Supramolecular silk from a peptide hydrogel†
Jie
Zhan‡
ab,
Yanbin
Cai‡
ab,
Shenglu
Ji
ab,
Yuna
Shang
ab,
Jie
Gao
*b and
Zhimou
Yang
*ab
aCollege of Life Sciences, Nankai University, Tianjin 300071, P. R. China. E-mail: yangzm@nankai.edu.cn
bState Key Laboratory of Medicinal Chemical Biology, Key Laboratory of Bioactive Materials, Ministry of Education, Nankai University, Tianjin 300071, P. R. China. E-mail: chemgaojie@nankai.edu.cn
First published on 7th December 2016
Abstract
A macroscopic silk-like fiber can be drawn directly from a hydrogel self-assembled from peptides, which offers an easy and versatile method for generating materials with hierarchically ordered structures over different scales and extends the utility of peptide hydrogels.
Sophisticated structures made from well-ordered and long-range aligned building blocks are ubiquitous in nature, especially in living systems, such as the feathers of birds, scales on the wings of a butterfly, and woody fibers of trees. The alignment in these structures is not an aesthetics requirement but is essential for the maintenance of their functions. For instance, microfibrils in type I collagen align together laterally with a good order to serve as the structural element in an extracellular matrix. Cellular alignment is also widely found in tissues such as heart muscle, and in the neural system. Cardiomyocytes form elongated and aligned cell bundles, creating an anisotropic syncytium that provides the heart muscle with unique mechanical properties. Axon tracts in nerves, the spinal cord and the brain, rely on highly ordered and aligned bundles of axons to function. It is therefore highly interesting for researchers to learn from nature and create aligned materials from nanostructures.1–9 Several strategies have been developed and applied to fabricate self-assembled supramolecular nanostructures into highly ordered materials, such as spinning of aligned supramolecular nanotubes,10 axial unidirectional growth of self-assembled nanotubes,11,12 solvent thermal annealing,13 covalent crosslinking of peptide fibrils,14 magnetic field guidance15,16etc.
Peptide-based supramolecular hydrogels, consisting of networks of nanostructures from self-assembling short peptides, have shown great potential in biomedical applications including drug controlled release,17–26 sensing and imaging,27–32 regenerative medicine,33–38 and immune modulation.39–44 Though peptide molecules in nanostructures self-assemble with good order at a nanoscale level, to align these nanostructures at a macroscopic scale level remains a great challenge, mainly due to the flexibility of soft peptide assemblies and the weak and complicated non-covalent interactions between them. A few approaches to generate peptide hydrogels with macroscopically aligned structures have been recently reported. For instance, Stupp and co-workers have developed a thermal pathway that can convert isotropic solutions of peptide amphiphiles to a strongly birefringent hydrogel containing large arrays of aligned nanofibers,45 and prepare noodle-like fibers to guide neurite outgrowth and cell migration.46 Xu and co-workers have used increased aromatic–aromatic interactions between nanofibers of peptides to enhance interfiber contacts, thus enabling an enzymatic formation of an anisotropic hydrogel with spontaneously aligned nanofibers.47,48 Bai, Ulijn and co-workers applied ultrasound to trigger the self-assembly and alignment of tripeptide nanostructures, resulting in thermodynamically stable anisotropic hydrogels.49 The shear-flow50 and magnetic field51 have also been used to obtain ordered macroscopic structures within peptide hydrogels. In this report, a supramolecular silk-like material using spontaneously aligned nanotubes from a peptide hydrogel is introduced. The peptide hydrogel is anisotropic when formed, but the nanotubes inside the gel can rearrange and align along the direction of external force, thus resulting in an anisotropic supramolecular silk with well-ordered structures over a macroscopic scale.
Liu’s group has recently reported a supramolecular yarn from a solution of an amino acid derivative.10 Our group focuses on the preparation of short-peptide based hydrogels, and Liu’s results inspired us to test whether we could obtain similar yarns or silk-like materials from our hydrogels. We occasionally found that silk-like fibers could be drawn out using a pipet tip or syringe needle from one of our prepared hydrogels, the gel of Nap-FGG (naphthalene-phenylalanine-glycine-glycine). The compound Nap-FGG was prepared via standard Fmoc- solid phase peptide synthesis and its chemical structure is illustrated in Fig. 1A. Nap-FGG was dissolved in hot phosphate buffer saline (2.0 wt% in PBS, pH = 7.4, around 90 °C), the solution was then allowed to cool back to room temperature one hour later, shown as the inset in Fig. 1A. The results obtained via a dynamic frequency sweep showed that the G′ value was only 3–6 times bigger than the G′′ value (Fig. 1B), suggesting a mechanically weak hydrogel of Nap-FGG.
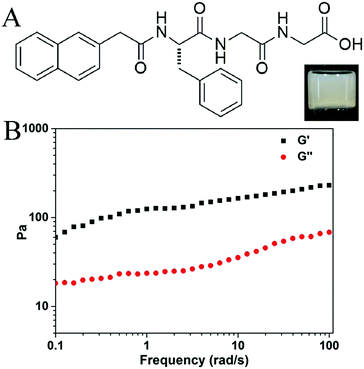 | ||
| Fig. 1 (A) Chemical structure of Nap-FGG, inset: optical image of gel containing 2.0 wt% of Nap-FGG and (B) dynamic frequency sweep of the gel. | ||
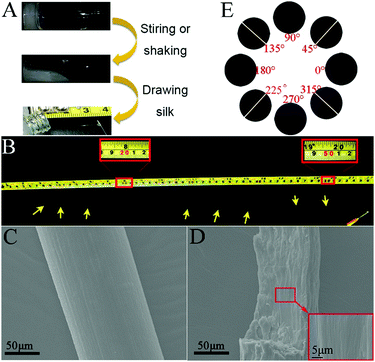
We found that the hydrogel could be turned into a sticky liquid after shaking or stirring at room temperature. To our surprise, a macroscopic silk-like material could be directly drawn out from the sticky liquid using a pipet tip (Fig. 2A). When using a syringe needle gently and carefully to draw, the silk could be up to more than 50 cm in length (Fig. 2B). However, we could only obtain very short fibers (shorter than 10 cm) from the gel without external forces. We then investigated the microscopic structures of the silk using scanning electron microscopy (SEM). As shown in Fig. 2C, the silk was about 130 μm in diameter and has a smooth appearance at its surface. The magnified view indicates that the cross section of the silk was actually constructed by many well aligned nanofibers (Fig. 2D). A polarization microscope was then used to investigate the orientation of the nanofibers in the silk. As shown in Fig. 2E, when a free-standing fiber was aligned 45° to the polarizer, the anisotropy birefringence would come to the maximum; while the silk was placed parallel to the polarizer, the birefringence became very weak. These results clearly indicated that the anisotropic existed throughout the silk, which further demonstrated that the nanostructures in the silk were well aligned and had orientation.
To understand the dynamic self-assembly and characteristics of the nanostructures in the gel, we used transmission electron microscopy (TEM) to examine the nanostructures of the gel at different time points. At the time when the hydrogel did not form and no silk fibers could be drawn out from the system (5 minutes after the heating), we observed vesicles (Fig. 3A) with diameters ranging from 32 to 45 nm in the warm solution. Interestingly, 1 hour after hydrogelation (2 hours after heating), we observed that the vesicles aggregated and their membranes tended to fuse with each other to form nanotubes (Fig. 3B), and very short silk fibers could be drawn out from this sample. After 3 days, when very long and fine silk fibers could be made, we performed TEM measurements for gels without and with the application of external force (stirring or shaking). As shown in Fig. S15 (ESI†), we observed uniform nanotubes in the gel at day 3. The diameter of the nanotubes was about 22 nm and they entangled with each other to form networks. Surprisingly, a well-arranged alignment of nanotubes was discovered when external forces were applied to the gel (Fig. 3C). The external force of shearing and gentle shaking has been widely applied to form aligned nanostructures.1,45,52–54 It was therefore assumed that shaking or gently stirring would help the uniform nanotubes to align in parallel and form small bundles. The small bundles of nanotubes would stick to each other upon being drawn from the solution. We could therefore obtain fibers with well-aligned nanotubes.
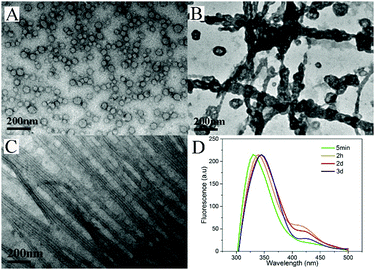 | ||
| Fig. 3 TEM images of the solution or gel taken (A) 5 minutes, (B) 2 hours and (C) 3 days after heating; (D) emission spectra of the gel (λexc = 300 nm) | ||
We also used fluorescence spectroscopy to characterize the emission spectra of the gels to understand the molecular arrangement in the nanostructures. As shown in Fig. 3D, the emission peaks of the samples showed continuous red-shifts from 5 min to 2 days (from 330 to 345 nm), suggesting more and more efficient π–π stacking between naphthalene groups, and more and more ordered molecular packing in the nanostructures. There were no big differences between the samples at day 2 and day 3, suggesting that the self-assembly reached equilibrium after about 2 days. Based on these results and the TEM pictures, we draw a hypothesis on the molecular arrangement of Nap-FGG in the nanostructures, which is illustrated in Scheme 1. The hydrophobic interactions between the aromatic fragments and hydrogen bonds assisted the molecules to firstly self-assemble into meta-stable vesicles, which would fuse with each other to form uniform and long nanotubes. The nanotubes randomly dispersed and entangled with each other to form hydrogels. When external forces were applied, the nanotubes lined up along the direction of the forces and held together side by side through hydrogen bonding and hydrophobic interactions between neighbouring outer layers. We therefore observed small bundles of nanotubes.
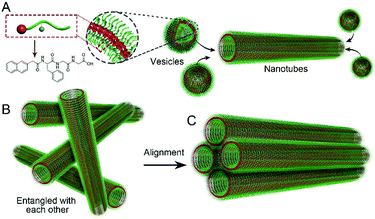 | ||
| Scheme 1 Illustration of (A) probable molecular arrangement of Nap-FGG in vesicles and nanotubes, and the formation of the (B) hydrogel and (C) microscope silk. | ||
To investigate whether the concentration of Nap-FGG could affect the formation of the hydrogel and silk-like fibers, different amounts of Nap-FGG (0.5, 1.0, 3.0 and 4.0 wt%) were dissolved in hot PBS (pH = 7.4) and all samples formed transparent solutions. The solutions were then allowed to cool back to room temperature and those containing 1.0, 3.0 and 4.0 wt% of Nap-FGG formed hydrogels, while the one with 0.5 wt% could only form a clear solution. Optical images of hydrogels and solutions are shown in Fig. S16 (ESI†). No silk-like fibers were formed in the solution containing 0.5 wt% of Nap-FFG, and the length of silk-like fibers drawn out of hydrogels formed by 1.0, 3.0, and 4.0 wt% of Nap-FGG was about 20, 5, and 6 cm, respectively. These observations indicate that the concentration of the peptide in the gels affects the silk-like fiber forming properties probably due to the different interactions between nanofibers.
We also changed the number of glycine (G)/phenylalanine (F) to make Nap-FnGm (n and m represents the number of repeated amino acids in the sequence; n = 1, 2, 3; m = 1, 2, 3) in order to investigate whether other peptide sequences could also form silk-like structures. After the heating and cooling procedure, Nap-FG3, Nap-F2G and Nap-F2G2 could form stable hydrogels, while Nap-FG, Nap-FG4 and Nap-F3G could only form viscous solutions (Fig. S17, ESI†). However, none of these samples could form long and stable silk-like fibers, only very short silk-like fibers (about 2–5 cm long) could be obtained and they broke very quickly after being drawn out of the samples. These results demonstrate that the silk-like fiber forming property is specific to the peptide sequence of Nap-FGG.
Morphologies of the nanostructures in the above mentioned samples after applying external forces were studied by TEM, and the images are shown in Fig. S18 (ESI†). Only nanostructures in the hydrogel of Nap-FFG (1.0 wt%) showed parallel alignment, while the others did not. This phenomenon is in agreement with the silk drawing experiments, in that the hydrogel of Nap-FFG could form the second longest silk-like fibers among all the samples. We assumed that the interaction between nanofibers might affect the rearrangement of the nanostructures under external forces, and thus affecting the formation of silk-like fibers.
Finally, the biocompatibility of Nap-FGG was studied. We used an MTT assay to examine the viability of NIH 3T3 mouse fibroblast cells in the presence of different concentrations of the compound. As shown in Fig. S19 (ESI†), the IC50 value was about 341 μM, far below the concentration needed for silk drawing (44 mM). The results indicate that the supramolecular silk of Nap-FGG has certain cytotoxicity and should be best used for in vitro applications such as biomineration.
Conclusions
A peptide hydrogel containing nanotubes self-assembled from a tripeptide derivative (Nap-FGG) was prepared. The nanotubes randomly dispersed and physically entangled with each other in the hydrogel and therefore showed an isotropy property. After external forces were applied, the nanotubes could stick with each other to form small bundles and therefore supramolecular silk-like fibers could be drawn directly from the hydrogel. TEM results clearly show that the nanotubes rearranged and aligned along the direction of the forces, giving the silk anisotropy on a macroscopic scale. The study on the silk-like fiber formation of analogues of Nap-FGG showed that the interactions between nanotubes were crucial for the formation of supramolecular silk. The interaction should be neither so strong that it hinders the rearrangements, nor too weak that it fails to maintain the alignment of the nanotubes. Our study offers an easy and versatile method for generating materials with hierarchically ordered structures over different scales and extends the utility of peptide hydrogels.We acknowledge the financial support from the International S&T Cooperation Program of China (ISTCP, 2015DFA50310), the National Natural Science Foundation of China (51373079 and 51403105) and the Program for Changjiang Scholars and Innovative Research Team in University (IRT13023).
Notes and references
- M. Zhu, Z. Wang, J. Zhang, L. Wang, X. Yang, J. Chen, G. Fan, S. Ji, C. Xing, K. Wang, Q. Zhao, Y. Zhu, D. Kong and L. Wang, Biomaterials, 2015, 61, 85–94 CrossRef CAS PubMed.
- S. Choi, W. Kang, J. Lee, C. Najeeb, H. Chun and J. Kim, Langmuir, 2010, 26, 15680–15685 CrossRef CAS PubMed.
- B. H. Kim, D. O. Shin, S.-J. Jeong, C. M. Koo, S. C. Jeon, W. J. Hwang, S. Lee, M. G. Lee and S. O. Kim, Adv. Mater., 2008, 20, 2303–2307 CrossRef CAS.
- D. Kohler, M. Schneider, M. Krueger, C.-M. Lehr, H. Moehwald and D. Wang, Adv. Mater., 2011, 23, 1376–1379 CrossRef CAS PubMed.
- J. R. Cox, J. H. Simpson and T. M. Swager, J. Am. Chem. Soc., 2013, 135, 640–643 CrossRef CAS PubMed.
- J. Hoogboom and T. M. Swager, J. Am. Chem. Soc., 2006, 128, 15058–15059 CrossRef CAS PubMed.
- T. Kato, T. Yasuda, Y. Kamikawa and M. Yoshio, Chem. Commun., 2009, 729–739 RSC.
- T. Kato, N. Mizoshita and K. Kanie, Macromol. Rapid Commun., 2001, 22, 797–814 CrossRef CAS.
- Z. L. Wu, T. Kurokawa, S. Liang, H. Furukawa and J. P. Gong, J. Am. Chem. Soc., 2010, 132, 10064–10069 CrossRef CAS PubMed.
- Y. Liu, T. Wang, Y. Huan, Z. Li, G. He and M. Liu, Adv. Mater., 2013, 25, 5875–5879 CrossRef CAS PubMed.
- M. C. Vasudev, H. Koerner, K. M. Singh, B. P. Partlow, D. L. Kaplan, E. Gazit, T. J. Bunning and R. R. Naik, Biomacromolecules, 2014, 15, 533–540 CrossRef CAS PubMed.
- M. Reches and E. Gazit, Nat. Nanotechnol., 2006, 1, 195–200 CrossRef CAS PubMed.
- Y. Xuehai, L. Junbai and M. H. Helmuth, Adv. Mater., 2011, 23, 2796–2801 CrossRef PubMed.
- X. Yan, Y. Su, J. Li, J. Früh and H. Möhwald, Angew. Chem., Int. Ed., 2011, 50, 11186–11191 CrossRef CAS PubMed.
- B. Cao, Y. Zhu, L. Wang and C. Mao, Angew. Chem., Int. Ed., 2013, 52, 11750–11754 CrossRef CAS PubMed.
- R. J. A. Hill, V. L. Sedman, S. Allen, P. M. Williams, M. Paoli, L. Adler-Abramovich, E. Gazit, L. Eaves and S. J. B. Tendler, Adv. Mater., 2007, 19, 4474–4479 CrossRef CAS.
- C. Yang, Z. Wang, C. Ou, M. Chen, L. Wang and Z. Yang, Chem. Commun., 2014, 50, 9413–9415 RSC.
- Y. Kuang, J. Shi, J. Li, D. Yuan, K. A. Alberti, Q. Xu and B. Xu, Angew. Chem., Int. Ed., 2014, 53, 8104–8107 CrossRef CAS PubMed.
- H. Wang, J. Wei, C. Yang, H. Zhao, D. Li, Z. Yin and Z. Yang, Biomaterials, 2012, 33, 5848–5853 CrossRef CAS PubMed.
- S. Soukasene, D. J. Toft, T. J. Moyer, H. Lu, H.-K. Lee, S. M. Standley, V. L. Cryns and S. I. Stupp, ACS Nano, 2011, 5, 9113–9121 CrossRef CAS PubMed.
- A. G. Cheetham, P. Zhang, Y.-a. Lin, L. L. Lock and H. Cui, J. Am. Chem. Soc., 2013, 135, 2907–2910 CrossRef CAS PubMed.
- A. Altunbas, S. J. Lee, S. A. Rajasekaran, J. P. Schneider and D. J. Pochan, Biomaterials, 2011, 32, 5906–5914 CrossRef CAS PubMed.
- J. Gao, W. Zheng, J. Zhang, D. Guan, Z. Yang, D. Kong and Q. Zhao, Chem. Commun., 2013, 49, 9173–9175 RSC.
- S. Ray, A. K. Das and A. Banerjee, Chem. Mater., 2007, 19, 1633–1639 CrossRef CAS.
- X. D. Xu, L. Liang, C. S. Chen, L. Bo, N. L. Wang, F. G. Jiang, X. Z. Zhang and R. X. Zhuo, ACS Appl. Mater. Interfaces, 2010, 2, 2663–2671 CAS.
- B. Job, K. Mathijs, T. A. Wezendonk, E. Rienk and J. H. van Esch, J. Am. Chem. Soc., 2012, 134, 12908–12911 CrossRef PubMed.
- T. Hayashi, Y. Yasueda, T. Tamura, Y. Takaoka and I. Hamachi, J. Am. Chem. Soc., 2015, 137, 5372–5380 CrossRef CAS PubMed.
- T. Yoshii, S. Onogi, H. Shigemitsu and I. Hamachi, J. Am. Chem. Soc., 2015, 137, 3360–3365 CrossRef CAS PubMed.
- C. Ren, J. Zhang, M. Chen and Z. Yang, Chem. Soc. Rev., 2014, 43, 7257–7266 RSC.
- Y. Cai, Y. Shi, H. Wang, J. Wang, D. Ding, L. Wang and Z. Yang, Anal. Chem., 2014, 86, 2193–2199 CrossRef CAS PubMed.
- C. Ren, H. Wang, D. Mao, X. Zhang, Q. Fengzhao, Y. Shi, D. Ding, D. Kong, L. Wang and Z. Yang, Angew. Chem., Int. Ed., 2015, 54, 4823–4827 CrossRef CAS PubMed.
- S. M. Bromfield and D. K. Smith, J. Am. Chem. Soc., 2015, 137, 10056–10059 CrossRef CAS PubMed.
- J. Boekhoven and S. I. Stupp, Adv. Mater., 2014, 26, 1642–1659 CrossRef CAS PubMed.
- M. T. He, J. B. Li, S. Tan, R. Z. Wang and Y. Zhang, J. Am. Chem. Soc., 2013, 135, 18718–18721 CrossRef CAS PubMed.
- R. J. Williams, A. M. Smith, R. Collins, N. Hodson, A. K. Das and R. V. Ulijn, Nat. Nanotechnol., 2009, 4, 19–24 CrossRef CAS PubMed.
- K. M. Galler, L. Aulisa, K. R. Regan, R. N. D'Souza and J. D. Hartgerink, J. Am. Chem. Soc., 2010, 132, 3217–3223 CrossRef CAS PubMed.
- J. H. Collier and T. Segura, Biomaterials, 2011, 32, 4198–4204 CrossRef CAS PubMed.
- G. F. Liu, D. Zhang and C. L. Feng, Angew. Chem., Int. Ed., 2014, 53, 7789–7793 CrossRef CAS PubMed.
- S. H. Medina, S. Li, O. M. Z. Howard, M. Dunlap, A. Trivett, J. P. Schneider and J. J. Oppenheim, Biomaterials, 2015, 53, 545–553 CrossRef CAS PubMed.
- Y. Tian, H. Wang, Y. Liu, L. Mao, W. Chen, Z. Zhu, W. Liu, W. Zheng, Y. Zhao, D. Kong, Z. Yang, W. Zhang, Y. Shao and X. Jiang, Nano Lett., 2014, 14, 1439–1445 CrossRef CAS PubMed.
- J. S. Rudra, T. Sun, K. C. Bird, M. D. Daniels, J. Z. Gasiorowski, A. S. Chong and J. H. Collier, ACS Nano, 2012, 6, 1557–1564 CrossRef CAS PubMed.
- J. S. Rudra, Y. F. Tian, J. P. Jung and J. H. Collier, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 622–627 CrossRef CAS PubMed.
- F. Zhao, J. Li, N. Zhou, J. Sakai, Y. Gao, J. Shi, B. Goldman, H. M. Browdy, H. R. Luo and B. Xu, Bioconjugate Chem., 2014, 25, 2116–2122 CrossRef CAS PubMed.
- G. A. Hudalla, S. Tao, J. Z. Gasiorowski, H. Huifang, Y. F. Tian, A. S. Chong and J. H. Collier, Nat. Mater., 2014, 13, 829–836 CrossRef CAS PubMed.
- S. Zhang, M. A. Greenfield, A. Mata, L. C. Palmer, R. Bitton, J. R. Mantei, C. Aparicio, M. O. de La Cruz and S. I. Stupp, Nat. Mater., 2010, 9, 594–601 CrossRef CAS PubMed.
- E. J. Berns, S. Sur, L. Pan, J. E. Goldberger, S. Suresh, S. Zhang, J. A. Kessler and S. I. Stupp, Biomaterials, 2013, 35, 185–195 CrossRef PubMed.
- F. Zhao, Y. Gao, J. Shi, H. M. Browdy and B. Xu, Langmuir, 2011, 27, 1510–1512 CrossRef CAS PubMed.
- J. Zhou, X. Du, Y. Gao, J. Shi and B. Xu, J. Am. Chem. Soc., 2014, 136, 2970–2973 CrossRef CAS PubMed.
- C. G. Pappas, P. W. Frederix, T. Mutasa, S. Fleming, Y. M. Abul-Haija, S. M. Kelly, A. Gachagan, D. Kalafatovic, J. Trevino and R. V. Ulijn, Chem. Commun., 2015, 51, 8465–8468 RSC.
- B. D. Wall, S. R. Diegelmann, S. Zhang, T. J. Dawidczyk, W. L. Wilson, H. E. Katz, H.-Q. Mao and J. D. Tovar, Adv. Mater., 2011, 23, 5009–5014 CrossRef CAS PubMed.
- M. Yoshio, Y. Shoji, Y. Tochigi, Y. Nishikawa and T. Kato, J. Am. Chem. Soc., 2009, 131, 6763–6767 CrossRef CAS PubMed.
- Z. L. Wu, D. Sawada, T. Kurokawa, A. Kakugo, W. Yang, H. Furukawa and J. P. Gong, Macromolecules, 2011, 44, 3542–3547 CrossRef CAS.
- M. Arifuzzaman, Z. L. Wu, T. Kurokawa, A. Kakugo and J. P. Gong, Soft Matter, 2012, 8, 8060–8066 RSC.
- M. Lescanne, A. Colin, O. Mondain-Monval, K. Heuzé, F. Fages and J.-L. Pozzo, Langmuir, 2002, 18, 7151–7153 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6qm00233a |
| ‡ The authors contributed equally to this work. |
| This journal is © the Partner Organisations 2017 |