linked Fe3O4@TbBd nanospheres. As presented in Fig. 3(A), the adsorption bands of Bd at 3328 cm−1, 3392 cm−1, and 3438 cm−1 were ascribed to the N–H stretch modes (Spectrum A). The peak at 1698 cm−1, and the bands at 1450 cm−1 and 1501 cm−1 were assigned to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching and aromatic C–C ring stretching of Tb, respectively (Spectrum B). The band at 587 cm−1 was assigned to the characteristic Fe–O–Fe stretch mode and the peaks at 1396 cm−1 and 1645 cm−1 were assigned to the carbonyl stretching of the carboxyl groups on the surface of Fe3O4 MNPs (Spectrum C), demonstrating the existence of carboxyl groups. In comparison to the spectrum of Fe3O4 MNPs, similar adsorption bands were also observed in the spectrum of the Fe3O4@TbBd nanospheres. Furthermore, the emerging stretch at 1621 cm−1 was also found, which was attributed to the C
O stretching and aromatic C–C ring stretching of Tb, respectively (Spectrum B). The band at 587 cm−1 was assigned to the characteristic Fe–O–Fe stretch mode and the peaks at 1396 cm−1 and 1645 cm−1 were assigned to the carbonyl stretching of the carboxyl groups on the surface of Fe3O4 MNPs (Spectrum C), demonstrating the existence of carboxyl groups. In comparison to the spectrum of Fe3O4 MNPs, similar adsorption bands were also observed in the spectrum of the Fe3O4@TbBd nanospheres. Furthermore, the emerging stretch at 1621 cm−1 was also found, which was attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretch mode. The result indicated that the COF shells of TbBd have been successfully covered on the surface of Fe3O4 MNPs via Schiff-base condensation reaction (Spectrum D).
N stretch mode. The result indicated that the COF shells of TbBd have been successfully covered on the surface of Fe3O4 MNPs via Schiff-base condensation reaction (Spectrum D).
Thermogravimetric analysis (TGA) revealed the thermal stability and mass ratios of different components (Fig. 3(B)). As for the bare Fe3O4 MNPs, a distinct weight-loss step of 12% up to ca. 300 °C was observed which was attributed to the loss of guest molecules (e.g. H2O) from the surface or unreacted species (e.g. trisodium citrate dihydrate) from the Fe3O4 MNPs. In contrast, the Fe3O4@TbBd nanospheres displayed a sharp weight-loss profile in the range of 350–550 °C and there was approximately 34% weight loss in the studied range, implying the weight loss of COF shells. It is noteworthy that a long plateau was observed under 350 °C, demonstrating the high thermal stability of the Fe3O4@TbBd nanospheres.
Brunauer–Emmett–Teller (BET) gas sorptometry measurements were performed to evaluate the pore properties of the Fe3O4@TbBd nanospheres. As presented in Fig. 3(C), the isotherm of Fe3O4 basically belonged to IUPAC type II, indicating the obviously nonporous structure of the solid sphere. In contrast, the Fe3O4@TbBd nanospheres exhibited a typical type IV isotherm, validating a mesoporous characteristic. The pore-size distribution (the inset of Fig. 3(C)) revealed that the Fe3O4@TbBd nanospheres contained an average pore size of 2.8 nm with a narrow size distribution, which was well in accordance with the theoretical value of the bulk COFs (2.6 nm).43 In addition, The specific surface area and the pore volume of the Fe3O4@TbBd nanospheres were calculated to be 196.21 m2 g−1 and 0.63 cm3 g−1, respectively. The high external surface area and mesoporous structure endowed the Fe3O4@TbBd nanospheres with high adsorption capacity and excellent selectivity for peptides.
The crystalline structure and chemical stability of the Fe3O4@TbBd nanospheres were further examined using powder X-ray diffraction (XRD) analysis and the result is shown in Fig. 3(D). Wide-angle XRD of the Fe3O4@TbBd nanospheres exhibited 6 peaks with 2θ at 30.28°, 35.46°, 43.40°, 53.28°, 57.14° and 62.76°, corresponding to (220), (311), (400), (422), (511) and (440), which matched well with those from the JCPDS card (75-1609) for magnetite, indicating that the products were well crystallized and had high crystallinity even after coating COF shells. Moreover, the comparison of the Fe3O4 MNPs and Fe3O4@TbBd nanospheres also suggested that the broad diffraction peak at ∼3.5° might be due to the low crystallinity of the COF shells (the inset of Fig. 3(D)). To examine the chemical stability of the Fe3O4@TbBd nanospheres, the products were dispersed in ACN, DMF, 10 mM HCl, 1 mM NaOH and water overnight and then measured using small-angle XRD and FT-IR analysis. Surprisingly, the results indicated that all the products exhibited similar diffraction peaks in XRD patterns and adsorption bands in FT-IR spectra (Fig. S1 and S2, ESI†), indicating the good chemical stability of the Fe3O4@TbBd nanospheres. Apparently, the stable C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N covalent bond assisted imine-linkage networks contributed to the good chemical stability.
N covalent bond assisted imine-linkage networks contributed to the good chemical stability.
The magnetic properties of the Fe3O4@TbBd nanospheres were further characterized with a superconductive quantum interference device (SQUID) at 300 K. The magnetic hysteresis curve (Fig. 4) showed that the Fe3O4@TbBd nanospheres had superparamagnetic properties possessing a saturated magnetization value of 41.5 emu g−1, just a little drop compared with the bare Fe3O4 MNPs (54.7 emu g−1). The high saturation magnetization endowed the Fe3O4@TbBd nanospheres with fast response to an external magnetic field. As presented in the inset of Fig. 4, the Fe3O4@TbBd nanospheres were well dispersed in water and presented a brown suspension. However, rapid aggregation (∼1 min) of the Fe3O4@TbBd nanospheres from the homogeneous suspension was obtained with the help of an external magnet and thus the dispersed solution became clear.
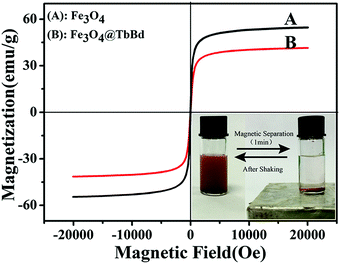 | ||
| Fig. 4 The magnetic hysteresis curve of the Fe3O4 MNPs and Fe3O4@TbBd nanospheres and the inset shows the magnetic separation behavior of the Fe3O4@TbBd nanospheres in aqueous solution. | ||
Adsorption capacity and kinetics of the Fe3O4@TbBd nanospheres
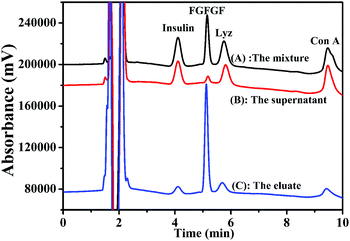
The feasibility of the Fe3O4@TbBd nanospheres as an adsorbent for peptide enrichment was evaluated by using four representative peptides (FGFGF, GGFGG, YGGFM, and YGGFL) as test compounds, which have different polarities, molecular weights and phenyl groups. Fig. 5(A) shows a baseline separation of the four peptide standards before treatment with the Fe3O4@TbBd nanospheres. After treatment, analysis of the supernatant revealed that nearly 89% of FGFGF, but only 28% of YGGFL, 27% of YGGFM, and 6% of GGFGG were captured by the Fe3O4@TbBd nanospheres from the mixture (Fig. 5(B)). It should be mentioned that the extraction efficiencies of the Fe3O4@TbBd nanospheres for the four peptides were ranked in descending order, FGFGF > YGGFL > YGGFM > GGFGG, which matched well with their GRAVY values. Apparently, van der Waals forces and strong π–π interactions between the COF shells and the phenyl groups of peptides can contribute to the hydrophobic peptide extraction and enrichment. Subsequently, the peptide-captured Fe3O4@TbBd nanospheres were eluted with 50% ACN aqueous solution, and more than 60% FGFGF was recovered in the eluent (Fig. 5(C)).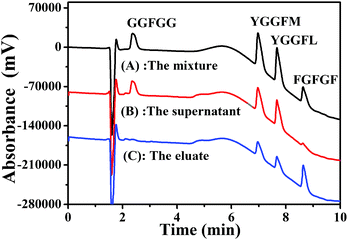 | ||
| Fig. 5 (A–C) Separation of four peptides before and after treatment with the Fe3O4@TbBd nanospheres: (A) the peptide standards; (B) the supernatant; (C) the eluate. | ||
Accordingly, the adsorption capacities of the Fe3O4@TbBd nanospheres for the four peptides were further investigated and the result is presented in Fig. 6(A). It was clearly observed that the adsorption capacity of FGFGF on the Fe3O4@TbBd nanospheres remarkably increased with an increasing concentration of FGFGF and reached equilibrium over 70 μg mL−1. In this case, the corresponding experimental saturated adsorption capacity was estimated to be 29.7 mg g−1, much higher than those obtained for the other three peptides, which were 6.6 mg g−1 for YGGFL, 6.4 mg g−1 for YGGFM, and 4.2 mg g−1 for GGFGG, respectively. The results were well coincident with those obtained in chromatographic separation, demonstrating the high selectivity and adsorption capacity of the Fe3O4@TbBd nanospheres for hydrophobic peptides. In addition, the adsorption kinetics of the peptides on the Fe3O4@TbBd nanospheres was also studied and the result (Fig. 6(B)) showed that the Fe3O4@TbBd nanospheres took up 98% of FGFGF within 5 min, and the equilibrium time was less than 10 min. Obviously, the strong interaction between the COF shells and the peptides, and the high porosity of the Fe3O4@TbBd nanospheres with low mass-transfer resistance can respond to the fast adsorption kinetics. In addition, the reusability of the Fe3O4@TbBd nanospheres was examined and the result (Fig. S3, ESI†) demonstrated less than 2.5% loss in adsorption capacity of FGFGF after 30 adsorption–regeneration cycles. The superior reusability of the Fe3O4@TbBd nanospheres was mainly due to the excellent chemical stability of the COF shells.
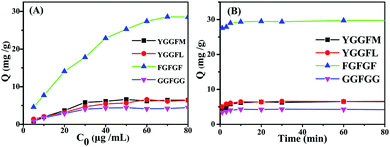 | ||
| Fig. 6 (A) Adsorption isotherms and (B) adsorption kinetics of peptides on the Fe3O4@TbBd nanospheres. | ||
Size-exclusion effect and protein repellency of the Fe3O4@TbBd nanoparticles
A distinctive feature of the Fe3O4@TbBd nanospheres is the size-exclusion effect due to its unique mesoporous structure. Hereto, three proteins with different sizes and molecular weights, including insulin (4.6 × 6.2 × 5.6 nm, Mw: 5.8 kDa), Lyz (7.7 × 7.7 × 3.7 nm, Mw: 14 kDa) and Con A (6.3 × 8.6 × 8.9 nm, Mw: 25.5 kDa) were mixed with FGFGF and adopted as test compounds to evaluate the cutoff mechanism. Fig. 7(A) presents the baseline separation of FGFGF and proteins. After treatment with the Fe3O4@TbBd nanospheres, approximately 90% of FGFGF was captured from the mixture. However, more than 95% of insulin, Lyz and Con A remained in the supernatant (Fig. 7(B)). This result can be explained by the fact that the Fe3O4@TbBd nanospheres have a mesoporous structure with an average pore diameter of 2.8 nm, and therefore the proteins with large sizes were excluded from the mesoporous channels. In contrast, the small-size peptides could easily enter into the mesoporous channels and be captured based on the van der Waals forces and strong π–π interactions. In addition, it was found in the eluate that ∼65% of FGFGF with less than 3% of insulin, Lyz and Con A were obtained (Fig. 7(C)).Selective enrichment of peptide from spiked human serum
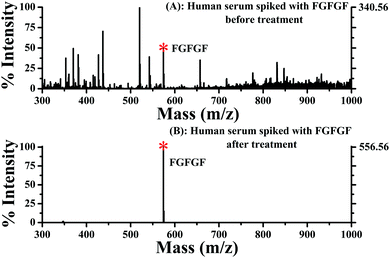
The good performance of the Fe3O4@TbBd nanospheres encouraged us to examine its capacity for selective enrichment of low-abundance peptides from complex biological samples. Herein, a 50-fold diluted human serum spiked with 25 μg mL−1 FGFGF was analyzed using an HPLC-UV/Vis detector. It was observed from Fig. S4 (ESI†) that the coexistence of multiple high-abundance proteins such as albumin and immunoglobulins in human serum complicated the separation and detection of FGFGF. When treated with Fe3O4@TbBd nanospheres, however, most FGFGF was captured from the human serum sample. As a result, no obvious chromatographic peak of FGFGF was detected in the supernatant. Meanwhile, some major proteins still remained in the supernatant without obvious changes in their peak intensities, indicating that these proteins were not captured using the Fe3O4@TbBd nanospheres. Similarly, 60% of FGFGF could be recovered in the eluate, confirming again the feasibility of the Fe3O4@TbBd nanospheres as an absorbent for selective and efficient enrichment of hydrophobic peptides.To gain a better understanding of the overwhelming selectivity of the sorbent, 0.5 ng μL−1 of FGFGF was spiked into 100-fold diluted human serum as a mixture solution and further analyzed using the direct Q-TOF/MS method. As shown in Fig. 8(A), the complicated mass spectra obtained from the mixture solution without treatment make it rather difficult for direct identification of FGFGF, in which the protein-related ions with high background noise dominated the spectra in the m/z 300–1000 region. In comparison with Fig. 8(A), Fig. 8(B) exhibited a clean background in the studied mass region and sensitive detection of FGFGF (more than 3-fold higher signal intensities compared with those obtained without treatment) was achieved after treatment with the Fe3O4@TbBd nanospheres, demonstrating the superior selectivity and high efficiency of the Fe3O4@TbBd nanospheres for hydrophobic peptides.
Analysis of human serum tryptic digestion
The practicability of the Fe3O4@TbBd nanospheres for selective enrichment of hydrophobic peptides was further evaluated by analyzing human serum digest (5 ng μL−1 and 0.5 ng μL−1, respectively) before and after treatment with the nanospheres. As for 5 ng μL−1 human serum tryptic digestion, a total of 29 hydrophobic peptides assigned to 12 proteins with higher signal intensities were identified after treatment with the Fe3O4@TbBd nanospheres, much more than those obtained before treatment (Tables S1 and S2, ESI†). In addition, the limit of detection (LOD) for serum digest after enrichment by the Fe3O4@TbBd nanospheres was also examined. With the decrease of serum concentration to 0.5 ng μL−1, ten peptides (Tables S3 and S4, ESI†) could be detected before enrichment, however, the MS signal intensities of these peptides were rather low because of the serious background interference. In contrast, seven peptides with greatly enhanced signal-to-noise (S/N) ratio were unambiguously observed after enrichment. It should be mentioned that six out of seven peptides were hydrophobic. These results can bear a comparison with our previous work,40 validating the good performance of the Fe3O4@TbBd nanospheres in selective enrichment of hydrophobic peptides.Conclusions
In summary, we developed a facile approach for rapid room-temperature synthesis of core–shell structured Fe3O4@TbBd composite nanospheres. The synthetic method was simple, efficient and time-saving, which was quite different from the traditional solvothermal method. The resultant Fe3O4@TbBd nanospheres possessed high saturation magnetization, large surface area, uniform mesoporous structure, excellent chemical stability and selectivity. Taking these advantages together, the Fe3O4@TbBd nanospheres could be applied to selectively enrich hydrophobic peptides with high adsorption capacity, fast binding kinetics, and simultaneous exclusion of proteins. In addition, their successful application in the selective enrichment of hydrophobic peptides from tryptic digestion of human serum demonstrated their promising potential in proteomic analysis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21375018, 31401017 and 21675025) and the Program for Changjiang Scholars and Innovative Research Team in University (No. IRT15R11).Notes and references
- M. Mann, P. Hojrup and P. Roepstorff, Biol. Mass Spectrom., 1993, 22, 338–345 CrossRef CAS PubMed.
- J. R. Yates, S. Speicher, P. R. Griffin and T. Hunkapiller, Anal. Biochem., 1993, 214, 397–408 CrossRef CAS PubMed.
- V. C. Wasinger, S. J. Cordwell, A. Cerpa-poljak, J. X. Yan, A. A. Gooley, M. R. Wilkins, M. W. Duncan, R. Harris, K. L. Williams and I. Humphery-smith, Electrophoresis, 1995, 16, 1090–1094 CrossRef CAS PubMed.
- S. Fields, Science, 2001, 291, 1221–1224 CrossRef CAS PubMed.
- A. Abbott, Nature, 2001, 409, 747 CrossRef CAS PubMed.
- H. M. Chen, X. Q. Xu, N. Yao, C. H. Deng, P. Y. Yang and X. M. Zhang, Proteomics, 2008, 8, 2778–2784 CrossRef CAS PubMed.
- N. Lv, Z. G. Wang, W. Z. Bi, G. M. Li, J. L. Zhang and J. Z. Ni, J. Mater. Chem. B, 2016, 4, 4402–4409 RSC.
- S. F. Ren and Y. L. Guo, J. Am. Soc. Mass Spectrom., 2006, 17, 1023–1027 CrossRef CAS PubMed.
- H. M. Chen, C. H. Deng and X. M. Zhang, Angew. Chem., Int. Ed., 2010, 49, 607–611 CrossRef CAS PubMed.
- Y. H. Zhang, X. Y. Wang, W. Shan, B. Y. Wu, H. Z. Fan, X. J. Yu, Y. Tang and P. Y. Yang, Angew. Chem., Int. Ed., 2005, 44, 615–617 CrossRef CAS PubMed.
- W. W. Shen, H. M. Xiong, Y. Xu, S. J. Cai, H. J. Lu and P. Y. Yang, Anal. Chem., 2008, 80, 6758–6763 CrossRef CAS PubMed.
- M. Zhao, C. H. Deng and X. M. Zhang, ACS Appl. Mater. Interfaces, 2013, 5, 13104–13112 CAS.
- X. N. Sun, X. L. Liu, J. N. Feng, Y. Li, C. H. Deng and G. L. Duan, Anal. Chim. Acta, 2015, 880, 67–76 CrossRef CAS PubMed.
- J. R. Whiteaker, L. Zhao, H. Y. Zhang, L. C. Feng, B. D. Piening, L. Anderson and A. G. Paulovich, Anal. Biochem., 2007, 362, 44–54 CrossRef CAS PubMed.
- L. Zhang, S. Wu, C. Li and Q. Yang, Chem. Commun., 2012, 48, 4190–4192 RSC.
- H. Q. Qin, P. Gao, F. J. Wang, L. Zhao, J. Zhu, A. Q. Wang, T. Zhang and H. F. Zou, Angew. Chem., Int. Ed., 2011, 50, 12218–12221 CrossRef CAS PubMed.
- Z. Y. Gu, Y. J. Chen, J. Q. Jiang and X. P. Yan, Chem. Commun., 2011, 47, 4787–4789 RSC.
- H. Wan, H. Q. Qin, Z. C. Xiong, W. B. Zhang and H. F. Zou, Nanoscale, 2013, 5, 10936–10944 RSC.
- M. Zhao, C. H. Deng, X. M. Zhang and P. Y. Yang, Proteomics, 2013, 13, 3387–3392 CrossRef CAS PubMed.
- H. M. Chen, S. S. Liu, H. L. Yang, Y. Mao, C. H. Deng, X. M. Zhang and P. Y. Yang, Proteomics, 2010, 10, 930–939 CAS.
- G. T. Zhu, X. Chen, X. M. He, H. Wang, Z. Zhang and Y. Q. Feng, Chem. – Eur. J., 2015, 21, 4450–4456 CrossRef CAS PubMed.
- F. Liu, P. Yuan, J. J. Wan, K. Qian, G. F. Wei, J. Yang, B. H. Liu, Y. H. Wang and C. Z. Yu, J. Nanosci. Nanotechnol., 2011, 11, 5215–5222 CrossRef CAS PubMed.
- J. N. Zheng, Y. Xiao, L. Wang, Z. A. Lin, H. H. Yang, L. Zhang and G. N. Chen, J. Chromatogr. A, 2014, 1358, 29–38 CrossRef CAS PubMed.
- A. P. Cote, A. I. Benin, N. W. Ockwig, M. Okeeffe and A. J. Matzger, Science, 2005, 310, 1166–1170 CrossRef CAS PubMed.
- S. Y. Ding, J. Gao, Q. Wang, Y. Zhang, W. G. Song, C. Y. Su and W. Wang, J. Am. Chem. Soc., 2011, 133, 19816–19822 CrossRef CAS PubMed.
- X. R. Wang, X. Han, J. Zhang, X. W. Wu, Y. Liu and Y. Cui, J. Am. Chem. Soc., 2016, 138, 12332–12335 CrossRef CAS PubMed.
- H. Xu, J. Gao and D. L. Jiang, Nat. Chem., 2015, 7, 905–912 CrossRef CAS PubMed.
- Z. P. Li, Y. W. Zhang, H. Xia, Y. Mu and X. M. Liu, Chem. Commun., 2016, 52, 6613–6616 RSC.
- R. Gomes and A. Bhaumik, RSC Adv., 2016, 6, 28047–28054 RSC.
- S. Dalapati, S. B. Jin, J. Gao, Y. H. Xu, A. Nagai and D. L. Jiang, J. Am. Chem. Soc., 2013, 135, 17310–17313 CrossRef CAS PubMed.
- M. Dogru, M. Handloser, F. Auras, T. Kunz, D. Medina, A. Hartschuh, P. Knochel and T. Bein, Angew. Chem., Int. Ed., 2013, 52, 2920–2996 CrossRef CAS PubMed.
- S. Wan, J. Guo, J. Kim, H. Ihee and D. L. Jiang, Angew. Chem., Int. Ed., 2008, 47, 8826–8830 CrossRef CAS PubMed.
- M. Dogru and T. Bein, Chem. Commun., 2014, 50, 5531–5546 RSC.
- M. G. Rabbani, A. K. Sekizkardes, Z. Kahveci, T. E. Reich, R. Ding and H. M. EI-Kaderi, Chem. – Eur. J., 2013, 19, 3324–3328 CrossRef CAS PubMed.
- Y. H. Jin, Y. L. Zhu and W. Zhang, CrystEngComm, 2013, 15, 1484–1499 RSC.
- P. Mohanty, L. D. Kull and K. Landskron, Nat. Chem., 2011, 2, 401 Search PubMed.
- J. Tan, S. Namuangruk, W. F. Kong, N. Kungwan, J. Guo and C. C. Wang, Angew. Chem., Int. Ed., 2016, 128, 14185–14190 CrossRef.
- Y. Li, C. X. Yang and X. P. Yan, Chem. Commun., 2017, 53, 2511–2514 RSC.
- S. J. He, T. Zeng, S. H. Wang, H. Y. Niu and Y. Q. Cai, ACS Appl. Mater. Interfaces, 2017, 9, 2959–2965 CAS.
- G. Lin, C. H. Gao, Q. Zheng, Z. X. Lei, H. J. Geng, Z. A. Lin, H. H. Yang and Z. W. Cai, Chem. Commun., 2017, 53, 3649–3652 RSC.
- J. N. Zheng, Z. A. Lin, G. Lin, H. H. Yang and L. Zhang, J. Mater. Chem. B, 2015, 3, 2185–2191 RSC.
- J. Liu, Z. K. Sun, Y. H. Deng, Y. Zou, C. Y. Li, X. H. Guo, L. Q. Xiong, Y. Gao, F. Y. Li and D. Y. Zhao, Angew. Chem., Int. Ed., 2009, 121, 5989–5993 CrossRef.
- L. R. Xu, X. Zhou, Y. X. Yu, W. Q. Tian, J. Ma and S. B. Lei, ACS Nano, 2013, 7, 8066–8073 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7tb01807j |
| ‡ The first two authors contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2017 |