Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Identification of cadmium resistance and adsorption gene from Escherichia coli BL21 (DE3)†
Weitong Qin‡
,
Xiaoqing Liu‡,
Xiaoxia Yu,
Xiaoyu Chu,
Jian Tian * and
Ningfeng Wu
* and
Ningfeng Wu
Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, China. E-mail: tianjian@caas.cn; Tel: +86-10-82109864
First published on 6th November 2017
Abstract
Cadmium is recognized as one of the most toxic heavy metals, and chronic cadmium exposure threatens plant, animal and human health. Because certain bacteria can play an important role in remedying heavy metal pollution, it is essential to further our understanding and clone cadmium adsorption and resistance genes from microorganisms. In this study, the cadmium resistance of different Escherichia coli strains including BL21 (DE3), JM109, DH5α, Top10 and Mach was determined. Among those strains, the E. coli BL21 (DE3) exhibited the highest cadmium resistance. Sequence analyses of four cadmium resistant clones screened from the BL21 (DE3) fosmid library showed that the insertion of each fosmid plasmid contained a fragment located at the lambda phage DE3 region. Next, we overexpressed several genes of this fragment in BL21 (DE3) and identified that cadmium resistance of E. coli is acquired through the capB gene. Indeed, overexpression of the capB gene increased the adsorption rate of cadmium. Taken together, these results indicated that the capB gene confers E. coli BL21 (DE3) with increased cadmium resistance by adsorption of cadmium. This study provided new insight into the mechanism of microbial cadmium resistance and expanded our understanding of cadmium resistance, adsorption and bioremediation by microorganisms.
1. Introduction
Cadmium is a nonessential nutrient element for organisms and is regarded as one of most toxic heavy metals with an unknown biological function.1,2 Further, as a persistent toxic substance, cadmium pollution is irreversible and especially soil pollution is of great concern in many parts of the world.3–6 Chronic cadmium exposure can not only affect the growth, yield and quality of the crops,7 but cadmium can also enter the body through the food chain and consequently endanger human health; Itai-Itai disease, respiratory and kidney disease.8,9According to previous studies, bacteria evolved several cadmium resistance mechanisms:1,2,10–12 metal exclusion by a cellular permeability barrier such as the cell membrane or envelope; intracellular and extracellular sequestration through exopolysaccharide coating;1–3 reduced metal sensitivity of cellular targets and transport efflux pumps.13 Particularly the intracellular and extracellular binding of metal is of interest to researchers in the field of bioremediation as this method could be used to immobilize and adsorb target metals.
Escherichia coli, a common industrial Gram-negative bacterium and a model organism, is often used in studies concerning microbial remediation of cadmium pollution.14–21 Currently, most studies focus on the overexpression of cadmium resistant genes from Gram-positive bacteria to reverse cadmium pollution or to isolate strains in heavy metal contaminated areas:2 cadA from S. aureus,22 czc system from Alcaligenes eutrophus,23 and MTS16,23,24 and phsABC25 from Salmonella typhimurium. In fact, E. coli is intrinsically tolerant to high levels of cadmium up to 0.9–1.0 mM.16,25,26 However the exact mechanism of cadmium resistance in E. coli is not fully understood. To date, we know that ZntA, a Pb(II)-, Zn(II)-, and Cd(II)-transporting ATPases confer cadmium tolerance in E. coli.13,17–19 Interestingly, most cadmium resistance mechanisms currently known in microbiology are cadmium exclusion. Thus, the identification of cadmium resistant genes, especially cadmium adsorption genes, in E. coli would be of great importance for applications in remedying cadmium contamination.
In this study, we determined the cadmium resistance of different strains of E. coli including BL21 (DE3), JM109, DH5α, Top10, and Mach. We discovered that BL21 (DE3) had the highest cadmium resistance, up to 1.0 mM. Consequently, we constructed a BL21 (DE3) fosmid library to look for genes conferring the high resistance of E. coli to cadmium. Four clones with higher cadmium resistance than the wild type were identified, and the insertion fragments of the four strains were overlapped. Through overexpression of different genes in this section, we were eventually able to identify the gene related to cadmium resistance and absorption. Based on our knowledge, this is the first study to report the key gene related to cadmium absorption of BL21 (DE3). This work has paved the way for bioremediation and increasing cadmium resistance.
2. Material and methods
2.1 Bacterial strains and growth conditions
The plasmids and bacterial strains used in this study are listed in Table 1; EPI300-T1R came with the Fosmid Library Construct Kit. All the strains were grown in Luria Bertani (LB) medium (10 g L−1 trypoton, 5 g L−1 yeast extract, 10 g L−1 NaCl) overnight at 37 °C with shaking at 200 rpm. All reagents used in this study were purchased from Sigma-Aldrich (St. Louis, Missouri, USA).| Strains and plasmids | Description | Resources |
|---|---|---|
| BL21 (DE3) | F-,ompT,hsdS(rBB-mB-),gal,dcm(DE3) | CWBIO |
| DH5α | F-,ϕ80dlacZΔM15,Δ(lacZYA-agF)U169,deoR,ecA1,endA1,hsdR17(k−,mk+),phoA,supE44,λ-,thi-1,gyA96,elA1 | CWBIO |
| JM109 | ecA1,endA1,gyA96,thi-1,hsdR17,supE44,elA1,Δ(lac-poAB)F′[taD36,poAB+,lacIq,lacZΔM15] | CWBIO |
| Top10 | F-,mcAΔ(m-hsdRMS-mcBC),ϕ80,lacZΔM15,Δlacx74,ecA1,aaΔ139Δ(aa-leu)7697,galU,galK,ps,(St)endA1,nupG | Our lab |
| Mach | recA,tonA,endA1,hsdR, lacZΔM15 | ThermoFisher |
| BL21-pUC19-GFP | Control strain with empty pUC19-GFP plasmid | Our lab |
| BL21-pUC19-capA-GFP | capA gene was overexpressed | In this study |
| BL21-pUC19-capB-GFP | capB gene was overexpressed | In this study |
| BL21-pUC19-capJ-GFP | capJ gene was overexpressed | In this study |
| BL21-pUC19-regI-GFP | I region was overexpressed | In this study |
| BL21-pUC19-regII-GFP | II region was overexpressed | In this study |
| BL21-pUC19-regIII-GFP | III region was overexpressed | In this study |
| pUC19-GFP | Expression vector | Our lab |
2.2 Determination of minimum inhibitory concentration (MIC) of cadmium
The MIC values27 were assessed by measuring the growth rate of different strains estimated from the optical density at 600 nm (OD600). For the assay, the overnight culture of the test strains was transferred into fresh LB medium and incubated at 37 °C until an OD600 of 0.6–0.8 was reached. Next, the different strains (1%) were inoculated into 96-well plates containing different concentrations of CdCl2 (0–1.4 mM) in triplicate. After incubation at 37 °C for 20–24 hours, the OD600 was measured with a fluorescence absorbance cuvette.2.3 Construction and screening of BL21 (DE3) genomic fosmid library
The BL21 (DE3) total genomic DNA was extracted with the Bacterial Genome Rapid Extraction Kit (BioTeKe, China), according to the manufacturer's protocol. DNA concentration and quality were checked by absorbance cuvette (A260/A280 ∼ 1.8) and gel electrophoresis (1%). Approximately 0.5 μg μL−1 DNA extract was used to construct a fosmid library with the CopyControl HTP Fosmid Library Production Kit and pCC1FOS Vector (Epicentre, Madison, WI, USA), according to the manufacturer's instructions. Twelve fosmid clones were randomly selected for evaluating the insert size by BamHI digestion and agarose gel electrophoresis analysis (Fig. S1†). A total of 1443 fosmid clones (insert size range 25–40 kb) were obtained and stored at −70 °C in 40% glycerol.To screen for clones with cadmium resistance, all fosmid clones were replicated in 96-well plates containing LB broth supplemented with chloramphenicol (12.5 μg mL−1) and cultivated at 37 °C for 8–12 h. The cultures were then transferred onto solid LB medium supplemented with chloramphenicol (12.5 μg mL−1) containing 1.2 mM CdCl2 and incubated at 37 °C for 36 h.
Fosmid DNA was isolated from the positive fosmid clones with the High Purity Plasmid Extraction Kit (TIANGEN, China) and sequenced from both ends with an ABI 3730 DNA analyzer and the pCC1FOS vector universal primers fosmid-T7 and fosmid-RP (Table. S1†). The sequences were analyzed by BlastN (NCBI) to determine the location of the inserted fragments in the entire genome.
2.4 Gene overexpression in E. coli
Genes containing their promoters predicted using Web Promoter Scan Services (https://www-bimas.cit.nih.gov/molbio/proscan/) were amplified by PCR from the positive plasmids with the primers listed in Table S1,† and digested with KpnI and XbaI. These fragments were inserted into the KpnI/XbaI double digest of pUC19-GFP, containing a reporter green fluorescent protein (GFP) gene gfp, to create plasmids pUC19-capA-GFP, pUC19-capB-GFP, pUC19-capJ-GFP, pUC19-regI-GFP, pUC19-regII-GFP and pUC19-regIII-GFP (Table. S1†). Each plasmid was verified by DNA sequencing and transformed into E. coli BL21 (DE3) to express the GFP-tagged proteins CapA, CapB, CapJ, CapC, CapD, CapE, CapFi, CapFii, CapV, CapQ and CapT, respectively. All the host cells harboring the recombinant vectors were grown in LB at 37 °C for 20 h. To confirm gene expression, fluorescence of the cultures was measured with a fluorescence absorbance cuvette. Fluorescence in the wells was measured at an excitation/emission wavelength of 484/507 nm, using a microplate reader SpectraMax M2 (Molecular Devices, USA). BL21 (DE3) cells harboring the empty pUC19-GFP expression vector were used as the negative controls.2.5 Cd2+ removal by protein CapB
The capB overexpression strains and control strains (BL21 (DE3)) with empty vector pUC19-GFP were cultivated in LB broth supplemented with ampicillin (100 μg mL−1) for 12 h in a shaking incubator (200 rpm), before transferring them into 3 mL LB supplemented with 0.1 mM Cd2+ or 0.05 mM Cd2+ for 24 h. Then the cultures were transferred into 1.5 mL tubes and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. The supernatant was separated and diluted 100 folds using 0.6 M HCl and the Cd2+ concentration was determined with an atomic absorption spectrophotometer (AAS).28,29 Based on the mean absorbance, concentration of Cd2+ was determined by comparing with mean absorbance of the standard Cd2+. The experiments were performed in triplicate.
000g for 10 min. The supernatant was separated and diluted 100 folds using 0.6 M HCl and the Cd2+ concentration was determined with an atomic absorption spectrophotometer (AAS).28,29 Based on the mean absorbance, concentration of Cd2+ was determined by comparing with mean absorbance of the standard Cd2+. The experiments were performed in triplicate.
3. Results and discussion
3.1 Resistance of E. coli stains to Cd2+
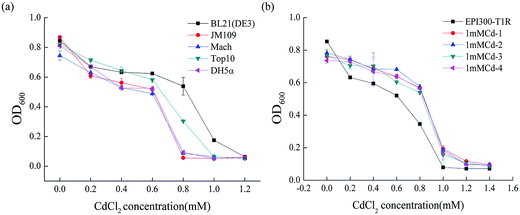
To determine the degree of Cd2+ resistance of E. coli, we examined the growth of different E. coli stains including BL21 (DE3), JM109, DH5α, Top10 and Mach in LB liquid medium with various Cd2+ concentrations. As depicted in Fig. 1a, the strain BL21 (DE3) resisted Cd2+ concentrations of up to 1.0 mM. Strain Top10 resisted up to 0.8 mM Cd2+ and Mach, JM109 and DH5α up to 0.6 mM Cd2+. Clearly, from all the E. coli stains used in this study, BL21 (DE3) had the highest cadmium resistance (1.0 mM ± 0.02). The evident difference between BL21 (DE3) and the other four genetically engineered E. coli stains is that BL21 (DE3) carries a lysogenic lambda phage DE3 that expresses T7 RNA polymerase under the control of a lacUV5 promoter.30 So we speculated that this specific DE3 sequence, located at 748![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 396–791
396–791![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320 in the BL21 (DE3) genome, might confer the higher resistance to cadmium.
320 in the BL21 (DE3) genome, might confer the higher resistance to cadmium.
3.2 Screening of the Cd2+ resistant DNA fragments
To figure out the gene(s) involved in the Cd2+ resistance of BL21 (DE3), a fosmid library from genomic DNA of BL21 (DE3) was constructed in the E. coli strain EPI300-T1R and screened for Cd2+ resistant clones. Out of the 1443 clones, four clones (1mMCd-1, 1mMCd-2, 1mMCd-3 and 1mMCd-4) exhibited higher Cd2+ resistance than the host strain EPI300-T1R (Fig. 1b). The fosmid DNA isolated from these four clones was sequenced and by aligning the sequences with the BL21 (DE3) genome (accession number: NC_012892), we found that the inserted fragment from the four plasmids all contained a DNA fragment located between 768![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 481 and 801
481 and 801![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 476 (Table 2). Indeed, this region overlapped with the aforementioned DE3 sequence. The result confirmed that the DE3 sequence is important for the cadmium resistance of BL21 (DE3).
476 (Table 2). Indeed, this region overlapped with the aforementioned DE3 sequence. The result confirmed that the DE3 sequence is important for the cadmium resistance of BL21 (DE3).
| Plasmid number | The location of the insertion in BL21 (DE3) genome | Overlap section |
|---|---|---|
| 1mMCd-1 | 768![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 481–825 481–825![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 247 247 |
768![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 481–801 481–801![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 476 476 |
| 1mMCd-2 | 768![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 435–801 435–801![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 476 476 |
|
| 1mMCd-3 | 768![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 464–823 464–823![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 194 194 |
|
| 1mMCd-4 | 768![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 403–824 403–824![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 390 390 |
3.3 Overexpression of protein CapB can improve the cadmium resistance of BL21 (DE3)
To identify the cadmium resistant gene(s) in the DE3 sequence, we further analyzed the genetic composition and gene function of the fragment locating between 768![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 481 and 791
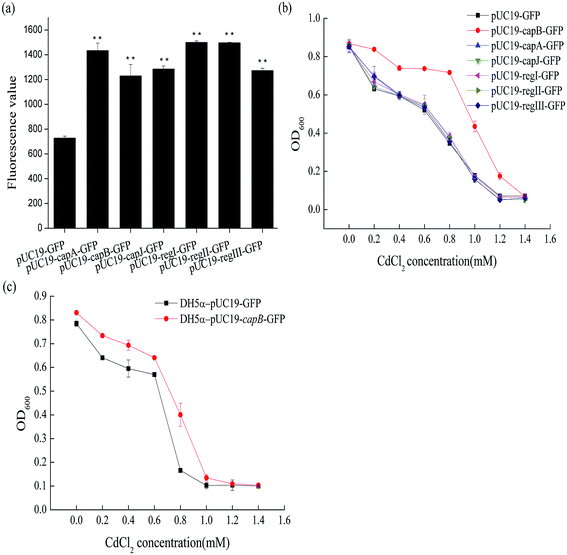
481 and 791![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320. It contained 24 genes coding for DNA packing proteins, capsid components and tail components (Fig. 2). In order to identify the target genes conferring the cadmium resistance, some of these 24 genes were overexpressed in BL21 (DE3) to see whether they could increase the cadmium resistance of BL21 (DE3). The selected genes were fusion expressed with GFP to conveniently detect the expression of target genes by measuring fluorescence. As shown in Fig. 3a, the genes capA, capB and capJ were successfully overexpressed. However, only overexpression of capB could increase the MIC of Cd2+ for BL21 (DE3) from 1.0 mM to 1.2 mM (Fig. 3b). In order to confirm that the CapB protein is expressed under its own promoter, we also performed SDS-PAGE (Fig. S2†). In addition, overexpression of capB in DH5α was also shown to improve the Cd2+ resistance of DH5α (Fig. 3c).
320. It contained 24 genes coding for DNA packing proteins, capsid components and tail components (Fig. 2). In order to identify the target genes conferring the cadmium resistance, some of these 24 genes were overexpressed in BL21 (DE3) to see whether they could increase the cadmium resistance of BL21 (DE3). The selected genes were fusion expressed with GFP to conveniently detect the expression of target genes by measuring fluorescence. As shown in Fig. 3a, the genes capA, capB and capJ were successfully overexpressed. However, only overexpression of capB could increase the MIC of Cd2+ for BL21 (DE3) from 1.0 mM to 1.2 mM (Fig. 3b). In order to confirm that the CapB protein is expressed under its own promoter, we also performed SDS-PAGE (Fig. S2†). In addition, overexpression of capB in DH5α was also shown to improve the Cd2+ resistance of DH5α (Fig. 3c).
CapB encodes a capsid component of the lambda phage DE3 and the isoelectric point (pI) of this protein was 5.31. In the neutral pH environment in the cell, CapB has a negative charge that could bind to the positive ion of cadmium. Similarly, also the pIs of the proteins CapC, CapD, CapE, CapFi, CapFii, CapV, CapG and CapT were low (Table S2†). We therefore also selected these to be overexpressed in BL21 (DE3) in the hope to identify more Cd2+ resistance related genes. Based on the promoter and position of these genes, we expressed three fragments separately: region I (containing genes capC and capD), region II (containing genes capE, capFi and capFii) and region III (containing genes capV, capG and capT) (Fig. 2). Although the proteins were successfully expressed in BL21 (DE3) (Fig. 3a), none of the fragments changed its Cd2+ resistance (Fig. 3c). Taken together, the results suggested that the CapB protein encoded by capB contributes to the Cd2+ resistance of E. coli.
3.4 Absorption of cadmium ions by the protein CapB
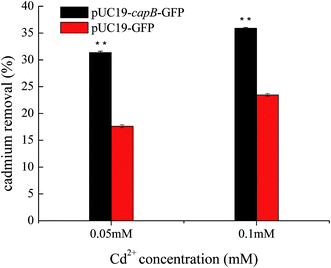
As CapB is not a membrane protein, we speculated that it may be expressed in cells to increase Cd2+ resistance by absorbing Cd2+. To verify this, the final concentration of Cd2+ was measured in culture media supernatant after cultivation of capB overexpressing strains. We found that the Cd2+ removal rate of capB overexpressing strains was significantly higher than the wild type BL21 (DE3) strain (Fig. 4). At 0.05 mM Cd2+, the Cd2+ removal rate of capB overexpressing strains was about two-fold greater than that of the wild type BL21 (DE3). At 0.1 mM Cd2+, the Cd2+ removal rate of capB overexpressing strains was also higher than that of the wild type (Fig. 4). It was speculated that such a high concentration of metal ions was toxic to cells and the gradual saturation of adsorption sites by cadmium ions would have limited the capacity of Cd2+ removal.31 The above results suggested that the CapB protein indeed increased cadmium resistance by absorbing more Cd2+. Unlike the earlier discovered metallothionein,10,16,32,33 CapB does not contain a lot of cysteine short peptides. The pI of CapB (5.31) was lower than the neutral pH, implying that the protein surface will have many negative charges that can bind the positive cadmium ions in the cell. Perhaps it is just those functional negative groups that chelate with cadmium ions.34 However, a low pI is just one of the characteristics of CapB since not all proteins with a low pI could adsorb cadmium. Therefore, in addition to the low pI, the special sequence or structure is also fundamental to conferring cadmium resistance. Our future studies will thus focus on the binding characteristics of the CapB protein.Our results reiterated that bacterial proteins could improve heavy metal resistance or adsorption. With advances in bioinformatics and the newly acquired knowledge on CapB, we may be able to improve upon the design of such proteins to remedy heavy metal pollution in the future.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC, Grant No. 31770124) and Science and Technology Innovation Project from the Chinese Academy of Agricultural Sciences (grant no. CAAS-XTCX2016018).References
- M. R. Bruins, S. Kapil and F. W. Oehme, Ecotoxicol. Environ. Saf., 2000, 45, 198–207 CrossRef CAS PubMed.
- Z. Khan, A. Rehman and S. Z. Hussain, Chemosphere, 2016, 159, 32–43 CrossRef CAS PubMed.
- Z. Li, Z. Ma, T. J. van der Kuijp, Z. Yuan and L. Huang, Sci. Total Environ., 2014, 468–469, 843–853 CrossRef CAS PubMed.
- G. N. Egwu and J. O. Agbenin, Arch. Agron. Soil Sci., 2013, 59, 875–887 CrossRef CAS.
- J. T. Li, J. W. Qiu, X. W. Wang, Y. Zhong, C. Y. Lan and W. S. Shu, Environ. Pollut., 2006, 143, 159–165 CrossRef CAS PubMed.
- S. Satarug, J. R. Baker, S. Urbenjapol, M. Haswell-Elkins, P. E. Reilly, D. J. Williams and M. R. Moore, Toxicol. Lett., 2003, 137, 65–83 CrossRef CAS PubMed.
- F. Villiers, V. Hugouvieux, N. Leonhardt, A. Vavasseur, C. Junot, Y. Vandenbrouck and J. Bourguignon, in Metal Toxicity in Plants: Perception, Signaling and Remediation, ed. D. Gupta and L. Sandalio, Springer, Berlin, Heidelberg, 2012, pp. 119–142 Search PubMed.
- K. Nogawa, M. Sakurai, M. Ishizaki, T. Kido, H. Nakagawa and Y. Suwazono, J. Appl. Toxicol., 2017, 37, 962–966 CrossRef CAS PubMed.
- S. Thijssen, J. Maringwa, C. Faes, I. Lambrichts and E. Van Kerkhove, Toxicology, 2007, 229, 145–156 CrossRef CAS PubMed.
- M. Mejare and L. Bulow, Trends Biotechnol., 2001, 19, 67–73 CrossRef CAS PubMed.
- C. Parsons, S. Lee, V. Jayeola and S. Kathariou, Appl. Environ. Microbiol., 2017, 83 Search PubMed.
- T. Limcharoensuk, N. Sooksawat, A. Sumarnrote, T. Awutpet, M. Kruatrachue, P. Pokethitiyook and C. Auesukaree, Ecotoxicol. Environ. Saf., 2015, 122, 322–330 CrossRef CAS PubMed.
- K. J. Tsai, C. M. Hsu and B. P. Rosen, Zool. Stud., 1997, 36, 1–16 CrossRef CAS PubMed.
- E. Anane, C. D. C. Lopez, P. Neubauer and M. N. C. Bournazou, Biochem. Eng. J., 2017, 125, 23–30 CrossRef CAS.
- H. Shi, Z. Z. Chen, D. Chen and J. Q. Kan, Food Control, 2017, 82, 190–195 CrossRef CAS.
- S. K. Kim, B. S. Lee, D. B. Wilson and E. K. Kim, J. Biosci. Bioeng., 2005, 99, 109–114 CrossRef CAS PubMed.
- M. R. B. Binet and R. K. Poole, FEBS Lett., 2000, 473, 67–70 CrossRef CAS PubMed.
- C. Rensing, B. Mitra and B. P. Rosen, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 14326–14331 CrossRef CAS.
- R. Sharma, C. Rensing, B. P. Rosen and B. Mitra, J. Biol. Chem., 2000, 275, 3873–3878 CrossRef CAS PubMed.
- L. Huang, Q. Wang, S. Jiang, Y. Zhou, G. Zhang and Y. Ma, Bioprocess Biosyst. Eng., 2016, 39, 1679–1687 CrossRef CAS PubMed.
- J. Tian, N. Wu, J. Li, Y. Liu, J. Guo, B. Yao and Y. Fan, Appl. Environ. Microbiol., 2007, 73, 2364–2368 CrossRef CAS PubMed.
- A. Alonso, P. Sanchez and J. L. Martinez, Antimicrob. Agents Chemother., 2000, 44, 1778–1782 CrossRef CAS PubMed.
- C. Grosse, A. Anton, T. Hoffmann, S. Franke, G. Schleuder and D. H. Nies, Arch. Microbiol., 2004, 182, 109–118 CrossRef CAS PubMed.
- E. Tokuda, E. Okawa, S. Watanabe and S. Ono, Hum. Mol. Genet., 2014, 23, 1271–1285 CrossRef CAS PubMed.
- S. W. Bang, D. S. Clark and J. D. Keasling, Biotechnol. Lett., 2000, 22, 1331–1335 CrossRef CAS.
- R. A. Laddaga and S. Silver, J. Bacteriol., 1985, 162, 1100–1105 CAS.
- X. C. Chen, J. Y. Shi, Y. X. Chen, X. H. Xu, S. Y. Xu and Y. P. Wang, Can. J. Microbiol., 2006, 52, 308–316 CrossRef CAS PubMed.
- A. Vidhyaparkavi, J. Osborne and S. Babu, 3 Biotech, 2017, 7, 9 CrossRef PubMed.
- Z. G. Wei, J. W. Wong, H. Y. Zhao, H. J. Zhang, H. X. Li and F. Hu, Biol. Trace Elem. Res., 2007, 118, 146–158 CrossRef CAS PubMed.
- B. A. Moffatt, J. J. Dunn and F. W. Studier, J. Mol. Biol., 1984, 173, 265–269 CrossRef CAS PubMed.
- Z. Z. Yu, Q. F. Dang, C. S. Liu, D. S. Cha, H. F. Zhang, W. J. Zhu, Q. Q. Zhang and B. Fan, Carbohydr. Polym., 2017, 172, 28–39 CrossRef CAS PubMed.
- C. D. Klaassen, J. Liu and S. Choudhuri, Annu. Rev. Pharmacol. Toxicol., 1999, 39, 267–294 CrossRef CAS PubMed.
- C. D. Klaassen, J. Liu and B. A. Diwan, Toxicol. Appl. Pharmacol., 2009, 238, 215–220 CrossRef CAS PubMed.
- B. Liao, W. Y. Sun, N. Guo, S. L. Ding and S. J. Su, Carbohydr. Polym., 2016, 151, 20–28 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra10656d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |