Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Characterization of the interaction between triclosan and catalase
Luyi Zoua,
Chenyu Mia,
Hongyan Yua,
Wenxiu Gub and
Yue Teng *a
*a
aJiangsu Key Laboratory of Anaerobic Biotechnology, School of Environmental and Civil Engineering, Jiangnan University, 1800# Lihu Avenue, Wuxi 214122, PR China. E-mail: tengyue@jiangnan.edu.cn
bSchool of Chemical and Material Engineering, Jiangnan University, 1800# Lihu Avenue, Wuxi 214122, PR China
First published on 27th January 2017
Abstract
Triclosan (TCS), as a kind of broad spectrum antimicrobial agent, is widely used in personal care products. However, its effectiveness and safety are met with much skepticism. High exposure of TCS to the environment poses a potential threat to ecological safety and human health. In this article, we investigated the interaction between TCS and the antioxidant enzyme catalase (CAT) using spectroscopic techniques and molecular docking technologies. The experimental results showed that TCS could bind into the central cavity of CAT with one binding site to form a TCS–CAT complex, and the main force was the hydrophobic effect. The interaction made some changes to the conformation and micro-environment of CAT and inhibited the activity of CAT. This work explored the interaction between TCS and CAT at the molecular level, which could help us to understand the toxicity mechanism of TCS and provide a reference for pollutant toxicity assessments.
1. Introduction
In recent years, public and scientific concern has heightened interest in the occurrence of pharmaceuticals and personal care products (PPCPs) in the environment. As one kind of PPCPs, triclosan (2,4,4′-trichloro-2′-hydroxy diphenyl ether, TCS) is commonly used as a broad-spectrum antimicrobial agent in personal care products, including toothpastes, soaps, cosmetics, and medical disinfectants, as well as in household items.1,2 On September 2, 2016, the U.S. Food and Drug Administration issued a final rule on TCS, which triggered a heated discussion about the safety of TCS once again. Many studies have reported environmental exposure and risk assessments of TCS. Research has shown that more than 95% of antimicrobial consumer products are washed down after use and treated in municipal sewage treatment facilities.1,2 However, the sludge adsorption removal of TCS is about 27%, and then TCS is released from the sludge, which implies the continual release of these compounds into the environment.1 TCS has been found in aquatic environments, terrestrial environments, organisms, and has even been detected in human milk.3 Studies have shown that TCS has a negative impact on the soil nitrogen cycle and biological community structure.4,5 In animal experiments, low doses of TCS were ecotoxic to aquatic organisms and also had endocrine disrupting effects in amphibians and fishes.6–9 Furthermore, research has shown that TCS can cause DNA damage in human normal stem cells, and has an obvious time–effect relationship.10 These studies concluded that risk assessments and toxicity information for TCS were significant and urgently needed. Researchers have put much effort into animal level and cell level experiments to explore the toxicity effects of TCS, however, studies at the molecular level for the evaluation of TCS are not clear yet, and this needs to be further researched.A pollutant’s exposure to the environment could cause diseases, and many studies have shown that oxidative stress is related to diseases. In contrast, an antioxidant system could repair the oxidative damage and maintain the dynamic balance of redox reactions in an organism.11 Investigating the interactions of pollutants and antioxidants has been an important topic. Chen et al. have investigated the interaction between TCS and human serum albumin.12 But research on TCS and antioxidases hasn’t been carried out. Catalase (CAT) is one kind of antioxidant enzyme, and has the ability to decompose H2O2 into harmless water and oxygen,13 so as to avoid organism oxidation injury.14 In this work, the interaction mechanism of TCS with CAT was explored. The binding constants, the number of binding sites and the binding forces of the interaction were investigated by fluorescence measurements. Through molecular docking, spectroscopic analyses, and determination of enzyme activity, we can obtain information about the precise binding site of TCS on CAT, and the conformational and enzymatic changes of CAT by TCS. This work can provide the basic data for understanding the enzyme toxicity of TCS to CAT and the human health risk of TCS in vivo.
2. Experimental section
2.1. Reagents
CAT (bought from Beijing BioDee Biotechnology Co. Ltd, and extracted from a bovine liver) was dissolved in 100 mL of ultrapure water to form a 1.0 × 10−5 M solution, and then preserved at 0–4 °C and diluted as required.TCS (1 × 10−3 M) was prepared as a stock solution by dissolving 0.03 g of TCS (bought from Shanghai Macklin Biochemical Co., Ltd) in 100 mL of ultrapure water with the addition of NaOH (1.0 M) due to TCS being more soluble in alkaline solutions.
Phosphate buffer (0.2 M, a mixture of NaH2PO4·2H2O and Na2HPO4·12H2O, pH 7.6) was used to control the pH of the solutions. NaH2PO4·2H2O and Na2HPO4·12H2O were of analytical reagent grade, and obtained from Sinopharm Chemical Reagent Co., Ltd.
H2O2 (1 × 10−2 M) was prepared by pipetting 50 μL of 30% H2O2 (10 M, obtained from Sinopharm Chemical Reagent Co., Ltd) and diluting it with ultrapure water to 50 mL.
Ultrapure water (18.25 MΩ) was used throughout the experiments.
2.2. Apparatus and methods
All fluorescence spectra were taken using an RF-5301 PC fluorescence spectrophotometer (SHIMADZU, Japan) equipped with a 1 cm cell.UV-visible absorption spectra were recorded on a UV-2700 spectrophotometer (SHIMADZU, Japan) with 1 cm quartz cells.
Circular dichroism (CD) measurements were performed on a MOS 450 spectrometer (Bio Logic, France) in a 1 cm cell.
The absorbance of H2O2 was measured on a UV-1100 model spectrophotometer (MAPADA, China) with 1 cm quartz cells.
The pH values were measured with a STARTER 300 OHAUS.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 energy evaluations, and the population size was set to 150. Finally, we selected the lowest binding energy conformation to analyze further.
000 energy evaluations, and the population size was set to 150. Finally, we selected the lowest binding energy conformation to analyze further.
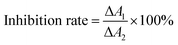 | (1) |
3. Results and discussion
3.1. Characterization of the interaction of TCS with CAT by fluorescence measurements
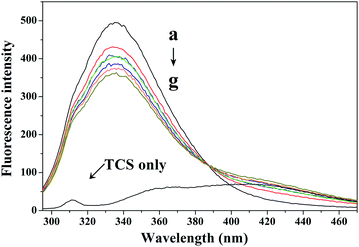 | ||
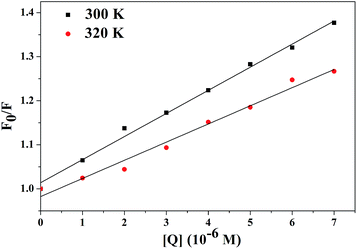
| Fig. 1 Effect of TCS on the fluorescence of CAT. Conditions: CAT: 1 × 10−7 M; TCS (×10−6 M): (a) 0, (b) 1, (c) 2, (d) 3, (e) 4, (f) 5, and (g) 6. TCS only: 1 × 10−4 M; pH 7.4; T = 300 K. | ||
A quenching mechanism includes static and dynamic quenching. For dynamic quenching, a higher temperature results in a larger diffusion coefficient and an increase in the dynamic quenching constants. In contrast, an increased temperature can decrease the stability of complexes, and lower the value of the static quenching constants.19
To confirm the quenching mechanism, the fluorescence quenching data were analyzed according to the Stern–Volmer equation.20
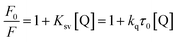 | (2) |
The Stern–Volmer plots are presented in Fig. 2, and the fluorescence data were analyzed according to F0/F versus [Q] at 300 and 320 K. We can determine Ksv by a linear regression plot of F0/F against [Q] using eqn (2). Because the fluorescence lifetime of the biopolymer (τ0) is 1 × 10−8 s,21 the value of kq can be obtained.
The maximum scatter collision quenching constant of various quenchers with a biopolymer is 2.0 × 1010 L mol−1 s−1.22 As is shown in Table 1, the Ksv values decreased with increasing temperature and kq was greater than 2.0 × 1010 L mol−1 s−1. So we can conclude that overall quenching was dominated by a static quenching mechanism forming a TCS–CAT complex.
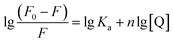 | (3) |
| T (K) | Ka (×105 M−1) | n | Ra | ΔH (kJ mol−1) | ΔS (J mol−1 K−1) | ΔG (kJ mol−1) |
|---|---|---|---|---|---|---|
| a R: the correlation coefficient. | ||||||
| 300 | 0.501 | 0.8575 | 0.9820 | 213.45 | 787.83 | −22.9 |
| 320 | 0.455 | 1.3118 | 0.9814 | 786.09 | −38.1 | |
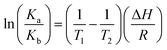 | (4) |
ΔG = ΔH − TΔS = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka Ka
| (5) |
3.2. Investigation of CAT conformation changes
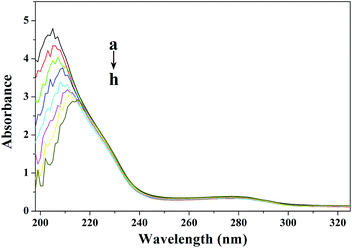
The strong absorption peak at about 205 nm reflects the framework conformation of the protein.26 With the increasing concentration of TCS, the absorbance of CAT decreased and the maximum peak position of TCS–CAT was red-shifted, indicating that the interaction between TCS and CAT resulted in the loosening and unfolding of the protein skeleton.
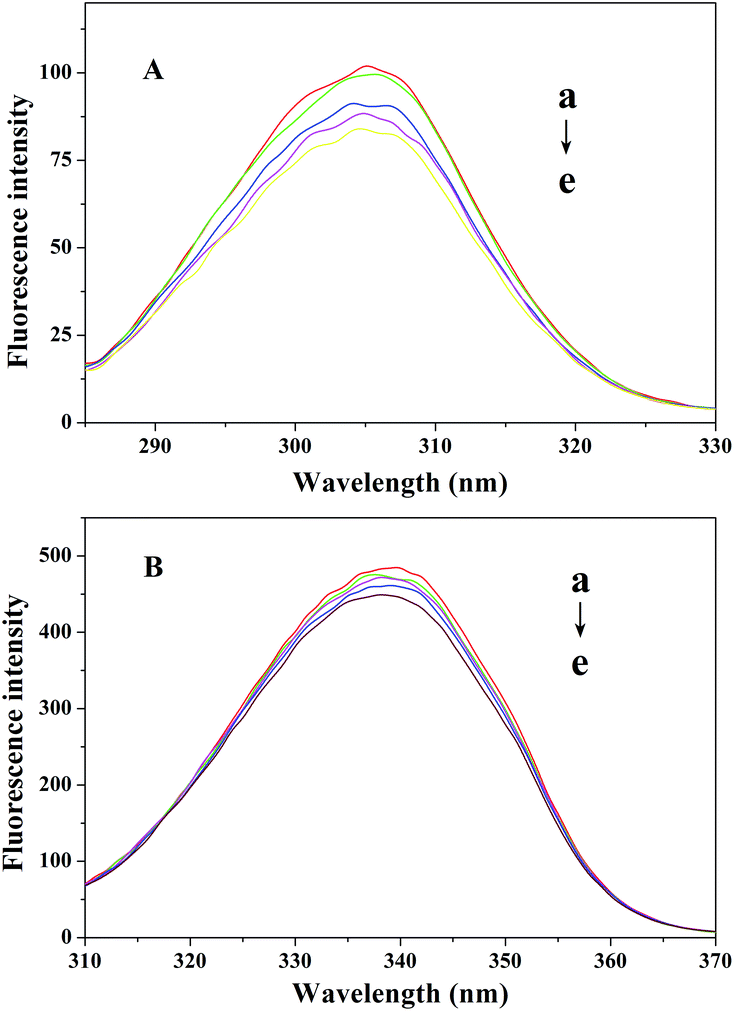
The synchronous fluorescence spectra of CAT at two different wavelength intervals are shown in Fig. 5. It can be seen in Fig. 5A that the maximum emission wavelength had a small blue shift when Δλ was 15 nm, revealing that the hydrophobicity of the Tyr residues increased, and the polarity of the microenvironment around the Tyr residues decreased. In Fig. 5B, the position of the maximum emission of the Trp residues doesn’t change obviously, which indicates that TCS had little effect on the microenvironment of the Trp residues in CAT.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 25
1 and 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), the α-helix content of CAT decreased from 34.3% to 20.6% and 20.4%, and the β-sheet content increased from 11.3% to 24.5% and 25.3%. Obviously, TCS transformed some α-helices into β-sheets and random coils and TCS induced these changes in the secondary structure of CAT.
1), the α-helix content of CAT decreased from 34.3% to 20.6% and 20.4%, and the β-sheet content increased from 11.3% to 24.5% and 25.3%. Obviously, TCS transformed some α-helices into β-sheets and random coils and TCS induced these changes in the secondary structure of CAT.
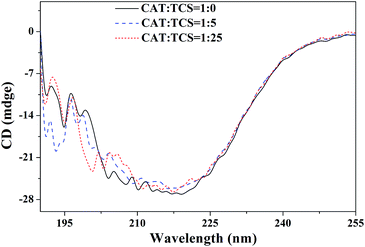 | ||
| Fig. 6 CD spectra of CAT and the CAT–TCS system. Conditions: CAT: 2 × 10−7 M; TCS concentrations in the CAT–TCS system were 0, 1 × 10−6 M, and 5 × 10−6 M. | ||
| Molar ratio CAT to TCS | Secondary structural elements in CAT | |||
|---|---|---|---|---|
| α-Helix (%) | β-Sheet (%) | β-Turns (%) | Unordered (%) | |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
34.3 | 11.3 | 23.3 | 32.3 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
20.6 | 24.5 | 19.3 | 38.6 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 25 |
20.4 | 25.3 | 19.3 | 39.2 |
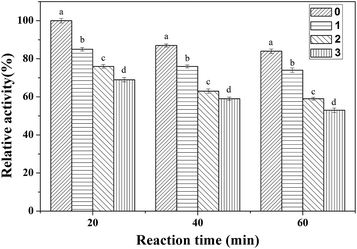
3.3. Effect of TCS on CAT activity
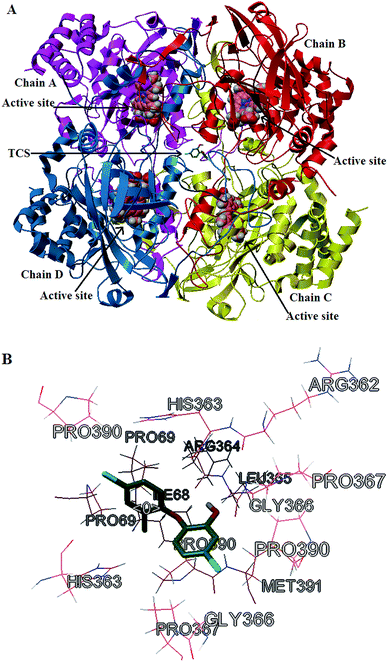
The effect of different concentrations of TCS on the relative CAT activity is shown in Fig. 7. We can see that the relative CAT activities are reduced to 85%, 76%, and 74% after reaction with 1, 2 and 3 (×10−5 M) TCS for 20 minutes. As the reaction happened, the relative activities were reduced to 74%, 59% and 53% after 1 h of reaction. It’s revealed that both the reaction time and the concentration of TCS had a notable impact on the CAT activity. Combined with the previous experimental results and molecular docking analysis, we can conclude that although the TCS molecule did not directly bind into the CAT active site, it influenced the microenvironment around the active site, resulting in the reduction of CAT activity.4. Conclusions
In this paper, we simulated physiological conditions to explore the interactions between the widely used broad-spectrum antimicrobial agent TCS and the antioxidant enzyme CAT. The spectroscopic analysis and thermodynamic results indicated that the TCS molecule can spontaneously bind with CAT to form a TCS–CAT complex with one binding site mainly through hydrophobic interaction forces. TCS could also obviously change the skeleton and the secondary structure of CAT. According to molecular docking simulation analysis and enzyme activity experiments, we found that the exact binding site of TCS with CAT was the central cavity, and TCS influenced the microenvironment of the active site and further induced the inhibition of CAT activity. This research can provide data support for the comprehensive evaluation of TCS and act as a reference for other environmental pollutant toxicity mechanism research.Acknowledgements
The work is supported by the National Nature Science Foundation of China (NSFC, 21307043), the China Postdoctoral Science Foundation (2016M590411), the Postdoctoral Science Foundation of Jiangsu Province (1601230C) and the Independent Research Project of Jiangnan University (JUSRP11525).References
- K. Bester, Water Res., 2003, 37, 3891–3896 CrossRef CAS PubMed.
- D. Sabaliunas, S. F. Webb, A. Hauk, M. Jacob and W. S. Eckhoff, Water Res., 2003, 37, 3145–3154 CrossRef CAS PubMed.
- M. Adolfsson-Erici, M. Pettersson, J. Parkkonen and J. Sturve, Chemosphere, 2002, 46, 1485–1489 CrossRef CAS PubMed.
- N. J. Waller and R. S. Kookana, Environ. Toxicol. Chem., 2009, 28, 65–70 CrossRef CAS PubMed.
- H. Svenningsen, T. Henriksen, A. Priemé and A. R. Johnsen, Environ. Pollut., 2011, 159, 1599–1605 CrossRef CAS PubMed.
- R. Reiss, N. Mackay, C. Habig and J. Griffin, Environ. Toxicol. Chem., 2002, 21, 2483–2492 CrossRef CAS PubMed.
- C. M. Foran, E. R. Bennett and W. H. Benson, Mar. Environ. Res., 2000, 50, 153–156 CrossRef CAS PubMed.
- S. L. Fraker and G. R. Smith, Environ. Toxicol., 2004, 19, 250–256 CrossRef CAS PubMed.
- N. Matsumura, H. Ishibashi, M. Hirano, Y. Nagao, N. Watanabe, H. Shiratsuchi, T. Kai, T. Nishimura, A. Kashiwagi and K. Arizono, Biol. Pharm. Bull., 2005, 28, 1748–1751 CAS.
- L. Linpeng, M. Huimin, H. Junjie, L. Yuhua, W. Yipei, S. Guoying and F. Mojia, Journal of Ecological Environment, 2010, 19, 2897–2901 Search PubMed.
- J. Limón-Pacheco and M. E. Gonsebatt, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2008, 674, 137–147 Search PubMed.
- J. Chen, X. Zhou, Y. Zhang, Y. Zi, Y. Qian, H. Gao and S. Lin, Environ. Sci. Pollut. Res. Int., 2011, 19, 2528–2536 CrossRef PubMed.
- D. Giustarini, I. Dalledonne, D. Tsikas and R. Rossi, Crit. Rev. Clin. Lab. Sci., 2009, 46, 241–281 CrossRef CAS PubMed.
- X. Zhang and J. Jin, J. Mol. Struct., 2008, 882, 96–100 CrossRef CAS.
- L. Whitmore and B. A. Wallace, Biopolymers, 2008, 89, 392–400 CrossRef CAS PubMed.
- S. Ardissone, E. Laurenti, P. Frendo, E. M. Ghibaudi and A. Puppo, JBIC, J. Biol. Inorg. Chem., 2005, 10, 813–826 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- J. J. Irwin, T. Sterling, M. M. Mysinger, E. S. Bolstad and R. G. Coleman, J. Chem. Inf. Model., 2012, 52, 1757–1768 CrossRef CAS PubMed.
- D. Lu, X. Zhao, Y. Zhao, B. Zhang, B. Zhang, M. Geng and R. Liu, Food Chem. Toxicol., 2011, 49, 3158 CrossRef CAS PubMed.
- S. Soares, A. Nuno Mateus and V. D. Freitas, J. Agric. Food Chem., 2007, 55, 6726–6735 CrossRef CAS PubMed.
- J. R. Lakowicz and G. Weber, Biochemistry, 1973, 12, 4161–4170 CrossRef CAS PubMed.
- Z. Chi, R. Liu and H. Zhang, Biomacromolecules, 2010, 11, 2454–2459 CrossRef CAS PubMed.
- X. Zhao, R. Liu, Z. Chi, Y. Teng and P. Qin, J. Phys. Chem. B, 2010, 114, 5625–5631 CrossRef CAS PubMed.
- Z. Chi and R. Liu, Biomacromolecules, 2011, 12, 203–209 CrossRef CAS PubMed.
- P. D. Ross and S. Subramanian, Biochemistry, 1981, 20, 3096–3102 CrossRef CAS PubMed.
- D. Li, B. Ji and J. Jin, J. Lumin., 2008, 128, 1399–1406 CrossRef CAS.
- J. Zhu, X. Zhang, D. Li and J. Jin, J. Mol. Struct., 2007, 843, 38–44 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2017 |