Open Access Article
Open Access ArticleApplying target data screening followed by characteristic fragment filtering for the comprehensive screening and identification of alkaloids in Corydalis yanhusuo W. T. Wang by UPLC-Q-TOF/MSE†
Meiling Wang ,
Yuehong Liu,
Shuang Fu,
Qingqing Zhang,
Qing Wang and
Xiaoyan Gao*
,
Yuehong Liu,
Shuang Fu,
Qingqing Zhang,
Qing Wang and
Xiaoyan Gao*
School of Chinese Pharmacy, School of Chinese Materia Medica, Beijing University of Chinese Medicine, East of Baiyang Road, Liangxiang Town, Fangshan District, Beijing 102488, China. E-mail: gaoxiaoyan0913@sina.com
First published on 21st November 2017
Abstract
A target data screening strategy followed by characteristic fragment filtering by UPLC-Q-TOF/MSE was developed for rapidly and comprehensively identifying alkaloids in Corydalis yanhusuo W. T. Wang (Yanhusuo). The proposed strategy consisted of the following four steps. (1) Fragmental patterns and characteristic fragments of various types of alkaloids were summarized based on the reference compounds and previous reference literature. (2) Target data screening was conducted and data screening tables involving various types of alkaloids were constructed referring to the structural characteristics of alkaloids, the type and number of substituents, and characteristic fragments. This data screening table includes all possible molecular weights of various types of alkaloids. (3) The raw data detected by UPLC-Q-TOF/MSE were screened preliminarily using data screening tables, then characteristic fragment filtering was used to rapidly recognize various types of alkaloids. (4) A comparison was made with retention time, accurate mass, MS/MS fragmentation and online databases, as well as a reference to related literature to validate the data. As a result, a total of 86 compounds were identified or characterized, including 24 tetrahydroprotoberberine alkaloids, 22 protoberberine alkaloids, 6 protopine alkaloids, 12 aporphine alkaloids and 22 other compounds. Among them, 8 were potentially new compounds, and 5 components were discovered in Yanhusuo for the first time. The method proposed in this paper was proved to be an efficient data processing approach to rapidly discover and characterize chemical constituents from complicated herbal extracts, without the help of standard substances. Furthermore, this research enriched the material basis of Yanhusuo and provided meaningful guidance for the discovery of potentially new compounds.
1. Introduction
The dry rhizoma of Corydalis yanhusuo W. T. Wang (Papaveraceae) is an analgesic in Traditional Chinese Medicine (TCM) with a Chinese name “Yanhusuo”. A picture of Corydalis yanhusuo W. T. Wang (Papaveraceae) is shown in Fig. 1. It was found to promote blood circulation, promote qi and relieve pain.1,2 Modern pharmacological experiments showed that Yanhusuo provides better functions such as analgesic, sedative, antihypertensive and antiarrhythmic functions.3,4 The main active ingredients of Yanhusuo were alkaloid compounds based on the related literature reviews. The reported alkaloids in Yanhusuo mainly included tetrahydroprotoberberine alkaloids, protoberberine alkaloids, protopine alkaloids and aporphine alkaloids.2–6 Comprehensive ingredient identification from Yanhusuo is critical for the understanding of the therapeutic material basis and for establishing its quality control protocols.With the continuous development of new technologies, high-resolution mass spectrometry (HR-MS) technology has become an essential tool for qualitative identification of Traditional Chinese Medicine (TCM).7–9 However, the data information of samples obtained by HR-MS was extremely complex. Therefore, data mining and analyses were frequently laborious and time-consuming. In addition, the structural complexity, large content variations and lack of standard substances led to challenges in identifying compounds.10 Therefore, it is urgent to establish an effective screening and identification method to comprehensively identify and characterize the chemical composition of Traditional Chinese Medicines.
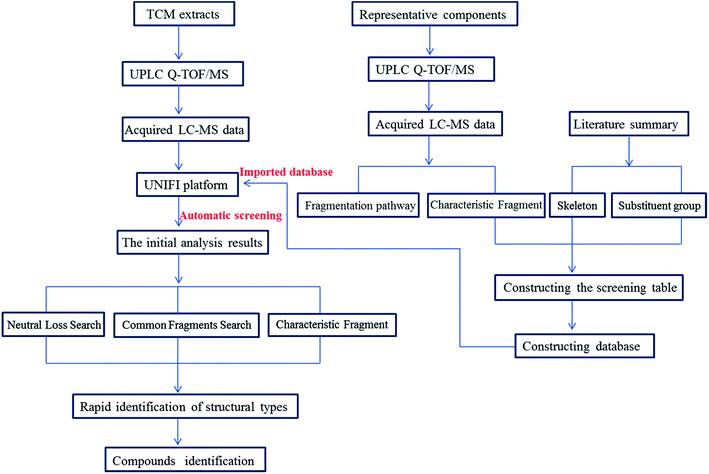
In this study, a target data screening strategy followed by characteristic fragment filtering by UPLC-Q-TOF/MSE was proposed. The specific research process of the approach was summarized in Fig. 2. The data screening tables contained all possible molecular weights of the main alkaloid types to facilitate the discovery of potentially new compounds. Characteristic fragment ions were invoked as criteria for judging the type of compounds to rapidly identify them. Compared to identifying compounds by traditional manual identification methods, the Natural Product Application Solution with UNIFI and databases based on data screening tables can automatically find useful information from the ocean of mass spectrometry information, significantly reduce researcher workload and make identification of compounds more accurate and comprehensive. This study provided a reference for the rapid identification of other TCMs and provided meaningful guidance for the discovery of potentially new compounds.
2. Experimental
2.1 Materials and reagents
Herbal materials of Yanhusuo was purchased from Beijing Tong Ren Tang Group Co. Ltd. (Beijing, China) and identified by Professor Yaojun Yang, School of Chinese Materia Medica, Beijing University of Chinese Medicine. The voucher specimens were deposited at the School of Chinese Materia Medica, Beijing University of Chinese Medicine, China. Allocryptopine, protopine, tetrahydropalmatine, berberine, tetrahydroberberine and palmatine were purchased from Chengdu Biopurify Phytochemicals CO. Ltd. (Sichuan, China); dehydrocorydaline was purchased from Shanghai Nature Standard Biotechnology CO. Ltd. (Shanghai, China). The seven reference compounds were determined to be greater than 98% by HPLC peak area. LC-MS grade acetonitrile, methanol, and formic acid were purchased from Fisher Scientific (Fisher, Fair Lawn, NJ, USA). Distilled water was available from the A.S. Watson Group Ltd. (Hong Kong, China).2.2 Sample and standard solution preparation
Corydalis yanhusuo was powdered by a grinder and sieved through a 40 mesh sieve; 2.0 g of powder was accurately weighed and placed into a 50 mL conical flask. Then, 20 mL of water/ethanol 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 (v/v) was added into the flask and ultrasonically extracted for 1 h at room temperature. The extracted solution was centrifuged at 12
70 (v/v) was added into the flask and ultrasonically extracted for 1 h at room temperature. The extracted solution was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min, and liquid supernatant was collected and stored at 4 °C. The stored solution was diluted ten times prior to being injected for liquid chromatography-tandem mass spectrometry (LC-MS) analysis. The appropriate amount of each standard was weighed and dissolved in methanol to make six individual stock solutions. The stock solutions were kept in the refrigerator of 4 °C and sunlight was avoided before analysis.
000 rpm for 10 min, and liquid supernatant was collected and stored at 4 °C. The stored solution was diluted ten times prior to being injected for liquid chromatography-tandem mass spectrometry (LC-MS) analysis. The appropriate amount of each standard was weighed and dissolved in methanol to make six individual stock solutions. The stock solutions were kept in the refrigerator of 4 °C and sunlight was avoided before analysis.
2.3 UPLC conditions
LC analyses were carried out on Waters ACQUITYTM UPLC I – Class system (Waters, USA) consisting of a binary solvent system, an autosampler and a column temperature controller. The system was operated under MassLynx V4.1 software (Waters, Manchester, U.K.). The chromatographic separation was carried out on an ACQUITY BEH C18 column (2.1 × 100 mm, 1.7 μm, Waters, UK). The mobile phase was composed of eluent A (0.1% formic acid in water) and eluent B (acetonitrile). The line gradient program was optimized as the following: 0–2 min, 1% B–10% B; 2–15 min, 10 B%–20% B; 15–22 min, 20% B–30% B; 22–27 min, 30% B–90% B; 27–29 min, 90% B; 29–29.1 min, 90% B–2% B; 29.1–31 min, 2% B–2% B. The column temperature was set at 40 °C, the mobile phase flow rate was set at 0.3 mL min−1, and the injection volume was 2 μL for each run.2.4 MS conditions
The MS analysis was carried out by a Waters SYNAPT G2-SI MS system (Waters, USA) equipped with electrospray ionization. The analysis was performed in the positive ion electrospray mode, and the source parameters were set as follows: the capillary voltage was 3.0 kV and the cone voltage was set at 40 V; the source temperature was 100 °C, and the desolvation gas temperature was 400 °C; the cone gas flow was 50 L h−1, and the desolvation gas flow was 800 L h−1; the low collision energy was 6 eV, and the high collision energy was 10–40 eV. The scan range was from m/z 100 to 1200 Da and the 3D data was collected in the continuum mode. The Leucine enkephalin (m/z 556.2771 in positive ion mode) was invoked as the lock mass solution to ensure accurate mass measurement. The mass spectrometry data was acquired by Waters MassLynx V4.1 software and processed by UNIFI™ 1.7 software (waters, Manchester, U.K.).3. Results and discussion
3.1 Optimization of UPLC-MS conditions
In this study, in order to achieve higher resolution and sensitivity of the mass spectrometry data, LC conditions and MS conditions were optimized. The detailed information of the optimization of UPLC-MS conditions was given in ESI.† In the positive ion mode, the base peak ion chromatogram (BPI) of 70% methanol extracts of Yanhusuo by UPLC/Q-TOF MSE was shown in Fig. 3.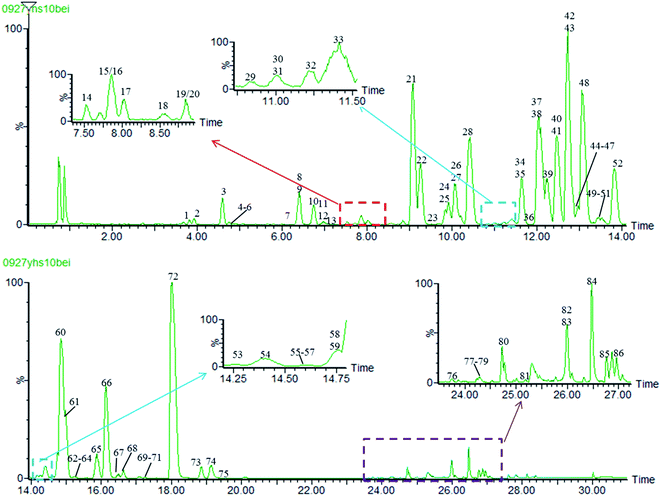 | ||
| Fig. 3 The base peak ion chromatogram (BPI) of 70% methanol extracts of Yanhusuo in positive ion mode. | ||
3.2 Alkaloid fragmentation pattern analysis and characteristic ion determination
From the research literature,2,4–6,11–14 the main components of Yanhusuo were alkaloids. Based on the alkaloid structures, the Yanhusuo alkaloids could be mainly classified as the following four types: tetrahydroprotoberberine-type, protoberberine-type, protopine-type and aporphine-type. The chemical structures of the four alkaloid types in Yanhusuo are shown in Fig. S1.† Most of these alkaloids possessed hydroxyl, methoxyl or methylenedioxyl groups at the C2–C3 and C9–C10 positions, respectively, which may be associated with the biosynthesis of alkaloids in Corydalis. The compounds with the same framework had similar cleavage pathways. Seven reference compounds were analyzed by UPLC-Q-TOF/MSE. The fragmentation patterns for different types of alkaloids were summarized to further determine the characteristic ions, which contributed to rapidly identify the alkaloid compounds in Corydalis. The detailed information of the fragmentation patterns and characteristic fragments of each type of alkaloids was given in ESI.† The characteristic fragments including common fragments and neutral loss of each type of alkaloids are shown in Fig. 4.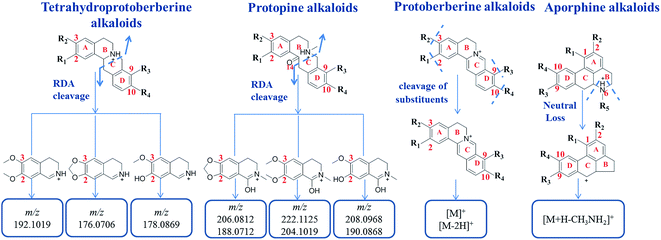 | ||
| Fig. 4 The characteristic fragments including common fragments and neutral loss of each type of alkaloids. | ||
3.3 Constructing a database based on data screening tables
According to the structure characteristics of alkaloids as well as the type and number of substituents and the fragmentation pathways of various alkaloids, a screening table of each type of alkaloid was established. Then, a database was constructed based on data screening tables. Summarizing the fragmentation pathway of the reference substances of each type of alkaloid, it was found that tetrahydroprotoberberine alkaloids and protopine alkaloids both easily undergo RDA cleavage to produce the highest intensity of nitrogen-containing fragments as characteristic fragments, which are related to the type of substituents at the C2–C3 positions. Therefore, data screening tables of these two types of alkaloids were constructed based on the characteristic ions. In the positive ion mode, based on characteristic ion m/z 192.1019, m/z 176.0706 and m/z 178.0869, the screening table of tetrahydroprotoberberine alkaloids was established. There were 40 molecular ions in the screening table (Table S1†), representing all possible molecular weights of tetrahydroprotoberberine alkaloids. Based on characteristic ion m/z 222.1125, m/z 206.0812 and m/z 208.0968, the screening table of protopine alkaloids was established. There were 40 molecular ions in the screening table (Table S2†), representing all possible molecular weights of protopine alkaloids. The C-ring structure of the protoberberine alkaloids is saturated, which makes it difficult to undergo RDA cleavage. The main cleavage is cleavage of the substituents. Therefore, a screening table of protoberberine alkaloids was constructed based on the number of substituents. There were 18 molecular ions in the screening table (Table S3†), representing all possible molecular weights of protoberberine alkaloids. Based on the presence or absence of (CH3)2NH or CH3NH2 substituents to construct a screening table of the aporphine alkaloids, a total of 54 compounds were designed (Table S4†), which included all possible compounds of aporphine alkaloid molecular weights.3.4 Applying target data screening followed by characteristic fragment filtering for characterization of the chemical compounds in the Yanhusuo herb
The raw data detected by UPLC-Q-TOF/MSE were screened preliminarily by data screening tables, then characteristic fragments filtering was used to rapidly recognize various types of alkaloids. The compounds were validated further by comparing with retention time, accurate mass, MS/MS fragmentation and online databases as well as referring to related literature. As a result, a total of 86 compounds were identified or characterized, including 24 tetrahydroprotoberberine alkaloids, 22 protoberberine alkaloids, 6 protopine alkaloids, 12 aporphine alkaloids and 22 other compounds. Meanwhile, 8 of them were potential new compounds, 5 components were discovered in the Yanhusuo for the first time (Fig. 3, Table S5†).Using the characteristic fragment of m/z 192.1019 to rapidly recognize tetrahydroprotoberberine-type alkaloids, which possessed two methoxyl groups on C2–C3 positions, 10 compounds were found, including peaks 3, 7, 15, 17, 22, 26, 27, 32, 42 and 60. The ten peaks generated the highest intensity of nitrogen-containing fragment ion at m/z 192.10 in their MS/MS spectra. Peak 42 was unambiguously identified as tetrahydropalmatine by being compared to the reference substance. Peaks 22, 26 and 27 were tentatively identified as the potentially new isomers of tetrahydropalmatine by the same quasi-molecular ion, the MS/MS fragment pattern as well as from previous studies,2,12,15 The quasi-molecular ion peaks of 7, 15 and 17 was m/z 342.17 [M + H]+ (C20H23NO4), which were 14 Da lower than tetrahydropalmatine. Their complementary fragment ion at m/z 151.07 [M + H − C11H13NO2]+ by RDA cleavage as well as other fragment ions on high charge-to-mass ratio area were also 14 Da lower than the corresponding fragment ions of tetrahydropalmatine, indicating the existence of one methoxyl and one hydroxyl substituents at the C9–C10 positions or two hydroxyl substitutions at the C9 and C10 positions and one methyl at the C13 position. Combined with information from the literature,2,3 peaks 7, 15 and 17 were deduced as corydalmine or its isomer. Peak 60 provided the protonated molecular ion at m/z 370.2018 [M + H]+, which was 14 Da (CH2) more than tetrahydropalmatine. Meanwhile, their complementary fragment ion at m/z 179.1074 [M + H − C11H13NO2]+ by RDA cleavage as well as other fragment ions on high charge-to-mass ratio area were also 14 Da more than the corresponding fragment ions of tetrahydropalmatine, indicating the existence of one methyl group at the C13 position. Therefore, peak 60 was assigned as corydaline.5,6,11,12 Peak 3 showed the protonated molecular ion at m/z 328.1538 [M + H]+ as well as the fragment ions at m/z 177.0544 [M + H − C8H8O2 − CH3]+ and m/z 313.1305 [M + H − CH3]+, etc., which were 28 Da lower than corresponding fragment ions of tetrahydropalmatine, suggesting there were two hydroxyls substituents at the C9 and C10 positions. Based on fragment ions and comparison with known compounds, peak 3 was tentatively identified as demethylcorydalmine.16 Peak 32 provided the protonated molecular ion at m/z 340.1548 [M + H]+ as well as the fragment ions at m/z 149.0605 [M + H − C11H13NO2]+, m/z 324.1209 [M + H − CH4]+, which was 16 Da lower than the corresponding fragment ions of tetrahydropalmatine, suggesting the methylenedioxyl substituent at C9–C10 positions. Compared with the previous reports and online database, one compound was matched with the molecular formula and structure feature of peak 32 (name, 8,9-dimethoxy-6,11,12,14-tetrahydro-6aH-[1,3]dioxolo[4,5-h]isoquinolino[2,1-b]isoquinoline, ID 4479103). Peak 32 was reported for the first time in Yanhusuo.
The identification process of characteristic fragments of m/z 178.0869 and m/z 176.0706 was same as above. The detailed identification information of the tetrahydroprotoberberine alkaloids was given in Table S5.†
For example, peaks 33, 37 and 41 had the same molecular ion at m/z 320.0925 [M]+, which were 16 Da lower than berberine. The main ions at m/z 320.0921 [M]+, m/z 318.0772 [M − 2H]+, m/z 292.0971 [M − CO]+, m/z 290.0815 [M − CO − 2H]+, m/z 264.1018 [M − CO − CO]+, m/z 262.0807 [M − CO − CO − 2H]+, m/z 277.0743 [M − CO − CH3]+ were also 16 Da lower than the corresponding fragment ions of berberine, indicating the existence of a methylenedioxyl group at the C9–C10 positions. Based on the above fragment information and comparison with structures in the literature, peaks 33, 37 and 41 were deduced as coptisine or pseudocoptisine.5,6
The identification process of other protoberberine alkaloids was same as above. The detailed identification information of the protoberberine alkaloids was given in Table S5.†
Using the characteristic fragment of m/z 206.0812 to rapidly recognize protopine-type alkaloids, which possessed a methylenedioxyl group on the C2–C3 positions, peaks 29 and 39 were found. Comparing the retention times and fragmentation information with reference standards, peaks 29 and 39 were unambiguously identified as protopine and allocryptopine, respectively. Using the characteristic fragment of m/z 222.1125 to rapidly recognize protopine-type alkaloids, which possessed two methoxyl groups on C2–C3 positions, no compounds were detected.
Using the characteristic fragment of m/z 208.0968 to rapidly recognize protopine-type alkaloids, which possessed one methoxyl and one hydroxyl group on the C2–C3 positions, a total of 4 compounds were found, including peaks 13, 18, 53 and 56. The formula of peaks 13, 18 and 56 was C21H25NO5. They had similar fragmentation behavior to protopine and allocryptopine, including the main ions at m/z 190.09 [M + H − C10H12O2 − H2O]+, m/z 208.09 [M + H − C10H12O2]+, m/z 354.17 [M + H − H2O]+, m/z 340.15 [M + H − CH3OH]+, m/z 165.09 [M + H − C11H14NO3]+, which could be deduced to possess two methoxyl groups on the C9–C10 positions or one methoxyl and one hydroxyl groups on the C9–C10 positions and one methyl on the C13 position. A comparison with the previous reports and online databases did not show any matching structures for the potentially new compounds. Peak 53 showed the protonated molecular ion at m/z 370.1654 [M + H]+ as well as the fragment ions at m/z 208.0968 [M + H − C10H10O2]+, m/z 190.0868 [M + H − C10H10O2 − H2O]+, m/z 352.1538 [M + H − H2O]+, m/z 163.0761 [M + H − C11H14NO3]+ indicating the existence of a methylenedioxyl group at the C9 and C10 positions and one methyl on the C13 position. Owing to no matching structures with the online databases, peak 53 was tentatively identified as a potential novel compound.
Using the characteristic fragment of 31.0422 Da to rapidly recognize aporphine-type alkaloids, which possessed (CH3)2NH group, peaks 1, 5, 8, 31, 48, 62, 82 and 83 were found. They all produced the highest intensity ion at [M + H − CH3NH2]+, corresponding to the neutral losses of 31 Da. They were tentatively identified as (+)-isoboldine or boldine,6,17,18, (+)-isoboldine or boldine,6,17,18 bulbocapnine,17 dehydroglaucine,20 nantenine3,21 or dicentrine,17 dehydroglaucine,20 and dehydronantenine,21 respectively. Furthermore, peak 43 displayed a molecular ion at m/z 382.1650 [M + H]+, based on fragment ions and the previous reports, peak 43 was deduced as 7-aldehyde dehydroglaucine. Peaks 49 and 74 displayed a molecular ion at m/z 352.1183 [M + H]+. Based on fragment ions and the previous reports, peaks 49 and 74 were deduced as oxoglaucine or its isomer.2,22 Peak 43 displayed a molecular ion at m/z 384.1806 [M + H]+, based on fragment ions and the previous reports, peak 79 was identified as 7-aldehyde glaucine.2 The detailed identification information of the aporphine alkaloids was given in Table S5.†
4. Conclusion
A target data screening strategy followed by characteristic fragment filtering by UPLC-Q-TOF/MSE was developed for rapidly and comprehensively identifying alkaloids in Corydalis yanhusuo W. T. Wang (Yanhusuo). A total of 86 compounds were identified or characterized. Among them, 8 were potential new compounds and 5 components were found in Yanhusuo for the first time. The construction of a database based on data screening tables was critical to the implementation of the method. Compared with other conventional database based reports in the published literature, the database based on data screening tables contained more comprehensive compounds. On the one hand, it enabled the supplementation of compounds due to inadequate literature research. On the other hand, it facilitated the discovery of potentially new compounds due to the design of all possible molecular weights. The results proved that the method was rapid and comprehensive for the identification and characterization of complex chemical compositions, but the method was still insufficient. The method was applicable to compounds with the same framework and was not suitable for compounds whose structural characteristics were not the same. This study provided a good reference for the rapid identification of other TCMs and provided meaningful guidance for the discovery of potentially new compounds.Conflicts of interest
The authors have declared no conflict of interest.Abbreviations
| UPLC Q-TOF/MS | Ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry |
| TCM | Traditional chinese medicine |
| BPI | Base peak ion chromatogram |
| ESI | Electro spray ionization |
| LC-MS | Liquid chromatography-tandem mass spectrometry |
Acknowledgements
This work was supported by the Key project of the National Natural Science Foundation of China (Grant Number: 81430094).References
- Chinese Pharmacopoeia Commission, Pharmacopoeia of the People's Republic of China, Chinese Medical Science and Technology Press, Beijing, 2015, vol. 1, pp. 139–140 Search PubMed.
- X. Yang, X. Yang and J. Liu, China J. Chin. Mater. Med., 2014, 39, 20–27 Search PubMed.
- Y. Q. Han, X. U. Jun, S. X. Gong, T. J. Zhang and C. X. Liu, Acta Pharm. Sin. B, 2016, 51, 1302–1308 Search PubMed.
- L. Yuan, J. Yin, M. Tian, J. Xie, Y. Wang, Z. Hou, Y. Li and Y. Zhang, Anal. Methods, 2016, 8, 2274–2281 RSC.
- M. Sun, J. Liu, C. Lin, M. Lan and L. Li, Acta Pharm. Sin. B, 2014, 4, 208–216 CrossRef PubMed.
- E. K. Jeong, S. Y. Lee, S. M. Yu, N. H. Park, H. S. Lee, Y. H. Yim, G. S. Hwang, C. Cheong, J. H. Jung and J. Hong, Rapid Commun. Mass Spectrom., 2012, 26, 1661 CrossRef CAS PubMed.
- Z. Jia-Yu, W. Zi-Jian, Z. Qian, W. Fang, M. Qun, L. Zhao-Zhou, L. Jian-Qiu and Q. Yan-Jiang, Talanta, 2014, 124, 111–122 CrossRef PubMed.
- F. X. Zhang, M. Li, L. R. Qiao, Z. H. Yao, C. Li, X. Y. Shen, Y. Wang, K. Yu, X. S. Yao and Y. Dai, J. Pharm. Biomed. Anal., 2016, 122, 59–80 CrossRef CAS PubMed.
- X. Qiao, R. Li, W. Song, W. J. Miao, J. Liu, H. B. Chen, D. A. Guo and M. Ye, J. Chromatogr. A, 2016, 1441, 83–95 CrossRef CAS PubMed.
- B. P. Ma, L. P. Kang, X. Pang and R. Y. Yan, Proceedings of the XIVth meeting of the European Society for Cognitive Psychology, ed. B. Hommel, G. P. Band, W. La Heij and G. Wolters, 2015, vol. 5, pp. 74–74 Search PubMed.
- H. Wu, K. Waldbauer, L. Tang, L. Xie, R. Mckinnon, M. Zehl, H. Yang, H. Xu and B. Kopp, Molecules, 2014, 19, 11487–11504 CrossRef PubMed.
- J. Zhang, Y. Jin, J. Dong, Y. Xiao, J. Feng, X. Xue, X. Zhang and X. Liang, Talanta, 2009, 78, 513–522 CrossRef CAS PubMed.
- Z. H. Cheng, Y. L. Guo, H. Y. Wang and G. Q. Chen, Anal. Chim. Acta, 2006, 555, 269–277 CrossRef CAS.
- X. Y. Cheng, Y. Shi, S. L. Zhen, H. Sun and W. Jin, J. Chromatogr. Sci., 2010, 48, 441–444 CAS.
- J. Zhang, Y. Jin, Y. Liu, Y. Xiao, J. Feng, X. Xue, X. Zhang and X. Liang, J. Sep. Sci., 2009, 2084–2089 CrossRef CAS PubMed.
- J. K. Lee, J. G. Cho, M. C. Song, J. S. Yoo, D. Y. Lee, H. J. Yang, K. M. Han, D. H. Kim, Y. J. Oh and T. S. Jeong, Appl. Biol. Chem., 2009, 52, 646–654 CAS.
- C. Stévigny, J. L. H. Jiwan, R. Rozenberg, E. D. Hoffmann and J. Quetin-Leclercq, Rapid Commun. Mass Spectrom., 2004, 18, 523–528 CrossRef PubMed.
- J. Schmidt, C. Boettcher, C. Kuhnt, T. M. Kutchan and M. H. Zenk, Phytochemistry, 2007, 68, 189–202 CrossRef CAS PubMed.
- Y. Zhang, Q. Shi, P. Shi, W. Zhang and Y. Cheng, Rapid Commun. Mass Spectrom., 2006, 20, 2328–2342 CrossRef CAS PubMed.
- J. Shi, X. Zhang, Z. Ma, M. Zhang and F. Sun, Molecules, 2010, 15, 3556–3566 CrossRef CAS PubMed.
- C. Y. Peng, J. Q. Liu, R. Zhang and J. C. Shu, Nat. Prod. Res., 2014, 28, 1159–1164 CrossRef CAS PubMed.
- Z. Lu, W. Sun, X. Duan, Z. Yang, Y. Liu and P. Tu, China J. Chin. Mater. Med., 2012, 37, 235 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra08720a |
| This journal is © The Royal Society of Chemistry 2017 |