Open Access Article
Open Access ArticlePhysicochemical characterization of high-quality bacterial cellulose produced by Komagataeibacter sp. strain W1 and identification of the associated genes in bacterial cellulose production†
Shan-Shan Wang‡
ab,
Yong-He Han‡ b,
Yu-Xuan Yec,
Xiao-Xia Shic,
Ping Xiang
b,
Yu-Xuan Yec,
Xiao-Xia Shic,
Ping Xiang c,
Deng-Long Chen*bd and
Min Li*a
c,
Deng-Long Chen*bd and
Min Li*a
aCollege of Life Science, Fujian Normal University, Fuzhou, 350108, China. E-mail: mli@fjnu.edu.cn
bQuangang Petrochemical Research Institute, Fujian Normal University, Quanzhou, 362801, China. E-mail: dlchen@fjnu.edu.cn; chendenglong@163.com
cState Key Laboratory of Pollution Control and Resource Reuse, School of the Environment, Nanjing University, Jiangsu 210023, China
dCollege of Environmental Science and Engineering, Fujian Normal University, Fuzhou, 350108, China
First published on 21st September 2017
Abstract
In the last few decades, bacteria capable of bacterial cellulose (BC) synthesis and the characterization of BC have been well-documented. In this study, a new BC-producing bacterial strain was isolated from fermented vinegar. The BC morphology, composition and diameter distribution, and the genes associated with BC production were analyzed. The results showed that one out of five isolates belonging to Komagataeibacter was a BC-producer, which mostly produced the typical cellulose I consisting of nanofibrils and had several functional groups similar to typing paper (i.e., plant cellulose). Several known genes such as glk, pgm and UPG2 in glucose metabolisms, bcsA, bcsB, bcsC and bcsD in BC synthesis and cmcax, ccpAx, bglxA and other genes in BC synthesis regulation or c-di-GMP metabolisms have also been found in the strain W1 based on genome sequencing and gene annotations. The functions of BcsX and BcxY might also be important for BC synthesis in Komagataeibacter sp. W1. Our study provided a new BC-producing bacterial strain that could be used to prepare high-quality BC and to study BC synthesis mechanisms.
1. Introduction
Cellulose is an organic compound that acts as a basic structural material of most plants and often combines with hemicellulose, lignin and pectin.1 Although plant-derived cellulose has been used in several areas such as paper-making, all-cellulose composite preparation and dietary fiber intake,2 it needs to be purified by enzymatic and/or mechanical treatments before further use.1,3 In addition to the high cost, these processes may also change the cellulose performance and limit its applications in several areas.1 Microorganisms including algae (e.g., Cladophora and Vallonia), fungi (e.g., Dictyostelium and Saprolegnia) and bacteria (e.g., Acetobacter, Achromobacter, Acidomonas, Aerobacter, Agrobacterium, Alcaligenes, Ameyamaea, Asaia, Gluconobacter, Gluconacetobacter, Granulibacter, Komagataeibacter, Kozakia, Neoasaia, Neokomagataea, Pseudomonas, Rhizobium, Saccharibacter, Sarcina, Swaminathania, Tanticharoenia and Zoogloea)4–6 also produce cellulose and have attracted people's attention due to its higher purity, crystallinity and water-holding capacity, lower production cost, thinner fiber diameter (i.e., ∼30–50 nm) and better biocompatibility as compared to plant cellulose.7,8 Now bacterial cellulose (BC) has been widely used as food material, paper-making material, acoustic or filter membrane, cosmetic and clothing material, medical material and electronic paper substrate.It is hypothesized that cellulose production in bacteria helps them to move to the oxygen-rich medium surface, protects them from ultraviolet light and retains moisture, and establishes close contact with a preferred host to facilitate efficient host–bacteria interactions.9,10 Based on molecular evidence, the synthesis of BC is a multistep process including two main mechanisms: the synthesis of uridine diphosphoglucose (UDPGlc), followed by the polymerization of glucose into long and unbranched chains, i.e., β-1,4-D-glucan, associated with hydrogen bonding.1,7 While the former mechanism is well known, the latter still needs exploring.
As the most two important members of BC producers, both Gluconacetobacter and Komagataeibacter are formerly grouped into Acetobacter and now regrouped into independent genera based on 16S rRNA gene sequences and phenotypic, ecologic and chemotaxonomic characteristics.11 Studies have shown that Gluconacetobacter is the most-well known genus due to its highest BC production ability among the reported BC-producers,1,7 while Komagataeibacter contributes mostly to the rapid pH decrease and BC production during vinegar fermentation.12,13 Although more than 14 species of Komagataeibacter have been regrouped from Gluconacetobacter,14,15 their BC production characteristics warrant further investigations. For example, most studied BC-encoding genes are bcsA, bcsB, bcsC and bcsD, but the functions of other genes such as bcsX and bcsY are little known.16 A recent study showed that gqqA encoded a novel protein involving in bacterial quorum quenching and cellulose formation, indicating more attention should be paid to BC synthesis mechanisms in bacteria.17
Since BC production is naturally occurring, most BC-producing bacteria are often isolated from the carbohydrate-containing substances such as rotten fruits, kombucha and vinegar.16,18,19 In the present study, a bacterium belonging to the genus Komagataeibater was firstly isolated from a famous vinegar factory located in Yongchun, Quanzhou, China. By subjecting the strain to Hestrin and Schramm (HS) medium, a typical BC membrane was found and characterized by scanning electron microscopy (SEM), X-ray diffraction (XRD) and Fourier transform infrared (FTIR) spectroscopy. Although the genomes of several species of Komagataeibacter have been sequenced, the gene information associated with BC synthesis and regulation is little known except for K. nataicola.6 To identify the genes involving in glucose utilization and BC synthesis, draft genome sequencing and associated annotations (e.g., COG functional prediction, NR annotation and Swissprot annotation) were also conducted in this study. The fully aim of this study was to give more precise work on understanding of BC synthesis mechanisms in bacteria and its importance in current and future applications.
2. Materials and methods
2.1. Sampling and bacteria isolation
To isolate BC-producing bacteria, both the vinegar and BC membranes were collected from Yongchun Laocu Industry Co., Ltd and stored in sterile plastic bottles (Corning, USA) in ice bath for taking back to lab. Before use, the BC membranes were washed carefully by sterile Milli-Q water to remove the surface bacteria, followed by soaking in 20 mL PBS containing 0.85% NaCl20 and shaking at 200 rpm for 30 min. After that, aliquots of vinegar and washing buffer were streaked onto HS agar medium consisting of 2% glucose, 0.5% peptone, 0.5% yeast extract, 0.68% Na2HPO4·12H2O, 0.051% MgSO4·7H2O, 0.115% C6H8O7·H2O and 1.8% agar (pH was adjusted to 6.0 by 1 M NaOH and HCl).21 All isolates were purified by re-streaking onto HS agar medium three times and subjected to HS liquid medium following 7 d incubation to pick out the bacteria capable of BC production. The obtained BC was pretreated as previously described and the BC-producing bacteria in the supernatant were mixed with 30% glycerol by 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) and stored at −80 °C or re-streaked onto HS agar medium following 7 d incubation and stored at 4 °C for further use.
1 (v/v) and stored at −80 °C or re-streaked onto HS agar medium following 7 d incubation and stored at 4 °C for further use.
2.2. Bacterial identification
All isolates that could grew on HS agar medium were collected and re-cultivated in HS liquid medium for 7 d, followed by centrifugation at 8000 × g for 5 min. The collected bacterial biomass was used to extract total DNA by using a FastDNA® SPIN Kit for Soil (MP Biomedicals, USA) according to the manufacturer's instructions. The 16S rRNA gene was amplified by the universal primers 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (ACGGCTACCTTGTTACGACTT) on a T100 Thermocycler (BioRad, USA).20 The polymerase chain reaction (PCR) mixtures consisted of 12.5 μL of 2 × Mix (Yifeixue Biotechnology, Nanjing, China), 1.5 μL of primer pair (10 μM), 10 μL of PCR degrade water and 1 μL of DNA template. The PCR programs for 16S rRNA amplification were: 5 min at 94 °C for pre-denaturation, 35 s at 94 °C for denaturation, 30 s at 55 °C for annealing and 1.5 min at 72 °C for extension (35 cycles), and 10 min at 72 °C for a final extension.20 The PCR products were purified and sequenced by GenScript Co., Ltd. (Nanjing, China). Based on the sequence analysis by BLAST similarity search in the NCBI database, the phylogenetic tree was constructed by using the neighbor-joining algorithm in the MEGA 5.0 program.To have an observation of bacterial morphology, the biomass was pretreated by a sequential dehydration procedure and freeze-dried for 24 h (FreeZone 6 plus, Labconco, USA) according to Wang et al.20 After a spray-gold treatment, the morphology was observed by SEM (Quanta™ 250 FEG, FEI, USA).
2.3. Bacterial cellulose characterization by SEM, XRD and FTIR
To observe the porous structure and fibril distribution on BC, one colony of the strain W1 was inoculated in HS liquid medium for 7 d, followed by washing and shaking treatments as shown above. After centrifugation at 8000 × g for 5 min, the bacterial biomass was re-suspended in 10 mL sterile Milli-Q water, aliquots of which were transferred to a new medium to a final OD600 at 0.01. After 7 d of incubation, the BC membranes were collected and washed by Milli-Q water, followed by boiling at 100 °C for 2 h by 0.1 M NaOH and another 2 h by Milli-Q water. The pre-treated BC was soaked in sterile Milli-Q water overnight at room temperature and then oven-dried at 50 °C. The dried BC was treated by spray-gold and observed by SEM. To evaluate the diameter distribution of BC fibrils, the SEM image of BC was analyzed on a NanoMeasurer 1.2 by calculating 100 nanofibrils randomly. In this study, the difference of each group of fibrils was 10 nm, and the results were presented as % of the total fibril numbers.Similar to SEM, all samples used for further characterization by XRD (Bruker D8 ADVANCE, Germany) and FTIR (Thermo Scientific Nicolet iS5, USA) were pre-treated as described. Of which, XRD pattern was obtained using nickel filtered copper Kα radiation, with 0.1° step, from 4° to 70° (2θ, angle), while FTIR analysis was conducted in the attenuated total reflection (ATR) mode with 32 scans per measurement between 400 and 4000 cm−1.
2.4. Draft genome sequencing and key functional genes annotation
To have an overview of BC synthesis mechanisms in the strain W1, the draft genome sequence and key functional genes were analyzed by Majorbio, Shanghai, China. Before sequencing, the total DNA was extracted as previously described and the DNA quality was evaluated by Nanodrop® ND-1000 (NanoDrop Technologies, Wilmington, DE, USA) based on DNA concentration and OD260/OD280. The established sample was fragmented to 400–500 bp by using M220 Focused-ultrasonicator™ (Covaris, Woburn, MA, USA), followed by library construction by using TruSeq™ DNA Sample Prep Kit (Illumina, Inc., USA) and sequencing on an Illumina Miseq (PE) platform by MiSeq Reagent KiT v3 (Illumina, Inc., USA). After data transformation by Base Calling, the raw sequences were saved as FASTQ and deposited in the NCBI-Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/Traces/sra) with the accession numbers PRJNA388252 (BioProject number), SAMN07173612 (BioSample number) and SRP108180 (SRA Study number).Before genome assembly, the low-quality data in the raw sequences were removed based on the following criteria: (1) the adapter sequences, (2) the sequences include A, G, C or T base in the 5′ terminal, (3) the terminal sequences with low quality values (<20), (4) the reads include ≥10% N and (5) the small sequences (<25 bp) after removing the adapter and low-quality data. The genome assembly was performed on a SOAP denovo platform (v2.04, http://soap.genomics.org.cn/) by multi-optimization of K-mer parameters, followed by a further base-proofreading by GapCloser (v1.12).22 The genome prediction was conducted by Glimmer (v3.02, http://www.cbcb.umd.edu/software/glimmer/). For functional gene annotation, all protein sequences in correspond to the predicted genes were analyzed by blastp research (BLAST 2.2.28+) against the known sequences in Nr, String, Genes and GO databases. Of which, the String annotation can obtain COG function analysis information, i.e., the protein function, classification and evolution status. Moreover, the blast results from Genes database can assign a certain gene to a KO number in KEEG pathway.
3. Results and discussion
3.1. Taxonomic characterization of BC-producing bacterial isolate
By using the typical HS agar medium for isolation of BC-producing bacteria, a total of five strains were obtained. However, only one (i.e., strain W1) produced BC in HS liquid medium. As shown in Fig. 1A, limited colonies of the strain W1 were found on HS agar medium, which might be due to the fact that BC-producing bacteria often use ∼50% energy to produce BC as compared to normal bacteria that use most energy to reproduce offspring.23 In addition, the strain W1 formed unregular colonies that might contain BC as the colonies were tough and smooth on agar medium (Fig. 1A). SEM observation found that the cells of strain W1 were short rod-shaped without flagellum (Fig. 1B). The size of each cell was 1.58 ± 0.11 μm long by 0.91 ± 0.09 μm wide (Fig. 1B).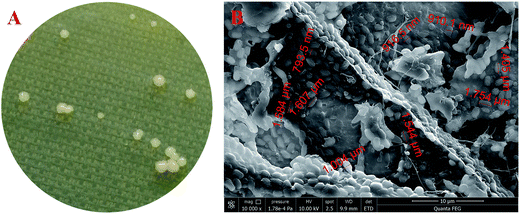 | ||
Fig. 1 Colony morphology (A) and SEM observation ((B), 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×) of Komagataeibacter sp. W1 isolated from spiced vinegar fermentation tank in Yongchun, Quanzhou, Fujian, China. 000×) of Komagataeibacter sp. W1 isolated from spiced vinegar fermentation tank in Yongchun, Quanzhou, Fujian, China. | ||
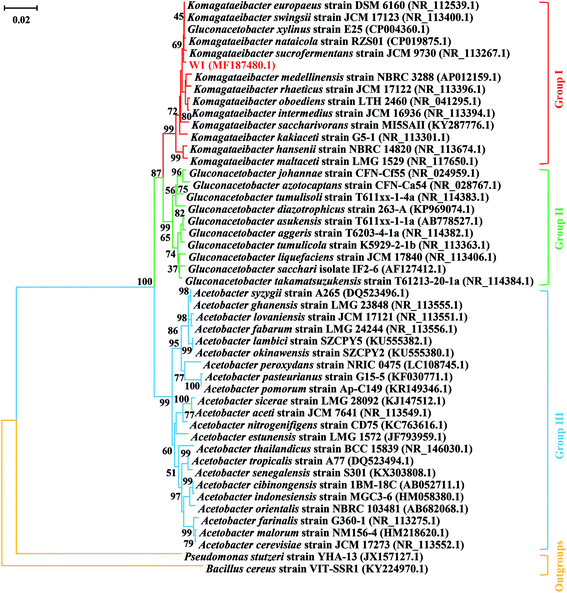
To further identify the strain W1, its 16S rRNA sequence (GenBank accession number MF187480) was aligned with the known sequences retrieved from the NCBI database. All three groups that were often confused with each other previously were listed. The results showed that although the 16S rRNA sequence of W1 shared high similarity with most reference sequences, it belonged to the Group I, i.e., the genus Komagataeibacter (Fig. 2). However, the strain hadn't been exactly classified at species level, thereby being named as Komagataeibacter sp. W1 (Fig. 2). Since Komagataeibacter is regrouped from the genus Gluconacetobacter, a typical and widely-studied BC-producing family, the strain isolated in this study could be a good candidate for BC preparation.14,15 Studies also showed that Komagataeibacter contributed mostly to pH decrease in vinegar production,12,13 indicating that the strain W1 was an important member in vinegar fermentation and BC production in the sampling site.
3.2. Morphological characteristics of cellulose produced by Komagataeibacter sp. strain W1
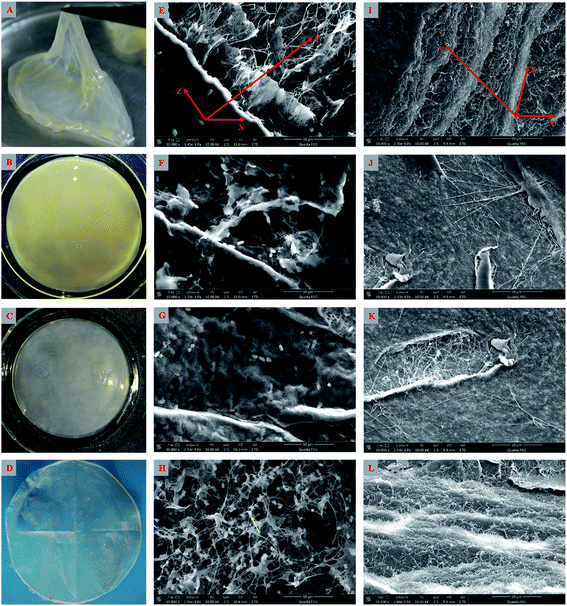
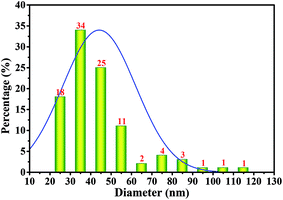
As shown above, the strain W1 produced BC membranes, which were pale yellow and transparent before and after treatment in 0.1 M NaOH bath, respectively (Fig. 3A–D). To have an in situ observation of the BC with or without NaOH treatment, all samples were oven-dried and BC images were taken by SEM. Results showed that the BC membranes had a three-dimensional structure and the bacteria distributed both on the surface of and inside BC membranes (Fig. 3E–L). It was also apparent that the bacteria produced fibrils on the cell surface (Fig. 3H), where several terminal complexes consist of BC synthase catalytic subunits, by a three-way branching pattern.23,24 In this case, a single cell can produce a ribbon of BC with 10–100 microfibrils.23 As a result of cell mitosis, the fibril assembly occurs, forming the glucan chain sheets by van der Waals bonding followed by stacking of the sheets by H-bonding to form the crystalline structure,25,26 which were supported by our data as shown in Fig. 3L. We hypothesized that the strain W1 produced a typical BC with high purity (Fig. 3I–L).To test the diameter distribution of the BC fibrils produced by Komagataeibacter sp. W1, 100 fibrils on SEM image were selected randomly and calculated by NanoMeasurer 1.2. As expected, 98% of the fibrils had the diameter <100 nm, 77% of which were <50 nm (Fig. 4), indicating that the BC obtained in this study consisted of nanofibrils. This was in line with previous studies that the visible BC fibrils consist of 40–60 nm cellulose ribbons, which are assembled by microfibrils consisting of 3–4 nm subelementary fibrils.27,28
3.3. XRD and FTIR analyses of cellulose produced by Komagataeibacter sp. strain W1
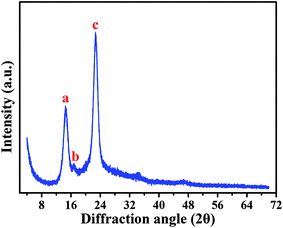
Based on a SEM observation, the BC produced by Komagataeibacter sp. strain W1 had some typical characteristics of the known BC, e.g., including nanofibrils and pellicles (Fig. 3). To further verify the BC compositions and purity, XRD and FTIR analyses were also performed.Fig. 5 shows XRD pattern of the BC produced by the strain W1. Similar to previous studies, three typical diffraction peaks were observed at 2θ 14.5°, 16.6° and 22.7° (Fig. 5), corresponding to (1![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) 0), (110), and (200) planes, respectively.29,30 Studies have shown that the broad peaks at 2θ 14.5° and 22.7° are associated with the presence of cellulose Iα and cellulose Iβ phases, i.e., 1001α, 1101β and 0101β planes at 14.5° and 1101α and 2001β at 22.7°.31,32 Thus, we concluded that W1-produced BC was pure. However, similar to other studies, the broad diffraction peaks with sharpness observed in this study indicated that the products included semi-crystalline BC (Fig. 5).29,30
0), (110), and (200) planes, respectively.29,30 Studies have shown that the broad peaks at 2θ 14.5° and 22.7° are associated with the presence of cellulose Iα and cellulose Iβ phases, i.e., 1001α, 1101β and 0101β planes at 14.5° and 1101α and 2001β at 22.7°.31,32 Thus, we concluded that W1-produced BC was pure. However, similar to other studies, the broad diffraction peaks with sharpness observed in this study indicated that the products included semi-crystalline BC (Fig. 5).29,30
In addition to XRD characterization, FTIR analysis is also an important alternative to identify the types and purity of BC.24,33 To have an comparative view on functional groups in cellulose, the FTIR spectrums of BC and typing paper (i.e., plant cellulose) were analyzed. Since the BC was difficult to be powdered, the ATR mode with 32 scans per measurement between 400 and 4000 cm−1 was used in this study according to previous studies.34,35 As shown in Table 1 and Fig. 6A, the W1-produced BC contained 17 functional groups, most of which were similar to those that in typing paper (Fig. 6B) and the BC produced by other bacteria.36,37 For example, the typical wavenumbers of BC produced by Komagataeibacter sp. W1 (i.e., 1426, 1335, 1314, 1160, 1108, 1054 and 1030 cm−1) were also found in A. xylinum ATCC 10245 (i.e., 1426, 1314, 1160 and 1053 cm−1), G. xylinus BCRC12335 (i.e., 1335, 1163, 1111, 1060 and 1035 cm−1) and K. saccharivorans PE5 (i.e., 1425, 1319, 1157 and 1023 cm−1).38,39
| Peak number | Wavenumber (cm−1) | Functional groupsa | |
|---|---|---|---|
| BC | Typing paper | ||
| a More information on the relationships between wavenumber and assignment can be found in Gea et al.66 and Moharram et al.41b Not detected.c Unknown. | |||
| 1 | 3338 | 3332 | O–H stretching vibration |
| 2 | 2895 | 2917 | C–H stretching of CH2 and CH3 groups |
| 3 | —b | 2865 | CH2 asymmetric stretching |
| 4 | 1642 | 1636 | H–O–H bending of absorbed water |
| 5 | 1426 | 1420 | CH2 symmetric bending or O–H in plane bending |
| 6 | 1361 | — | C–H bending |
| 7 | 1335 | 1335 | C–H deformation or O–H in-plane bending |
| 8 | 1314 | 1316 | Out-of-plane wagging of the CH2 groups |
| 9 | 1280 | — | C–H bending |
| 10 | 1204 | 1204 | |
| 11 | 1160 | 1160 | C–O–C antisymmetric bridge stretching of 1,4-β-D-glucoside |
| 12 | 1108 | 1105 | C–C bonds of the monomer units of polysaccharide or C–O bending vibration |
| 13 | 1054 | 1052 | The bending of C–O–H bond of carbohydrates or C–O–C pyranose ring skeletal vibration |
| 14 | 1030 | 1029 | |
| 15 | 899 | 897 | Antisymmetric out-of-phase ring stretching of β-glucosidic linkages between the glucose units |
| 16 | — | 872 | UKc |
| 17 | — | 712 | The monoclinic Iβ form of cellulose |
| 18 | 663 | 661 | O–H out-of-phase bending vibration |
| 19 | 598 | 600 | |
| 20 | 552 | 558 | |
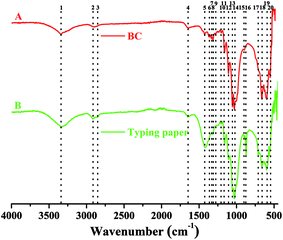 | ||
| Fig. 6 FTIR analysis of W1-produced bacterial cellulose on a Nicolet iS5 in the ATR mode with 32 scans per measurement between 400 and 4000 cm−1. | ||
Specifically, the typical absorption at around 3300 cm−1 (peak 1) corresponds to stretching vibration of intra and inter O–H in cellulose I.34,40,41 While the weak peak at around 2900 cm−1 (peaks 2 and 3) corresponds to C–H stretching of CH2 and CH3 groups or CH2 asymmetric stretching (Fig. 6), the latter is often found in plant cellulose (Fig. 6B).2,34 For both BC and typing paper, a peak at around 1650 cm−1 (peak 4) is observed, corresponding to H–O–H bending of absorbed water.2,40 Although it is thought that the peak at around 1430 cm−1 (peak 5) may originate from CH2 scissoring,42 most studies attribute it to CH2 symmetric bending or O–H in plane bending.2,43 The peak at around 1360 cm−1 corresponds to C–H bending, which also results in a typical absorption at 1280 and 1204 cm−1 (Fig. 6A).9,29,43 While the peak at 1335 cm−1 (peak 7) may correspond to C–H deformation or O–H in-plane bending,2,9 the absorption at around 1315 cm−1 (peak 8) is assigned to out-of-plane wagging of the CH2 groups.42 As a typical indicator of the presence of C–O–C antisymmetric bridge stretching of 1, 4-β-D-glucoside in BC, the absorption at around 1160 cm−1 is also observed.2,36 It has shown that the peaks at 1000–1100 cm−1 can be assigned to C–O stretching vibration in primary alcohol and C–O–C skeletal vibration.2,36,43 However, these hypotheses are controversial. For example, some studies attribute the absorption at 1030 and 1054 cm−1 to the bending of C–O–H bond of carbohydrates44–46 or C–O–C pyranose ring skeletal vibration.1,40,46 Moreover, Gao et al.47 showed that the peak at 1030 cm−1 might also be associated with the presence of OCH3, while the peak at 1108 cm−1 indicated C–C bonds of the monomer units of polysaccharide45 or C–O bending vibration,9 i.e., skeletal vibration. Furthermore, the peaks at 1162, 1107 and 1055 cm−1 can be assigned to C–C stretching vibration, skeletal vibration and ring vibration, respectively.48 Besides, we also found four peaks at 400–1000 cm−1 (Fig. 6A). Among of which, the peak at around 900 cm−1 corresponds to antisymmetric out-of-phase ring stretching of β-glucosidic linkages between the glucose units,2,43,49 which is designated as an “amorphous” absorption band, mainly contributing to the increase of BC intensity.45,50 For the peak at 712 cm−1, it is assigned to the monoclinic Iβ form of plant cellulose,9 but which is absent in BC (Fig. 6). Unlike the peak 17, the peaks 18–20 are present in both BC and typing paper, corresponding to O–H out-of-phase bending vibration.2
Since two weak peaks occur at around 1430 and 900 cm−1 (Fig. 6A), we hypothesized that the BC produced by Komagataeibacter sp. W1 mainly consisted of pure cellulose I,50–52 which was in accordance with XRD data (Fig. 5). Other peaks corresponding to cellulose I are also found in this study, e.g., 3338, 1160 and 899 cm−1, which have been well studied previously.53,54 However, another three weak peaks at 1335, 1314 and 1280 cm−1 and the blue-shift of wavenumber from 1430 to 1426 cm−1 give the evidence of the presence of cellulose II.53,54
3.4. Genome sequence identification in BC-producing bacterial isolate for cellulose synthesis and regulation
It is well-known that the BC production in bacteria is associated with several enzymes, including glucokinase (EC 2.7.1.2), phosphoglucomutase (EC 5.4.2.2), UTP-glucose-1-phosphate uridylyltransferase (EC 2.7.7.9) and cellulose synthases (EC 2.4.1.12).23 However, these enzymes vary in different genera of BC-producing bacteria.55 Moreover, the complete genomes of Komagataeibacter have only been sequenced in four species such as K. nataicola RZS01, K. xylinus E25, K. hansenii ATCC 23769 and K. medellinensis NBRC3288, while draft genomes are available for K. europaeus LMG18494, K. intermedius AF2, K. rhaeticus AF1, K. kakiaceti JCM 25156 and K. oboediens 174Bp2.6,16,56,57 To have an overview of the relevant enzymes in Komagataeibacter sp. W1, we also sequenced the draft genome and analyzed the functional genes and signaling pathways associated with BC synthesis.After removing the low-quality sequences, the total bases of clean data were about 1.32 Gb (Table S1†). The genome assembly showed that there were 168 scaffolds and 285 contigs in the draft genome (Table S2†). Moreover, a total of 3705 genes (i.e., open reading frames, orfs) were found based on sequence alignment with the known information in the KEEG database (Tables S3 and S4†). However, only 1783 and 1732 out of 3705 genes were annotated to COG functional categories and certain KEEG pathways, respectively (Fig. 7 and Table S5†). As shown in Fig. 7, there were 113 genes that corresponded to carbohydrate transport and metabolisms, of which 3 orfs were responsible for encoding glucokinase, phosphoglucomutase and UTP-glucose-1-phosphate uridylyltransferase, respectively, and 14 orfs were responsible for encoding cellulose synthases and associated co-enzymes (Table 2).
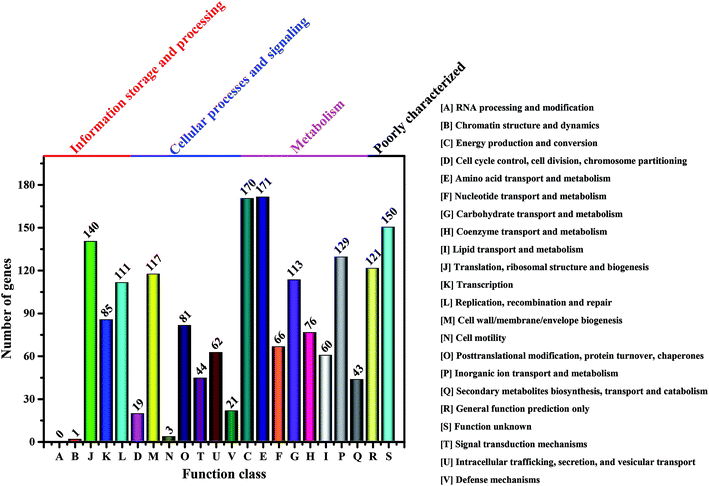 | ||
| Fig. 7 Number of genes associated with general COG functional categories based on the total number of protein coding genes in the genome. | ||
| Open reading frames number | Amino acid length (aa) | NR top hit | Similarity (%) | Swissprot top hit | Similarity (%) | Protein names |
|---|---|---|---|---|---|---|
| a The orfs that can be annotated to known pathways in KEEG pathway database as shown in Table 3.b The full sets of cellulose synthase genes in the bcs2 and bcs1 operons, respectively. | ||||||
| Glucose transformation | ||||||
| orf1235a | 322 | WP_019084814.1 | 100 | P21908.2 | 81 | Glucokinase [Komagataeibacter europaeus]; glucokinase = glucose kinase [Zymomonas mobilis subsp. mobilis ZM4 = ATCC 31821] |
| orf1086a | 555 | WP_019091484.1 | 100 | P38569.1 | 100 | Phosphoglucomutase [Komagataeibacter europaeus]; phosphoglucomutase = PGM = glucose phosphomutase [Komagataeibacter xylinus] |
| orf1122a | 280 | WP_019084882.1 | 100 | P27897.1 | 97 | UTP-glucose-1-phosphate uridylyltransferase [Komagataeibacter europaeus]; UTP-glucose-1-phosphate uridylyltransferase = alpha-D-glucosyl-1-phosphate uridylyltransferase = UDP-glucose pyrophosphorylas |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Cellulose synthesis | ||||||
| orf0140a | 1530 | — | — | Q9WX75.1 | 79 | Cellulose synthase catalytic subunit [UDP-forming] |
| orf0492b | 1328 | WP_026019532.1 | 100 | Q9WX71.1 | 88 | Cellulose synthase [Komagataeibacter europaeus]; cellulose synthase 2 operon protein C = flags: precursor [Komagataeibacter xylinus] |
| orf0493b | 386 | WP_019084639.1 | 100 | Q9WX70.1 | 94 | Acyltransferase [Komagataeibacter europaeus]; Putative membrane-bound transacylase BcsY [Komagataeibacter xylinus] |
| orf0494b | 223 | WP_019090487.1 | 100 | Q9WX69.1 | 99 | BcsX [Komagataeibacter europaeus]; protein BcsX [Komagataeibacter xylinus] |
| orf0495a,b | 1558 | — | — | Q9RBJ2.1 | 92 | Putative cellulose synthase 2 = cellulose synthase catalytic subunit [UDP-forming] |
| orf1576 | 321 | WP_019085254.1 | 92 | Q76KK0.1 | 60 | Hypothetical protein [Komagataeibacter europaeus]; cellulose-complementing protein [Komagataeibacter xylinus] |
| orf1578a,b | 691 | WP_026018529.1 | 100 | Q9WX61.1 | 96 | Cellulose synthase [Komagataeibacter europaeus]; cellulose synthase 1 catalytic subunit [Komagataeibacter xylinus] |
| orf1579a,b | 802 | — | — | Q9WX62.1 | 97 | Cyclic di-GMP-binding protein = CDGBP = cellulose synthase regulatory subunit = cellulose synthase protein B/flags: precursor |
| orf1580b | 1293 | WP_026019647.1 | 100 | Q9WX63.1 | 96 | Cellulose synthase 1 operon protein C/flags: precursor [Komagataeibacter xylinus]; cellulose synthase [Komagataeibacter europaeus] |
| orf1581b | 156 | WP_010507046.1 | 100 | Q9WX64.1 | 100 | Cellulose synthase operon protein D [Komagataeibacter xylinus]; cellulose synthase [Komagataeibacter europaeus] |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Cellulose synthesis regulation | ||||||
| orf1575 | 354 | WP_019085253.1 | 100 | P37696.1 | 86 | Endoglucanase [Komagataeibacter europaeus]; probable endoglucanase = cellulase = endo-1,4-beta-glucanase = flags: precursor [Komagataeibacter hansenii] |
| orf1582 | 734 | WP_010507044.1 | 100 | Q5BFG8.1 | 53 | Beta-glucosidase [Komagataeibacter europaeus]; beta-glucosidase B = beta-D-glucoside glucohydrolase B = cellobiase B = gentiobiase B |
| orf1669 | 325 | WP_019086736.1 | 100 | P58599.1 | 60 | Endoglucanase [Komagataeibacter europaeus]; endoglucanase = cellulase = endo-1,4-beta-glucanase = flags: precursor [Ralstonia solanacearum GMI1000] |
| orf2722 | 698 | WP_010508536.1 | 100 | Q9WX70.1 | 51 | Acyltransferase [Komagataeibacter europaeus]; putative membrane-bound transacylase BcsY [Komagataeibacter xylinus] |
By KEEG analysis, the enzymes encoded by orfs 1235, 1086 and 1122 were involved in glucose transformation in starch and sucrose metabolism pathway (Table 3 and Fig. S1†). All these enzymes co-produced the precursor (i.e., UDP-glucose) of BC (Fig. 8A), which was in accordance with the documented studies.23,27 Once UDP-glucose produced, the domain A encoded by bcsA and attached inner cell membrane is thought to catalyze the UDP-glucose to cellulose.27 This process also needs another core enzyme, i.e., BcsB, to accelerate the BC synthesis by combining to c-di-GMP.27,58,59 After that, the newly synthesized BC is crystallized with the aid of BcsD and then extruded by BcsC (Fig. 8B and C).60 As expected, all the genes associated with above proteins encoding were detected in the strain W1 and the corresponding amino acid sequences shared high similarity with K. europaeus and K. xylinus (Table 2). However, both BcsC and BcsD could not be annotated to certain KEEG pathways (Table 3). As stated by previous studies, these genes are located in bcs operon1 (bcs1).6,27 Similar to Zhang et al.,6 we also found two upstream genes cmcax (orf1575) and ccpAx (orf1576), and one downstream gene bglxA (orf1582) in bcs1 (Table 2 and Fig. 8A). While ccpAx encodes a cellulose-complementing protein,61,62 cmcax and bglxA encode endo-β-1,4-glucanase (EC 3.2.1.4) and β-glucosidase (EC 3.2.1.21), respectively, both of which assist cellulose biosynthesis by hydrolysing tangled glucan chains when a failure in chain arrangement occurs (Fig. S2†).6,63 It was interesting to note that another cmcax gene (orf 1669) was also found in Komagataeibacter sp. W1 (Table 2), which was different from K. nataicola6 but similar to K. europaeus (Table 2) and its functions in BC synthesis regulation warranted further studies. As shown in Table 2 and Fig. 8A, another bcs operon (bcs2) for BC synthesis was also found in this study. The bcs2 was composed of bcsA (orf0495), bcsX (orf0494), bcsY (orf0493) and bcsC (orf0492), of which bcsX and bcsY were located in the middle of bcsA and bcsC as previous report,6 but their potential roles in BC synthesis remained unknown. Morgan et al.64 showed that the protein encoded by bcsA contained the catalytically active subunit with a PilZ domain, which was responsive to c-di-GMP. Similar to Zhang et al.,6 we also found another bcsA (orf0140), which might be located on the reverse strand and its encoded protein did not contain a PilZ domain. Except for above genes, another bcsY (orf2722) was also located in the genome of Komagataeibacter sp. W1, which might involve in the expression of acyltransferase (Table 2).
| Open reading frames ID | Enzyme ID | Enzyme names | Ko ID (gene ID) | Ko names (gene names) | KEEG ID |
|---|---|---|---|---|---|
| orf1235 | 2.7.1.2 | Glucokinase | K12407; K00845 | GCK; glk | ko00010; ko00052; ko00500; ko00520; ko00521; ko00524 |
| orf1086 | 5.4.2.2 | Phosphoglucomutase | K01835 | pgm | ko00500; ko00520 |
| orf1122 | 2.7.7.9 | UTP-glucose-1-phosphate uridylyltransferase | K00963 | UGP2; galU; galF | ko00500 |
| orf0140, orf0495, orf1578, orf1579 | 2.4.1.12 | Cellulose synthase (UDP-forming) | K00694 | bcsA/B | ko00500 |
| orf1575, orf1669 | 3.2.1.4 | Endoglucanase | K01179 | cmcax | ko00500 |
| orf1582 | 3.2.1.21 | Beta-glucosidase | K01188 | bglxA | ko00500 |
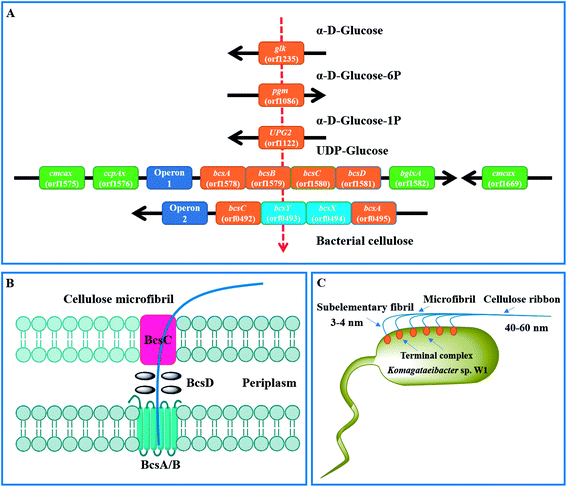 | ||
| Fig. 8 Mechanisms of BC synthesis in Komagataeibacter sp. W1. (A) genes associated with glucose transformation and BC synthesis; (B) proposed BC synthesis and excretion pathways by the regulation of bcs operon1;60 (C) BC assembly programs.27 | ||
As described, the PilZ domain is in response to the second messenger c-di-GMP. c-di-GMP is produced from 2 molecules of GTP by diguanylate cyclases (DGCs) and is broken down into 5′-phosphoguanylyl-(3′-5′)-guanosine by specific phosphodiesterases (PDEs).6 The c-di-GMP level can stimulate BC synthesis, leading to a 100-fold of BC increase.65 Similar to Zhang et al.,6 we found three cdg operons containing a c-di-GMP PDE gene (orf2263, orf2943 and orf3022) followed by a DGC gene (orf2264, orf2944 and orf3023). Besides, four standalone c-di-GMP PDE genes (orf1651, orf1917 and orf1918), one standalone cAMP DPE gene (orf2811) and three standalone DGC genes (orf0413, orf1556 and orf 2256) were also located in the genome of Komagataeibacter sp. W1 (Table S4†). Given the presence of two bcs operons in BC synthesis and three cdg operons in c-di-GMP regulation, it was possible that all these operons were attributed to the great ability of BC biosynthesis in Komagataeibacter sp. W1.
4. Conclusions
In this study, a BC-producing bacterium namely Komagataeibacter sp. W1 was isolated from a fermented vinegar. The SEM, XRD and FTIR analyses revealed that the pure BC produced by W1 was mostly cellulose I and composed of nanofibrils. By a draft genome sequencing, all typical genes associated with BC synthesis and regulation were found. However, the functional verification of some genes that might involve in BC synthesis co-regulation warranted future research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in party by the Science and Technology Program of Fujian Province (2017Y0027), the Education Department Fund of Fujian Province (JAT170144), the Research Project of Fujian Normal University (No. 5c1005), the Special Fund of Quangang Petrochemical Research Institute of Fujian Normal University (2016YJY01) and the Hong-Wu Weng Original Fund of Peking University.References
- C. Castro, A. Vesterinen, R. Zuluaga, G. Caro, I. Filpponen, O. J. Rojas, G. Kortaberria and P. Gañán, Cellulose, 2014, 21, 1745–1756 CrossRef CAS.
- A. Ashori, S. Sheykhnazari, T. Tabarsa, A. Shakeri and M. Golalipour, Carbohydr. Polym., 2012, 90, 413–418 CrossRef CAS PubMed.
- H. P. S. A. Khalil, A. H. Bhat and A. F. I. Yusra, Carbohydr. Polym., 2012, 87, 963–979 CrossRef.
- Y. Nishiyama, J. Wood Sci., 2009, 55, 241–249 CrossRef CAS.
- S. M. A. S. Keshk and A. F. El-Kott, in Science and Principles of Biodegradable and Bioresorbable Medical Polymers, ed. X. C. Zhang, Woodhead Publishing is an imprint of Elsevier, United Kingdom, 2017, pp. 295–319 Search PubMed.
- H. Zhang, X. Xu, X. Chen, F. Yuan, B. Sun, Y. Xu, J. Yang and D. Sun, Sci. Rep., 2017, 7, 4431 CrossRef PubMed.
- I. Reiniati, A. N. Hrymak and A. Margaritis, Crit. Rev. Biotechnol., 2017, 37, 510–524 CrossRef CAS PubMed.
- H. K. Uzyol and M. T. Saçan, Environ. Sci. Pollut. Res., 2017, 24, 11154–11162 CrossRef CAS PubMed.
- C. Castro, R. Zuluaga, J.-L. Putaux, G. Caro, I. Mondragon and P. Gañán, Carbohydr. Polym., 2011, 84, 96–102 CrossRef CAS.
- R. V. Augimeri, A. J. Varley and J. L. Strap, Front. Microbiol., 2015, 6, 1282 Search PubMed.
- K. Komagata, T. Iino and Y. Yamada, in The Prokaryotes, ed. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt and F. Thompson, Springer Berlin Heidelberg, Germany, Berlin, 2014, pp. 3–78 Search PubMed.
- F. Barja, C. Andrés-Barrao, R. O. Pérez, E. M. Cabello and M.-L. Chappuis, in Acetic Acid Bacteria, ed. K. Matsushita, H. Toyama, N. Tonouchi and A. Okamoto-Kainuma, Springer, Japan, 2016, pp. 201–221 Search PubMed.
- M. J. Valera, M. J. Torija, A. Mas and E. Mateo, Appl. Microbiol. Biotechnol., 2015, 99, 1349–1361 CrossRef CAS PubMed.
- Y. Yamada, Int. J. Syst. Evol. Microbiol., 2014, 64, 1670–1672 CrossRef PubMed.
- Y. Yamada, P. Yukphan, H. T. L. Vu, Y. Muramatsu, D. Ochaikul, S. Tanasupawat and Y. Nakagawa, J. Gen. Appl. Microbiol., 2012, 58, 397–404 CrossRef CAS PubMed.
- M. J. Valera, A. Poehlein, M. J. Torija, F. S. Haack, R. Daniel, W. R. Streit, E. Mateo and A. Mas, Genome Announc., 2015, 3, e01231-15 CrossRef PubMed.
- M. J. Valera, A. Mas, W. R. Streit and E. Mateo, Microb. Cell Fact., 2016, 15, 88 CrossRef PubMed.
- R. Jayabalan, R. V. Malbaša, E. S. Lončar, J. S. Vitas and M. Sathishkumar, Compr. Rev. Food Sci. Food Saf., 2014, 13, 538–550 CrossRef.
- J. K. Park, Y. H. Park and J. Y. Jung, Biotechnol. Bioprocess Eng., 2003, 8, 83–88 CrossRef CAS.
- S.-S. Wang, S.-L. Ye, Y.-H. Han, X.-X. Shi, D.-L. Chen and M. Li, RSC Adv., 2016, 6, 101153–101161 RSC.
- S. Hestrin and M. Schramm, Biochem. J., 1954, 58, 345–352 CrossRef CAS PubMed.
- R. Li, H. Zhu, J. Ruan, W. Qian, X. Fang, Z. Shi, Y. Li, S. Li, G. Shan, K. Kristiansen, S. Li, H. Yang, J. Wang and J. Wang, Genome Res., 2010, 20, 265–272 CrossRef CAS PubMed.
- I. M. Saxena and J. R. Malcolm Brown, in Bacterial NanoCellulose-A Sophisticated Multifunctional Material, ed. M. Gama, P. Gatenholm and D. Klemm, CRC Press, USA, Boca Raton, Florida, 2013, pp. 1–18 Search PubMed.
- Y. Nishi, M. Uryu, S. Yamanaka, K. Watanabe, N. Kitamura, M. Iguchi and S. Mitsuhashi, J. Mater. Sci., 1990, 25, 2997–3001 CrossRef CAS.
- S. K. Cousins and R. M. Brown Jr, Polymer, 1995, 36, 3885–3888 CrossRef CAS.
- S. K. Cousins and R. M. Brown Jr, Polymer, 1997, 38, 903–912 CrossRef CAS.
- Y. Amano, F. Ito and T. Kanda, J. Biol. Macromol., 2005, 5, 3–10 CAS.
- S. Yamanaka, K. Watanabe, N. Kitamura, M. Iguchi, S. Mitsuhashi, Y. Nishi and M. Uryu, J. Mater. Sci., 1989, 24, 3141–3145 CrossRef CAS.
- M. Ul-Islam, J. H. Ha, T. Khan and J. K. Park, Carbohydr. Polym., 2013, 92, 360–366 CrossRef CAS PubMed.
- H. Luo, G. Xiong, Z. Yang, S. R. Raman, H. Si and Y. Wan, RSC Adv., 2014, 4, 14369–14372 RSC.
- M. Wada and T. Okano, Cellulose, 2001, 8, 183–188 CrossRef CAS.
- A. C. O'Sullivan, Cellulose, 1997, 4, 173–207 CrossRef.
- K. Watanabe, M. Tabuchi, Y. Morinaga and F. Yoshinaga, Cellulose, 1998, 5, 187–200 CrossRef CAS.
- A. Tercjak, J. Gutierrez, H. d. S. Barud, R. R. Domeneguetti and S. J. L. Ribeiro, ACS Appl. Mater. Interfaces, 2015, 7, 4142–4150 CAS.
- H. Sai, R. Fu, L. Xing, J. Xiang, Z. Li, F. Li and T. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 7373–7381 CAS.
- M. C. I. M. Amin, N. Ahmad, N. Halib and I. Ahmad, Carbohydr. Polym., 2012, 88, 465–473 CrossRef.
- L. Hong, Y. L. Wang, S. R. Jia, Y. Huang, C. Gao and Y. Z. Wan, Mater. Lett., 2006, 60, 1710–1713 CrossRef CAS.
- E. A. Hassan, H. M. Abdelhady, S. S. A. El-Salam and S. M. Abdullah, Br. Microbiol. Res. J., 2015, 9, 1–13 CrossRef CAS.
- H.-H. Chen, L.-C. Chen, H.-C. Huang and S.-B. Lin, Cellulose, 2011, 18, 1573–1583 CrossRef CAS.
- F. Yassine, N. Bassil, A. Chokr, A. E. Samrani, A. Serghei, G. Boiteux and M. E. Tahchi, Cellulose, 2016, 23, 1087–1100 CrossRef CAS.
- M. A. Moharram and O. M. Mahmoud, J. Appl. Polym. Sci., 2008, 107, 30–36 CrossRef CAS.
- S. Dammström, L. Salmén and P. Gatenholm, Polymer, 2005, 46, 10364–10371 CrossRef.
- H. S. Barud, J. L. Souza, D. B. Santos, M. S. Crespi, C. A. Ribeiro, Y. Messaddeq and S. J. L. Ribeiro, Carbohydr. Polym., 2011, 83, 1279–1284 CrossRef CAS.
- D. Sun, J. Yang and X. Wang, Nanoscale, 2010, 2, 287–292 RSC.
- M. S. Dayal, N. Goswami, A. Sahai, V. Jain, G. Mathur and A. Mathur, Carbohydr. Polym., 2013, 94, 12–16 CrossRef CAS PubMed.
- C. Huang, X.-Y. Yang, L. Xiong, H.-J. Guo, J. Luo, B. Wang, H.-R. Zhang, X.-Q. Lin and X.-D. Chen, Lett. Appl. Microbiol., 2015, 60, 491–496 CrossRef CAS PubMed.
- W.-H. Gao, K.-F. Chen, R.-D. Yang, F. Yang and W.-J. Han, BioResources, 2011, 6, 144–153 CAS.
- D. Li, K. Ao, Q. Wang, P. Lv and Q. Wei, Molecules, 2016, 21, 618 CrossRef PubMed.
- T. Kondo and C. Sawatari, Polymer, 1996, 37, 393–399 CrossRef CAS.
- W. N. Goh, A. Rosma, B. Kaur, A. Fazilah, A. A. Karim and R. Bhat, Int. Food Res. J., 2012, 19, 153–158 CAS.
- X. Feng, N. Ullah, X. Wang, X. Sun, C. Li, Y. Bai, L. Chen and Z. Li, J. Food Sci., 2015, 80, E2217–E2227 CrossRef CAS PubMed.
- M. L. Nelson and R. T. O'Connor, J. Appl. Polym. Sci., 1964, 8, 1311–1324 CrossRef CAS.
- M. U. Rani, N. K. Rastogi and K. A. A. Appaiah, J. Microbiol. Biotechnol., 2011, 21, 739–745 CrossRef CAS PubMed.
- M. U. Rani, K. Udayasankar and K. A. A. Appaiah, J. Appl. Polym. Sci., 2011, 120, 2835–2841 CrossRef CAS.
- B. L. Quéré and J.-M. Ghigo, Mol. Microbiol., 2009, 72, 724–740 CrossRef PubMed.
- R. A. C. d. Santos, A. A. Berretta, H. d. S. Barud, S. J. L. Ribeiro, L. N. González-García, T. D. Zucchi, G. H. Goldman and D. M. Riaño-Pachón, Genome Announc., 2015, 3, e01404-15 CrossRef PubMed.
- R. A. C. d. Santos, A. A. Berretta, H. d. S. Barud, S. J. L. Ribeiro, L. N. González-García, T. D. Zucchi, G. H. Goldman and D. M. Riaño-Pachón, Genome Announc., 2014, 2, e00731-14 CrossRef PubMed.
- O. Omadjela, A. Narahari, J. Strumillo, H. Mélida, O. Mazur, V. Bulone and J. Zimmer, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 17856–17861 CrossRef CAS PubMed.
- J. B. McManus, Y. Deng, N. Nagachar, T.-h. Kao and M. Tien, Enzyme Microb. Technol., 2016, 82, 58–65 CrossRef CAS PubMed.
- P. R. Iyer, J. Catchmark, N. R. Brown and M. Tien, Cellulose, 2011, 18, 739–747 CrossRef CAS.
- S. Kawano, K. Tajima, Y. Uemori, H. Yamashita, T. Erata, M. Munekata and M. Takai, DNA Res., 2002, 9, 149–156 CrossRef CAS PubMed.
- N. Sunagawa, T. Fujiwara, T. Yoda, S. Kawano, Y. Satoh, M. Yao, K. Tajima and T. Dairi, J. Biosci. Bioeng., 2013, 115, 607–612 CrossRef CAS PubMed.
- T. Nakai, Y. Sugano, M. Shoda, H. Sakakibara, K. Oiwa, S. Tuzi, T. Imai, J. Sugiyama, M. Takeuchi, D. Yamauchi and Y. Mineyuki, J. Bacteriol., 2013, 195, 958–964 CrossRef CAS PubMed.
- J. L. W. Morgan, J. Strumillo and J. Zimmer, Nature, 2013, 493, 181–186 CrossRef CAS PubMed.
- P. Ross, R. Mayer and M. Benziman, Microbiol. Mol. Biol. Rev., 1991, 55, 35–58 CAS.
- S. Gea, C. T. Reynolds, N. Roohpour, B. Wirjosentono, N. Soykeabkaew, E. Bilotti and T. Peijs, Bioresour. Technol., 2011, 102, 9105–9110 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra08391b |
| ‡ These two authors contributed equally to this work and should be considered co-first authors. |
| This journal is © The Royal Society of Chemistry 2017 |