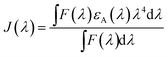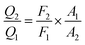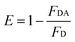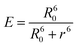DOI:
10.1039/C7RA06977D
(Paper)
RSC Adv., 2017,
7, 44348-44355
Conformational discrimination of three human centrins
Received
22nd June 2017
, Accepted 31st August 2017
First published on 14th September 2017
Abstract
Centrin belongs to the calcium-binding super-family, and is essential for the microtubule-organizing center (MTOC). With different locations and functions in the cell, three human centrin proteins have been identified. To obtain their structural basis, using 2-p-toluidinylnaphthalene-6-sulfonate (TNS) as a fluorescent probe, we investigated their conformational discrimination. The results suggest that the three human centrins contain large hydrophobic cavities, and the hydrophobic cavity of human centrin 1 (HsCen1) was the most pronounced. In addition, Tb3+ may induce centrin conformational changes and hydrophobic surface exposure. In 100 mM N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid (Hepes, pH 7.4) at different temperatures, thermodynamic data of centrin binding with TNS were measured. Based on Förster non-radiative energy transfer theory, the distances of TNS binding with human centrins HsCen1, HsCen2, and HsCen3 were measured. These results may provide some insight into structural information regarding why the three human centrins exhibit various biological functions.
1. Introduction
The animal microtubule-organizing center (MTOC), also known as the centrosome, is an exclusive structure found in eukaryotic cells. It consists of about 100 proteins, one of which has been identified as centrin or caltractin.1–3 Centrin belongs to the EF-hand super-family of calmodulin (CaM).4,5 Initially, it was identified in unicellular green algae such as Tetraselmis striata6 and Chlamydomonas reinhardtii.7 Later, it was shown to be a ubiquitous component of centrioles, centrosomes, and mitotic spindle poles.8 To date, homologous proteins have been identified in protozoa and yeast, as well as in plants and humans.9,10 There are three centrin homologs: centrin (1), centrin (2), and centrin (3). These centrins localize in different cellular compartments or are associated with specialized tissues with different biological functions. Human centrin (1) (HsCen1) is only expressed in male germ cells and localizes at the base of the flagella in the sperm.11 By contributing the mother centriole to the ovum allowing for centriole duplication, HsCen1 is involved in the first division of the zygote.12,13 HsCen2 and HsCen3 are expressed ubiquitously in somatic cells.14 HsCen2 has been found to take part in DNA excision repair within the nucleus15,16 and exporting mRNA from the cell nucleus. As for HsCen3, it has been reported to localize at the distal lumen of the centrioles and basal bodies.17,18 The centrins from humans, named as HsCen1-HsCen3, are small molecular mass (about 20 kDa), acid and calcium binding proteins that share common biochemical and structural properties with each other, as well as with Euplotes octocarinatus centrin (EoCen). The sequence alignments of centrins from different organisms with CaM are above 52%. Hence, it can be inferred that centrin is also composed of two independent globular domains connected by a flexible linker.1 Like other EF-hand proteins, centrins respond to cellular Ca2+ influx by selectively binding Ca2+ via highly conserved helix-loop-helix motifs. Through binding with Ca2+, structural rearrangement of α-helices occurs, resulting in the exposure of hydrophobic clefts.19 So far, the NMR structure (2joj.pdb) of the N-terminal domain on EoCen and the X-ray structure of HsCen2 bound with a peptide from xeroderma pigmentosum complementation group C (XPC) have been reported. Due to the flexibility and various conformations of centrins, no X-ray structures of a full centrin protein have been obtained.
Lanthanides (Ln) have attracted intensive research interest due to their diverse range of biological effects,20 and their applications in medicine.21 By virtue of holding similar ionic radii and coordination numbers, the lanthanides can substitute Ca2+ in proteins. Centrin may be a potential target of Ln via a calcium-dependent signal transduction pathway. Our previous work has also proved that Ln can bind with EF-hand motifs on EoCen at calcium-binding sites, promote the self-assembly of EoCen, contribute to protein kinase A phosphorylating EoCen at Ser166 and affect EoCen binding with melittin under physiological conditions.22 We also obtained evidence of the potential of centrins in reducing oxidative stress through binding with hemin.23 So, why do the three human centrins display different functions? Hardly any information regarding the structural basis of the three human centrins is available.
In this work, the three human centrins, HsCen1 (1–172 aa), HsCen2 (1–172 aa) and HsCen3 (1–167 aa), were constructed, expressed and purified using biological technology. Using fluorescence emission spectra, the interactions of the three proteins HsCen1, HsCen2, and HsCen3 with the hydrophobic probe 2-p-toluidinylnaphthalene-6-sulfonate (TNS) under neutral conditions were investigated. Our results revealed that HsCen1, HsCen2 and HsCen3 exhibit different hydrophobic surfaces to each other. Furthermore, metal ions may induce conformational changes in the three human centrins. However, the conformations of Tb3+-bound HsCen1, HsCen2, and HsCen3 were not completely identical. Under neutral conditions, the three proteins displayed different binding constants and different distances with TNS, which will shed light on the fundament of the biological functions of the three human centrins in the cell.
2. Materials and methods
2.1 Reagents
N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid (Hepes), disodium ethylenediaminetetraacetic acid (EDTA) buffer and 2-p-toluidinylnaphthalene-6-sulfonate (TNS) were purchased from Sigma. Hepes, TNS, salts, and other biological materials and chemicals ultilized in protein purification are of analytical grade.
Tryptone, yeast extract, ampicillin (Ampr) and isopropyl-β-D-thiogalactoside (IPTG) were purchased from Amresco Ltd. Other biochemical reagents used in the construction, expression and purification of proteins were purchased from TaKaRa.
2.2 Protein preparation
Three constructs of human centrin (1) (HsCen1), human centrin (2) (HsCen2) and human centrin (3) (HsCen3) were used in this paper. Target genes of the human centrins were obtained by PCR and then sub-cloned into the pGEX-6p-1 expression vector. The recombinant plasmids of HsCen1, HsCen2 and HsCen3 in the pGEX-6p-1 vector were constructed. After verification by DNA sequence analysis, the recombinant plasmids were transferred into E. coli (DE3), which was incubated at 37 °C. At an optical density of 0.4–0.6 (at 600 nm), protein synthesis was induced using isopropyl-β-D-thiogalactopyranoside (IPTG 0.5 mM) for 5 h. The HsCen1, HsCen2, and HsCen3 proteins were purified as GST fusion proteins through glutathione sepharose 4FF in PBS (KH2PO4 1.8, Na2HPO4 10, KCl 2.7 and NaCl 140 mM). The GST fusion proteins were then cleaved by PreScission Protease (PPase) at 4 °C and the cleavage protein fragments were further purified by HiLoid™ 16/60 Superdex™ 200.21 The purity of the intermediate and final samples was assessed by SDS-PAGE. After purification, the proteins were concentrated and kept at −80 °C. The proteins of HsCen1, HsCen2 and HsCen3 were soluble and their concentrations were measured using the absorption at 280 nm with an extinction coefficient of ε280(HsCen1) = 1490 M−1 cm−1, ε280(HsCen2) = 4350 M−1 cm−1 and ε280(HsCen3) = 8480 M−1 cm−1, respectively.
2.3 Metal removal
To remove contaminating bound cations, the protein samples were first pretreated with EDTA (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and then passed through a 60 cm × 16 cm Sephadex S200 column equilibrated in PBS buffer, at pH 7.4.
1) and then passed through a 60 cm × 16 cm Sephadex S200 column equilibrated in PBS buffer, at pH 7.4.
2.4 Interaction with hydrophobic probes
Binding of the human centrins with TNS was monitored by fluorescence spectrometer. After incubation of the human centrin samples (17 μM) with 80 μM TNS for 5 min in the presence or absence of metal ions, the mixed solutions were excited at 322 nm. The interaction of TNS with the human centrins at different temperatures has also been investigated under similar experimental conditions. A filter with a long pass of 390 nm was used to avoid secondary Rayleigh scattering.
3. Results and discussion
3.1 Expression and purification of centrins
The human centrins were GST-tagged at their N-termini. Engineered E. coli BL21 strains containing the recombinant plasmids pGEX-6p-HsCen1, pGEX-6p-HsCen2 or pGEX-6p-HsCen3 were cultured at 37 °C in 2YT medium for 3–4 h to produce the corresponding proteins. Following IPTG induction, soluble centrins were present in the supernatant of the bacterial lysate and then purified by glutathione sepharose 4FF as a GST fusion protein in PBS buffer (Fig. 1A). The GST fusion proteins were then cleaved by PreScission Protease (PPase) overnight at 4 °C, and the cleavage protein fragments of the human centrins were obtained. To obtain metal-free proteins, the human centrins were further treated with EDTA and then passed through a desalination column and a Sephadex S200 column (Fig. 1B).
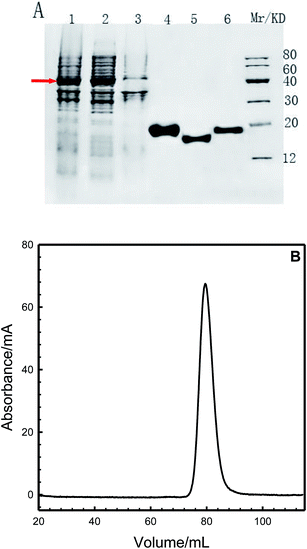 |
| | Fig. 1 (A): SDS-PAGE analysis of the expression and purification of human centrins in E. coli BL21 (DE3). (1) whole bacterial; (2) supernatant; (3) precipitate; (4) purified HsCen1; (5) purified HsCen2; (6) purified HsCen3 and marker. Arrow shows fusion proteins. (B): Elution profile of HsCen1 from the HiLoid™ 16/60 Sephadex gel filtration column of an AKTA explorer (Amersham). | |
3.2 Hydrophobic cavities of the human centrins
TNS is a useful probe for investigating hydrophobic cavities on proteins, due to its different fluorescence emission site and fluorescence intensity in polar and non-polar solutions. It is generally used to detect the exposed surfaces on proteins.24–26 It can be seen from Fig. 2A that TNS displayed only weak fluorescence emission at 500 nm in 100 mM Hepes and 150 mM NaCl at pH 7.4. With the addition of HsCen1, HsCen2 or HsCen3 protein, the fluorescence emission of TNS shifted to 430 nm, indicating that the environment of TNS changed from hydrophilic to hydrophobic. At the same time, the fluorescence intensity was increased significantly. However, HsCen1, HsCen2 and HsCen3 induced different degrees of fluorescence enhancement for TNS, whether the HsCen protein was in the metal-free state (apo state) or in the metal-saturated state (holo state) (Fig. 2). Following the addition of HsCen1, HsCen2 and HsCen3, TNS fluorescence emission was increased by 5.5-fold, 2.0-fold and 2.4-fold, respectively. Similar phenomena of fluorescence enhancement and fluorescence emission blue-shift have been reported by mixing TNS with centrin from Euplotes.8,22 Numerous studies have reported that upon Ca2+ binding, centrin undergoes a large conformational change from a closed state to an open state and in turn regulates a vast number of target proteins.27–29 After binding with metal ions, centrin indeed takes on a different conformation. As shown in Fig. 2B and C, TNS fluorescence was increased 12.5-fold, 5.5-fold and 6.4-fold with the addition of Tb3+-saturated HsCen1, HsCen2 and HsCen3, respectively. In the presence of a human centrin in the metal-free state (apoHsCen), the fluorescence emission of TNS was weaker than that in the presence of a metal ion-saturated protein (holoHsCen). Furthermore, HsCen1 exhibits more hydrophobic domains than HsCen2 or HsCen3, suggesting that HsCen1 may have a special biological function in regulating cell processes. The reported structures of HsCen2, truncated or in a complex with a peptide, proved that many hydrophobic cavities exist on centrin to stabilize the protein side chains, participating in hydrophobic packing or activating downstream target proteins and so on.3,14 Most of the residues (Phe, Ile, Leu, and Met etc.) forming the hydrophobic cores are conserved or conservatively substituted in other centrins or members of the CaM super-family. The conserved residues of Met145, Ile165, Met166, Ile126, Ile121 etc. (numbers from the HsCen2 sequence) in the three centrins form the hydrophobic core, through which peptides are bound. Sequence alignments indicated 84% sequence identity of HsCen2 with HsCen1 and 53% sequence identity of HsCen2 with HsCen3. It can be seen that some key residues were changed. By virtue of replacement of Ala149 in HsCen1 and HsCen2 with Phe149 in HsCen3, Val157 in HsCen1 and HsCen2 with Ile157 in HsCen3, and Phe113 in HsCen2 with Lys113 in HsCen1 and HsCen3, the binding affinities of the three human centrins with peptides were changed. In addition, Met97 and Phe62 from the N-terminal domain of HsCen2 may form a hydrophobic cap that controls whether the entrance into the internal core is open or closed. Conversion of Met97 to Val97 resulted in a change in the hydrophobic properties and protein anchoring. Based on the X-ray structure of HsCen2, homologous simulated structures of HsCen1 and HsCen3 are shown in Fig. 2. It can be seen that the hydrophobic characterization of HsCen3 is more obvious than that of HsCen1 or HsCen2. The three human centrins displayed various conformations. Thus, they showed different binding properties with TNS.
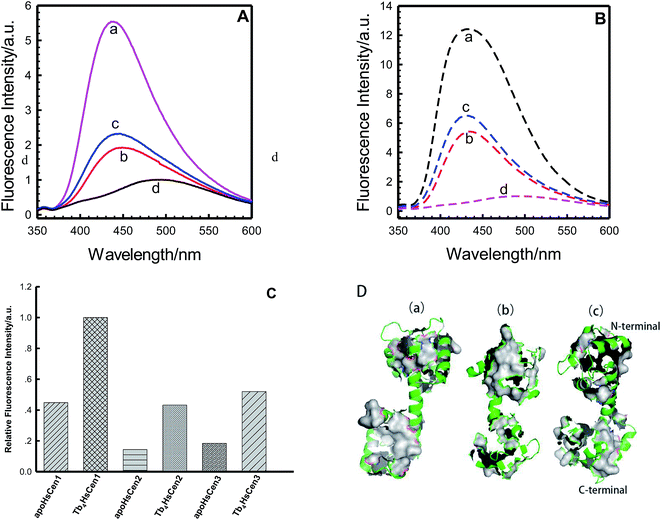 |
| | Fig. 2 (A): Fluorescence emission of TNS alone (d), and in the presence of HsCen1 (a), HsCen2 (b) or HsCen3 (c) in 100 mM Hepes, 150 mM NaCl at pH 7.4 and room temperature. The concentration of proteins was 17 μM. (B): Fluorescence emission of TNS alone (d), and in the presence of Tb3+-HsCen1 (a), Tb3+-HsCen2 (b) or Tb3+-HsCen3 (c) in 100 mM Hepes, 150 mM NaCl at pH 7.4 and room temperature. The concentration of proteins was 17 μM. (C): Fluorescence enhancement of TNS complexed with HsCen1, HsCen2 or HsCen3 in the metal ion-free state or metal ion-saturated state, compared with the fluorescence intensity of TNS alone. (D): Molecular simulation of complexes of HsCen1 (a) and HsCen3 (c), and the X-ray structure of HsCen2 (b). Backbone (green), hydrophobic surface (gray). | |
3.3 TNS binding with human centrins
In the presence of human centrins, the fluorescence intensity at 440 nm of TNS at each point in the titration was divided by the analytical concentration of TNS to eliminate the dilution effect and give the CPS value. Titration curves were prepared by plotting CPS vs. [TNS]/[HsCen] (Fig. 3). It can be seen that one inflexion appeared at about [TNS]/[HsCen] = 4.0 for the three human centrins, which confirmed the 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric ratio of TNS to the three proteins. HsCen1, HsCen2 and HsCen3 exhibit the same binding ratio with TNS. To determine the binding constant of TNS with the human centrins, it was assumed that n binding sites for TNS exist on each human centrin and that these binding sites are independent of each other. Thus, TNS binding can be described using the formula:
1 stoichiometric ratio of TNS to the three proteins. HsCen1, HsCen2 and HsCen3 exhibit the same binding ratio with TNS. To determine the binding constant of TNS with the human centrins, it was assumed that n binding sites for TNS exist on each human centrin and that these binding sites are independent of each other. Thus, TNS binding can be described using the formula:| | |
nTNS + HsCen → HsCen − TNSn
| (1) |
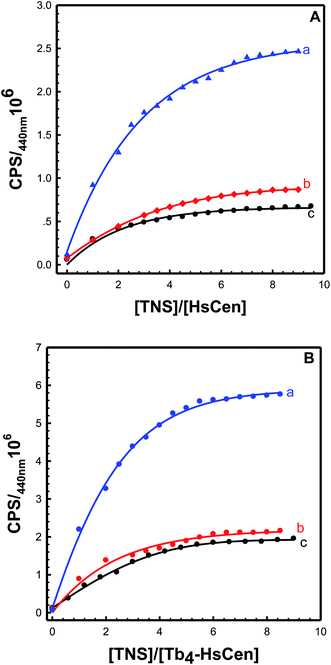 |
| | Fig. 3 Titration curves of TNS with (A) HsCen1 (a), HsCen2 (c) or HsCen3 (b), and (B) Tb3+-HsCen1 (a), Tb3+-HsCen2 (c) or Tb3+-HsCen3 (b) in 100 mM Hepes, and 150 mM NaCl at pH 7.4 and room temperature. The protein concentration was 17 μM. | |
At 440 nm, there is no fluorescence emission for the human centrins and only weak fluorescence emission for the TNS molecule. Assuming that the increase in fluorescence emission at a given value of [TNS]/[HsCen] is mainly attributed to a change from TNS to TNS–HsCen, the system can be described by mass balance equations for TNS and HsCen. The concentrations of different TNS and HsCen species can be calculated using eqn (2)–(5):8,30
| |
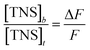 | (2) |
| |
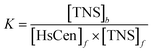 | (4) |
| |
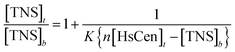 | (5) |
where [HsCen]
t and [HsCen]
b are the total and bound concentration of human centrin, respectively. [TNS]
t and [TNS]
b represent the total and bound concentration of the hydrophobic probe TNS, respectively, and
n is the binding number of TNS on the protein.
F and Δ
F represent the fluorescence intensity (CPS) at a given value of [TNS]/[HsCen] and the change in fluorescence intensity at 440 nm, respectively.
K is the binding constant of TNS with HsCen. By plotting
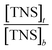 vs.
vs. 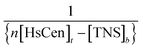
(
Fig. 5), the binding constant and binding ratio
n of TNS with the protein can be obtained (
Table 1). HsCen1 or Tb
3+-saturated HsCen1 binding with TNS displayed the most prominent constants compared to those of HsCen2 or HsCen3 or their corresponding Tb
3+-saturated proteins. Tb
3+-saturated HsCen exhibits a stronger binding affinity with TNS than the protein in the apo state.
Table 1 The binding constants and thermodynamic parameters between TNS and the human centrins (HsCen1, HsCen2 and HsCen3) at different temperatures
| |
T/K |
K (105) |
ΔG (kJ mol−1) |
ΔH (kJ mol−1) |
ΔS (J mol−1) |
| HsCen1 |
297 |
13.70 |
−35.01 |
−92.60 |
193.97 |
| 303 |
5.90 |
−33.47 |
| 310 |
3.14 |
−32.62 |
| Tb4-HsCen1 |
297 |
15.2 |
−35.27 |
−79.63 |
149.72 |
| 303 |
6.76 |
−33.82 |
| 310 |
4.27 |
−33.40 |
| HsCen2 |
297 |
1.19 |
−28.96 |
−21.13 |
26.23 |
| 303 |
1.03 |
−29.08 |
| 310 |
0.86 |
−29.27 |
| Tb4-HsCen2 |
297 |
3.23 |
−31.43 |
−58.25 |
89.80 |
| 303 |
2.34 |
−31.14 |
| 310 |
1.31 |
−30.37 |
| HsCen3 |
297 |
1.30 |
−29.17 |
−16.64 |
42.00 |
| 303 |
1.14 |
−29.33 |
| 310 |
1.00 |
−29.67 |
| Tb4-HsCen3 |
297 |
5.00 |
−32.51 |
−66.86 |
115.99 |
| 303 |
2.51 |
−31.33 |
| 310 |
1.71 |
−31.05 |
3.4 Thermodynamics of TNS binding with human centrins
To obtain thermodynamic data of TNS binding with centrin, TNS titration with HsCen in the apo or holo state was carried out at different temperatures (Fig. 4). In 100 mM Hepes at pH 7.4, in the presence of HsCen1, the fluorescence intensity of TNS at 440 nm decreased with increasing incubation temperature by virtue of collision quenching of two molecules. By plotting 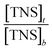 vs.
vs. 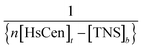 at different temperatures (Fig. 5) and combining with the Vant-Hoff eqn (6) and (7), the Gibbs free energy changes (ΔG), enthalpy changes (ΔH) and entropy changes (ΔS) of the conversion of TNS to TNS–HsCen were calculated. As shown in Table 1, the data of ΔG < 0, ΔH < 0, and ΔS > 0 indicated that TNS binding with the hydrophobic cavity on HsCen1 releases the water molecule that was previously located in the hydrophobic patch. From the thermodynamic data, it can also be inferred that this process is spontaneous and that the free energy change decreases. In addition, as ΔS > 0, and ΔH < 0, it can also be deduced that hydrophobic interactions may mainly contribute to the reaction of TNS with HsCen1, consistent with data previously reported.31,32 Table 1 showed that TNS binding with HsCen2 or HsCen3 was similar to that of HsCen1. The driving force may mainly be hydrophobic interactions, and their binding was also spontaneous. However, their different ΔG, ΔH and ΔS values proved that the three human centrins have different abilities to form complexes with TNS.
at different temperatures (Fig. 5) and combining with the Vant-Hoff eqn (6) and (7), the Gibbs free energy changes (ΔG), enthalpy changes (ΔH) and entropy changes (ΔS) of the conversion of TNS to TNS–HsCen were calculated. As shown in Table 1, the data of ΔG < 0, ΔH < 0, and ΔS > 0 indicated that TNS binding with the hydrophobic cavity on HsCen1 releases the water molecule that was previously located in the hydrophobic patch. From the thermodynamic data, it can also be inferred that this process is spontaneous and that the free energy change decreases. In addition, as ΔS > 0, and ΔH < 0, it can also be deduced that hydrophobic interactions may mainly contribute to the reaction of TNS with HsCen1, consistent with data previously reported.31,32 Table 1 showed that TNS binding with HsCen2 or HsCen3 was similar to that of HsCen1. The driving force may mainly be hydrophobic interactions, and their binding was also spontaneous. However, their different ΔG, ΔH and ΔS values proved that the three human centrins have different abilities to form complexes with TNS.| |
ΔG = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K K
| (7) |
where ΔG, ΔH and ΔS represent the free energy change, enthalpy change and entropy change, respectively.
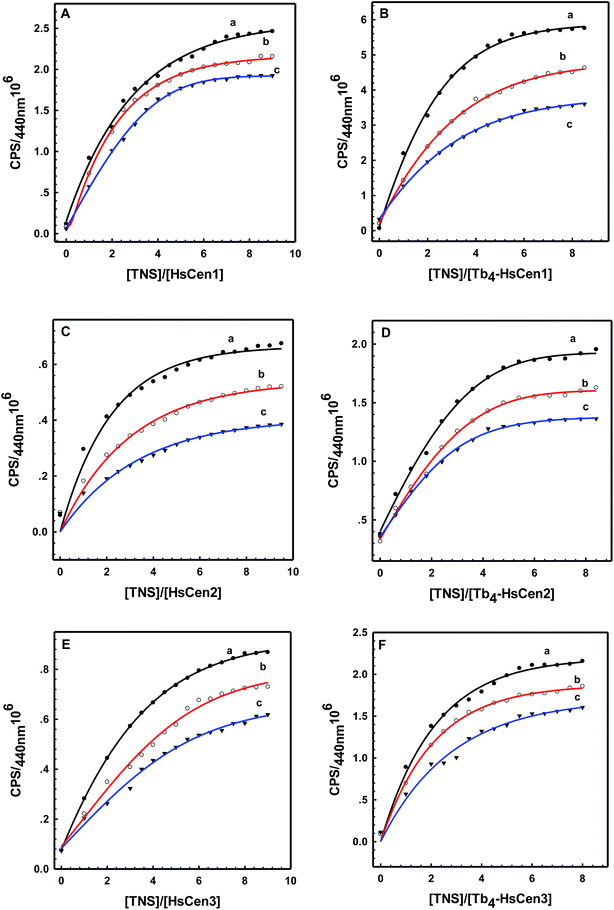 |
| | Fig. 4 The titration curves of HsCen1 (A), Tb3+-HsCen1 (B), HsCen2 (C), Tb3+-HsCen2 (D), HsCen3 (E), and Tb3+-HsCen3 (F) in 100 mM Hepes and 150 mM NaCl at pH 7.4 and different temperatures. a, b and c represent 298 K, 303 K and 313 K, respectively. [HsCen] = 17 μM, [TNS] = 85 μM. | |
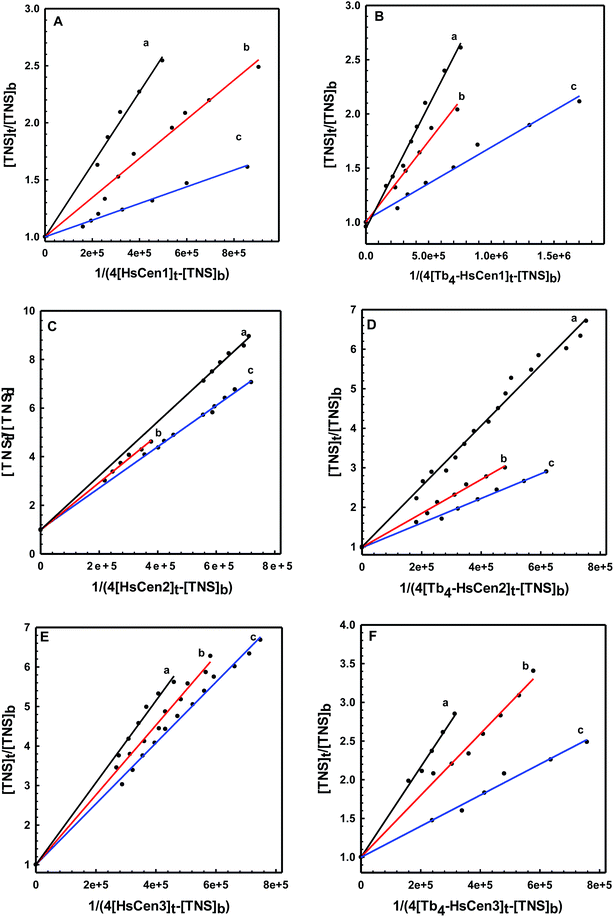 |
| | Fig. 5 The plot of ([TNS]t/[TNS]b) versus 1/(4[HsCen1]t-[TNS]b) (A), ([TNS]t/[TNS]b) versus 1/(4[Tb3+-HsCen1]t-[TNS]b) (B), ([TNS]t/[TNS]b) versus 1/(4[HsCen2]t-[TNS]b) (C), ([TNS]t/[TNS]b) versus 1/(4[Tb3+-HsCen2]t-[TNS]b) (D), ([TNS]t/[TNS]b) versus 1/(4[HsCen3]t-[TNS]b) (E) and ([TNS]t/[TNS]b) versus 1/(4[Tb3+-HsCen3]t-[TNS]b) (F). a, b and c represent 298 K, 303 K and 313 K, respectively. | |
3.5 Distance between TNS and the human centrins
According to Förster non-radiative energy transfer theory,19,20 energy transfer will occur if the donor can produce fluorescence emission, there is spectral overlap between the fluorescence emission spectrum of the donor and the UV-Vis absorbance spectrum of the acceptor and the distance between the donor and the acceptor is smaller than 7 nm. The distance between a protein and its target macromolecule can be estimated using eqn (8)–(12).18 Due to a Tyr residue, as a donor protein HsCen1 has intrinsic fluorescence emission, and the spectral overlap between the fluorescence emission of HsCen1 and the UV-Vis spectrum of TNS made it possible to calculate the distance between HsCen1 and TNS. Fig. 6 shows the overlap of the two molecules.| |
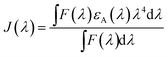 | (8) |
| |
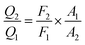 | (9) |
| | |
R60 = 8.79 × 10−25K2n−4QDJ(λ)
| (10) |
| |
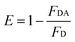 | (11) |
| |
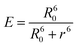 | (12) |
where r is the distance between the acceptor and donor, R0 is the critical distance at which the transfer efficiency is 50%, k is the spatial orientation factor of the dipole and n represents the refractive index of the buffer. Q1 is the quantum yield of the donor, which can be calculated to be 0.14 from the absolute quantum yield of Tyr in aqueous solution21 as a standard value. J(λ) is the overlap integral of the fluorescence emission spectrum of HsCen1 and the absorption spectrum of TNS. F(λ) is the fluorescence intensity of the fluorescent donor at a given wavelength λ, and εA(λ) is the molar extinction coefficient of the acceptor at a given wavelength λ. F1 and F2 represent the relative fluorescence intensity of HsCen1 in the absence and presence of the acceptor (TNS), respectively. The value of J(λ) can be evaluated to be 3.32 × 10−14 cm3 M−1 by integrating the spectra in Fig. 5 at λ = 310–400 nm. Furthermore, the characteristic distance R0 = 3.31 nm, using k2 = 2/3, and n = 1.4.21 The energy transfer effect E = 0.43, and the distance r between TNS and the Tyr residue in HsCen1 was calculated to be 3.1 nm. Using similar methods, the distance between TNS and HsCen3 was calculated to be 2.16 nm and that between TNS and HsCen2 was calculated to be 1.98 nm.
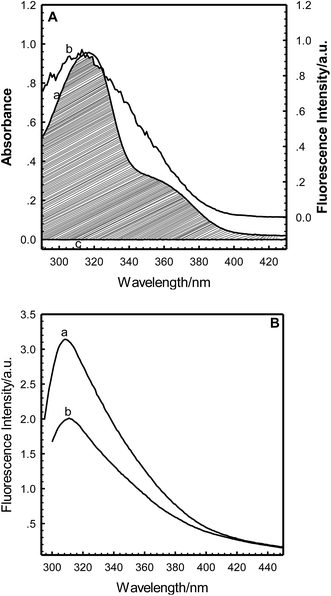 |
| | Fig. 6 (A): Overlap of the absorption spectrum of TNS–HsCen1 (a) and the fluorescence emission spectrum of HsCen1 (b) in 100 mM Hepes and 150 mM NaCl at pH 7.4. HsCen1: 20 μM, TNS: 190 μM. (B): The fluorescence spectra of HsCen1 (a) in the absence and (b) in the presence of TNS in 100 mM Hepes and 150 mM NaCl at pH 7.4 and room temperature. HsCen1: 20 μM, TNS: 190 μM. | |
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This work was supported by the National Natural Science Foundation of PR China (No. 20901048, No. 21201024 and No. 21571117) and the Natural Science Foundation of Shanxi Province (No. 2011021006-1 and No. 2012021009-1). We thank the Scientific Instrument Center of Shanxi University for the technical assistance.
References
- D. R. Kellogg, Nature, 1989, 340, 99 CrossRef CAS PubMed.
- D. R. Kellogg, M. Moritz and B. M. Alberts, Annu. Rev. Biochem., 1994, 63, 639 CrossRef CAS PubMed.
- J. L. Salisbury, Curr. Opin. Cell Biol., 1995, 7, 39 CrossRef CAS PubMed.
- H. Hu, J. H. Sheehan and W. J. Chazin, J. Biol. Chem., 2004, 279, 50895 CrossRef CAS PubMed.
- M. D. Tsai, T. Drakeberg, E. Thulin and S. Forsén, Biochemistry, 1987, 26, 3635 CrossRef CAS PubMed.
- J. L. Salisbury, A. Baron, B. Surek and M. Melkonian, J. Cell Biol., 1984, 99, 962 CrossRef CAS PubMed.
- B. Huang, A. Mengersen and V. D. Lee, J. Cell Biol., 1988, 107, 133 CrossRef CAS PubMed.
- Z. J. Wang, L. X. Ren, Y. Q. Zhao, G. T. Li, A. H. Liang and B. S. Yang, Spectrochim. Acta, Part A, 2007, 66, 1323 CrossRef PubMed.
- J. L. Salisbury, Curr. Opin. Cell Biol., 1995, 7, 39 CrossRef CAS PubMed.
- P. Baum, C. Furlong and B. Byers, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 5512 CrossRef CAS.
- V. D. Lee and B. Huang, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 11039 CrossRef CAS.
- G. Manandhar, C. Simerly, J. L. Salisbury and G. Schatten, Cell Motil. Cytoskeleton, 1999, 43, 137 CrossRef CAS PubMed.
- G. D. Palermo, H. Joris, P. Devroey and A. C. Van Steirteghem, Lancet, 1992, 340, 17 CrossRef CAS.
- R. Errabolu, M. A. Sanders and J. L. Salisbury, J. Cell Sci., 1994, 107, 9 CAS.
- M. Araki, C. Masutani, M. Takemura, A. Uchida, K. Sugasawa, J. Kondoh, Y. Ohkuma and F. Hanaoka, J. Biol. Chem., 2001, 276, 18665 CrossRef CAS PubMed.
- R. Nishi, Y. Okuda, E. Watanabe, T. Mori, S. Iwai, C. Masutani, K. Sugasawa and F. Hanaoka, Mol. Cell. Biol., 2005, 25, 5664 CrossRef CAS PubMed.
- O. Gavet, C. Alvarez, P. Gaspar and M. Bornens, Mol. Biol. Cell, 2003, 14, 1831 CrossRef PubMed.
- J. Laoukili, E. Perret, S. Middendorp, O. Houcine, C. Guennou, F. Marano, M. Bornens and F. Tournier, J. Cell Sci., 2000, 113, 1355 CAS.
- G. Kohsuke, Y. Akiko, O. Kazunori and T. Mihoko, Biochem. Biophys. Res. Commun., 2004, 323, 891 CrossRef PubMed.
- J. Hu, X. Jia, Q. Li, X. Yang and K. Wang, Biochemistry, 2004, 43, 2688 CrossRef CAS PubMed.
- W. Liu, L. Duan, B. Zhao, Y. Q. Zhao, A. H. Liang and B. S. Yang, Health, 2010, 2, 262 CrossRef.
- Y. Q. Zhao, X. J. Guo and B. S. Yang, RSC Adv., 2017, 7, 10206 RSC.
- Y. Q. Zhao, X. F. Chu and B. S. Yang, Bioelectrochemistry, 2017, 117, 15 CrossRef CAS PubMed.
- J. R. Lakowicz, Principles of fluorescence Spectroscopy, 3rd edn, 13, 2006 Search PubMed.
- W. O. McClure and G. M. Edelman, Biochemistry, 1966, 5, 1908 CrossRef CAS PubMed.
- E. Sentandreu, J. V. Carbonell and J. M. Sendra, Biotechnol. Bioeng., 2002, 78, 829 CrossRef CAS PubMed.
- J. J. Chou, S. Li, C. B. Klee and A. Bax, Nat. Struct. Biol., 2001, 8, 990 CrossRef CAS PubMed.
- M. Ikura, G. M. Clore, A. M. Gronenborn, G. Zhu, C. B. Klee and A. Bax, Science, 1992, 256, 632 CAS.
- M. Zhang, T. Tanaka and M. Ikura, Nat. Struct. Biol., 1995, 2, 758 CrossRef CAS PubMed.
- J. A. Cox, F. Tirone, I. Durussel, C. Firanescu, Y. Blouquit, P. Duchambon and C. T. Craescu, Biochemistry, 2005, 44, 840 CrossRef CAS PubMed.
- Z. J. Wang, L. X. Ren, Y. Q. Zhao, G. T. Li, L. Duan, A. H. Liang and B. S. Yang, Spectrochim. Acta, Part A, 2008, 70, 892 CrossRef PubMed.
- Y. Q. Zhao, X. L. Diao, J. Yan, Y. N. Feng, Z. J. Wang, A. H. Liang and B. S. Yang, J. Lumin., 2012, 132, 924 CrossRef CAS.
|
| This journal is © The Royal Society of Chemistry 2017 |
 Open Access Article
Open Access Article ,
Xiaofang Cui and
Binsheng Yang*
,
Xiaofang Cui and
Binsheng Yang*
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and then passed through a 60 cm × 16 cm Sephadex S200 column equilibrated in PBS buffer, at pH 7.4.
1) and then passed through a 60 cm × 16 cm Sephadex S200 column equilibrated in PBS buffer, at pH 7.4.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric ratio of TNS to the three proteins. HsCen1, HsCen2 and HsCen3 exhibit the same binding ratio with TNS. To determine the binding constant of TNS with the human centrins, it was assumed that n binding sites for TNS exist on each human centrin and that these binding sites are independent of each other. Thus, TNS binding can be described using the formula:
1 stoichiometric ratio of TNS to the three proteins. HsCen1, HsCen2 and HsCen3 exhibit the same binding ratio with TNS. To determine the binding constant of TNS with the human centrins, it was assumed that n binding sites for TNS exist on each human centrin and that these binding sites are independent of each other. Thus, TNS binding can be described using the formula: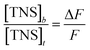
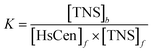
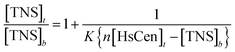
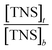 vs.
vs. 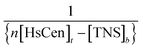 (Fig. 5), the binding constant and binding ratio n of TNS with the protein can be obtained (Table 1). HsCen1 or Tb3+-saturated HsCen1 binding with TNS displayed the most prominent constants compared to those of HsCen2 or HsCen3 or their corresponding Tb3+-saturated proteins. Tb3+-saturated HsCen exhibits a stronger binding affinity with TNS than the protein in the apo state.
(Fig. 5), the binding constant and binding ratio n of TNS with the protein can be obtained (Table 1). HsCen1 or Tb3+-saturated HsCen1 binding with TNS displayed the most prominent constants compared to those of HsCen2 or HsCen3 or their corresponding Tb3+-saturated proteins. Tb3+-saturated HsCen exhibits a stronger binding affinity with TNS than the protein in the apo state.
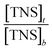 vs.
vs. 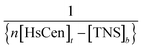 at different temperatures (Fig. 5) and combining with the Vant-Hoff eqn (6) and (7), the Gibbs free energy changes (ΔG), enthalpy changes (ΔH) and entropy changes (ΔS) of the conversion of TNS to TNS–HsCen were calculated. As shown in Table 1, the data of ΔG < 0, ΔH < 0, and ΔS > 0 indicated that TNS binding with the hydrophobic cavity on HsCen1 releases the water molecule that was previously located in the hydrophobic patch. From the thermodynamic data, it can also be inferred that this process is spontaneous and that the free energy change decreases. In addition, as ΔS > 0, and ΔH < 0, it can also be deduced that hydrophobic interactions may mainly contribute to the reaction of TNS with HsCen1, consistent with data previously reported.31,32 Table 1 showed that TNS binding with HsCen2 or HsCen3 was similar to that of HsCen1. The driving force may mainly be hydrophobic interactions, and their binding was also spontaneous. However, their different ΔG, ΔH and ΔS values proved that the three human centrins have different abilities to form complexes with TNS.
at different temperatures (Fig. 5) and combining with the Vant-Hoff eqn (6) and (7), the Gibbs free energy changes (ΔG), enthalpy changes (ΔH) and entropy changes (ΔS) of the conversion of TNS to TNS–HsCen were calculated. As shown in Table 1, the data of ΔG < 0, ΔH < 0, and ΔS > 0 indicated that TNS binding with the hydrophobic cavity on HsCen1 releases the water molecule that was previously located in the hydrophobic patch. From the thermodynamic data, it can also be inferred that this process is spontaneous and that the free energy change decreases. In addition, as ΔS > 0, and ΔH < 0, it can also be deduced that hydrophobic interactions may mainly contribute to the reaction of TNS with HsCen1, consistent with data previously reported.31,32 Table 1 showed that TNS binding with HsCen2 or HsCen3 was similar to that of HsCen1. The driving force may mainly be hydrophobic interactions, and their binding was also spontaneous. However, their different ΔG, ΔH and ΔS values proved that the three human centrins have different abilities to form complexes with TNS.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K
K