Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The formation of sesquiterpenoid presilphiperfolane and cameroonane metabolites in the Bcbot4 null mutant of Botrytis cinerea†
Gabriel
Franco dos Santos‡
 ab,
Javier
Moraga‡
ab,
Javier
Moraga‡
 a,
Jacqueline A.
Takahashi
a,
Jacqueline A.
Takahashi
 b,
Muriel
Viaud
b,
Muriel
Viaud
 c,
James R.
Hanson
d,
Rosario
Hernández Galán
c,
James R.
Hanson
d,
Rosario
Hernández Galán
 a and
Isidro G.
Collado
a and
Isidro G.
Collado
 *a
*a
aDepartamento de Química Orgánica, Facultad de Ciencias, Universidad de Cádiz, 11510 Puerto Real, Cádiz, Spain. E-mail: isidro.gonzalez@uca.es
bDepartamento de Química, Instituto de Ciências Exatas, Universidade Federal de Minas Gerais, 31270-901, Belo Horizonte, MG, Brazil
cUMR BIOGER, INRA, AgroParisTech, Université Paris-Saclay, 78850 Thiverval-Grignon, France
dDepartment of Organic Chemistry, University of Sussex, Brighton, Sussex BN1 9QJ, UK
First published on 15th June 2017
Abstract
Botrytis cinerea is a polyphagous fungal parasite which causes serious damage to more than 200 plant species and consequent economic losses for commercial crops. This pathogen produces two families of phytotoxins, the botryanes and botcinins, which are involved in the infection mechanism. The B. cinerea genome has provided a complete picture of the genes involved in the biosynthesis of its secondary metabolites. The botrydial biosynthetic gene cluster has been identified. This cluster consists of seven genes, where the genes BcBOT1, BcBOT3 and BcBOT4 encode three mono-oxygenases. A study of the Bcbot4Δ null mutant revealed that this mono-oxygenase was involved in the hydroxylation at C-4 of the probotryane skeleton (C-11 of the presilphiperfolane skeleton). A detailed study of the Bcbot4Δ null mutant has been undertaken in order to study the metabolic fate of the presilphiperfolan-8-ol intermediate biosynthesized by this organism and in particular by this strain. As a result three new presilphiperfolanes and three new cameroonanes have been identified. The results suggest that the absence of the oxygen function at C-11 of the presilphiperfolane skeleton permits rearrangement to a cameroonane whilst hydroxylation at C-11 precludes this rearrangement. It is possible that the interactions of the C-11 hydroxylated derivatives perturb the stereo-electronic requirements for the migration of the C-11:C-7 sigma bond to C-8.
Introduction
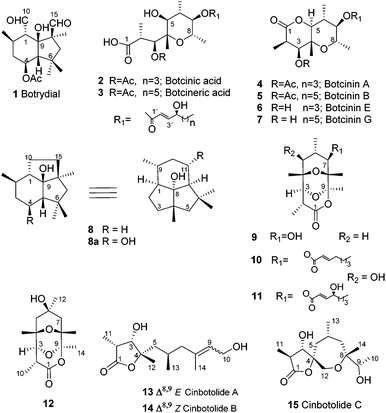
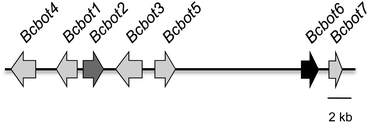
Fungi from the genus Botrytis are widespread, having been identified in cold regions such as Alaska and Canada as well as in sub-tropical regions such as Egypt. They are necrotrophic parasites which cause serious economic losses to commercial crops. The polyphagous species Botrytis cinerea has been identified as a pathogen of more than 200 plant species, often attacking damaged parts of the plant before spreading to healthy tissues. The most common symptoms involve the development of necrotic lesions on the leaves.1–4 The initiation and progress of the infection by B. cinerea involves a complex sequence of biological events including both enzymatic attack and the production of phytotoxic secondary metabolites.5,6 However in accordance with the broad range of host of this pathogen, there is no evidence for the production of host-specific toxins. The toxins produced by B. cinerea not only reproduce the symptoms of a fungal infection but have also been detected in plant tissues infected by B. cinerea.5,7–9 Two major families of phytotoxic metabolites are sesquiterpenes related to botrydial5,9 (1) and the polyketides, 3-acetylbotcinic and 3-acetylbotcineric acids (2, 3) and their botcinin (4, 5) relatives, Scheme 1.10–12 Botrydial (1) is produced during the infection of a plant and induces chlorosis and cell collapse.13 3-Acetylbotcinic acid (2) and its derivatives also induce chlorosis and necrosis and have some anti-fungal activity, (Scheme 1).12The sequencing of the B. cinerea genome has led to the identification of the botrydial biosynthetic gene cluster, Fig. 1.14,15 Such gene clusters are typical of the secondary metabolism in filamentous fungi.16,17 This cluster consists of seven genes (BcBOT1 to BcBOT7) coding for a sesquiterpene cyclase (BcBOT2), an acetyltransferase (BcBOT5), a transcription factor (BcBOT6), a putative dehydrogenase (BcBOT7) and three mono-oxygenases (BcBOT1, BcBOT3 and BcBOT4).17–19
The functional role of these genes in botrydial biosynthesis has been identified using mutants.15,18,19 The metabolites which were produced by the null mutants Bcbot3Δ and Bcbot4Δ were analysed after five days fermentation19 at the time of maximum botrydial production (Scheme 1).20 The fermentation of the BcBOT4 mutant revealed that the related mono-oxygenase was involved in the hydroxylation at C-11 (C-4 in botrydial numbering, see 8) of the presilphiperfolan-8-ol intermediate (8) in the biosynthesis. No other intermediates in the botrydial pathway were detected and, at this stage in the fermentation, compound 8 accumulated. When botrydial production was disrupted, larger quantities than normal of the polyketide metabolites were formed including compounds (2–7), the botrylactones (9–12) and the cinbotolides A–C (13–15).19
Since a large amount of compound 8 was isolated after the five-day fermentation of the Bcbot4Δ null mutant,19 it was of interest to study its metabolic fate after longer periods of fermentation. The structures of these metabolites form the subject of this paper.
Results and discussion
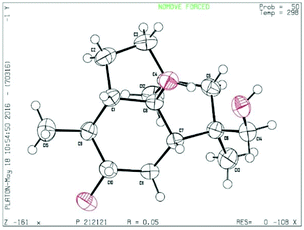
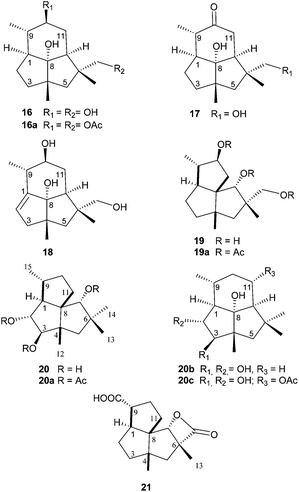
The mutant Bcbot4Δ was grown on a malt agar medium (1 L) for 10, 17 and 21 days. A detailed TLC analysis of the extracts revealed significant differences between the 10 and 17 day fermentations when compared to a 5 day fermentation. The mutant was then grown on a malt agar medium (6 L) for 17 days at 23 °C under white light. The metabolites were recovered from the medium by extraction with ethyl acetate and the constituents of the extract (6.7 g) were separated by chromatography. A small amount of compound 8 was obtained from the less polar fraction. The polyketides 2–7 and 9–15 were also isolated and identified by comparison of their spectroscopic data with those of authentic samples.19 Six new compounds (16–21) were isolated from the fractions of medium polarity and purified by further chromatography including HPLC. The 1H and 13C NMR spectra suggested that they possessed sesquiterpenoid structures.The HRMS and 13C NMR data for compound 16 which were obtained as a colourless crystalline solid, corresponded to a molecular formula of C15H26O3. It possessed IR absorption at 3430 cm−1 (OH). The 13C NMR spectrum, aided by DEPT and HSQC experiments, (see ESI, Table S1†) exhibited three methyl group signals, five methylenes including one hydroxymethyl group, four methines including one secondary alcohol and three quaternary carbons including a tertiary alcohol. The three methyl group signals in the 1H NMR spectrum were a doublet and two singlets, whilst the hydroxymethyl group signal only contained a geminal coupling. The secondary alcohol resonance was a doublet of double-doublets. These data suggested that compound 16 was a dihydroxypresilphiperfolan-8-ol. The HMBC spectrum revealed correlations between the signal for the secondary alcohol at δH 3.25 and C-7, C-9, C-11 and C-15 and between the hydroxymethyl group at δ 3.35 and C-6 thus establishing the location of the extra hydroxyl groups. The proposed structure for compound 16 was confirmed by extensive HMBC, HSQC and NOESY experiments (see Table S1 in the ESI†). In particular there was a NOESY correlation between the signal at δH 3.25 (H-10) and H-7α and H-15 indicating a β configuration for this hydroxyl group. Finally the structure was confirmed by X-ray crystallography (Fig. 2). On acetylation with acetic anhydride in pyridine, compound 16 formed a diacetate 16a (OAc δH 2.05 and 2.08) in which the signals for H-14 and H-10 were shifted to δH 4.0 and 4.6 respectively.
The minor metabolites 17 and 18 showed 1H NMR signals which were similar to those of compound 16 retaining the signals for the three methyl groups, the tertiary alcohol and the hydroxymethyl group. The principle difference in the spectra of 17 was the lack of the CH(OH) signal at δH 3.25 and the presence in the 13C NMR spectrum of a carbonyl signal at δC 213.7. The H-11 signals were deshielded to δH 2.34 and 2.52. Consequently compound 17 was the oxidation product of 16 at C-10, a conclusion supported by 2D COSY, HMBC and HSQC experiments (see Table S2, ESI†). The major difference in the NMR spectra of compound 18 compared to that of compound 16 was the presence of alkene signals at δH 4.94 (dd, J 2.1 and 2.5 Hz) and δC of 148.7 and 116.2 corresponding to a trisubstituted alkene. The location of the alkene between C-1 and C-2 was established by the deshielding of the methyl group doublet and the H-3 signals at δH 2.40 (1H, dd, J 17 and 2.1 Hz) and 2.25 (1H, dt, J 17 and 2.5 Hz). In the HMBC spectrum (see Table S3, ESI†) there were correlations between H-2 and C-3, C-4 and C-8 and between H-15 and C-1. Thus the structures of 16–18 were presilphiperfolan-8α,10β,14-triol (16), 8α,14-dihydroxypresilphiperfolan-10-one (17) and 8α,10β,14-trihydroxypresilphiperfol-1-ene (18).
The HRMS and 13C NMR data for compound 19 corresponded to a molecular formula, C15H26O3. The 1H and 13C NMR spectra, see Table S4,† revealed the presence of one secondary methyl group and two tertiary methyl groups, four methylene and one hydroxymethyl group, two secondary alcohols and two further methine carbons and three quaternary carbons. Whilst there were similarities to the spectra of the presilphiperfolantriol 16, there was a major difference in the absence of the tertiary alcohol. However the spectra were in accordance with a cameroonane carbon skeleton. Cameroonan-7α-ol has previously been obtained by the rearrangement of presilphiperfolan-8α-ol.21,22 The 1H NMR signal of one of the secondary alcohols was a singlet (δH 3.89) indicating that this alcohol was situated between two quaternary carbons typical of the cameroonan-7α-ol. The other secondary alcohol was a doublet of double-doublets (δH 3.72, ddd, J 11, 9.7 and 6.0 Hz). Two of these couplings were assigned to a methylene (δH 1.73 and 1.65). Apart from their geminal coupling (J 12.2 Hz), these resonances possessed no additional couplings. Together with the HMBC spectra, this was consistent with the location of this hydroxyl group at C-10 (see Table S4, ESI†). The stereochemistry of the alcohols was established by a NOESY correlation between the H-10 resonance and H-15, between H-7 and H-12 and H-9, and between H-14 and H-1. Furthermore when compared to the 13C NMR spectrum of cameroonan-7α-ol there were γ-gauche shieldings by the C-10 β hydroxyl group of the resonances of C-1, C-8 and C-15 and by the C-14 hydroxyl group of C-5. This compound formed a triacetate (19a), the spectra of which were consistent with the formulation of the parent alcohol as cameroonan-7α,10β,14-triol (19).
The 13C NMR spectrum (Table S5†) of compound 20 contained signals assigned to four methyl groups, three methylenes, five methines, three of which were secondary alcohols and three quaternary carbon atoms. The pattern of the 1H NMR signals was similar to that of compound 19 particularly in the presence of the singlet assigned to H-7. However the signal assigned to H-10 was replaced by a double-doublet (δ 3.49, J 8.6 and 9.1 Hz) and a doublet (δ 3.81, J 9.1 Hz) which were assigned to two secondary alcohols. Their vicinal relationship was revealed by the COSY spectrum whilst their location at C-2 and C-3 followed from HMBC correlations between H-2 and C-9 and C-3 and between H-3 and C-12 and C-5. Their stereochemistry followed from the magnitude of the vicinal coupling constants H-1:H-2 (J 8.6 Hz) and H-2:H-3 (J 9.1 Hz) which corresponded to trans relationships between these protons. There were nOe correlations between H-2 and H-9 and H-12 and between H-3 and H-1, H-5α and H-14. The stereochemistry of the alcohol at C-7 followed from the nOe enhancements of H-11β and H-13 on irradiation of H-7. Acetylation of the triol gave a triacetate (20a) with NMR data (see Experimental) that was consistent with the structure of cameroonan-2α,3β,7α-triol for the parent alcohol. It is interesting to note that the stereochemistry of this vicinal diol (20) corresponds to that formed from presilphiperfolan-8α-ol (20c) prior to the cleavage of the five-membered ring to form botrydial (1). The oxygenases that mediate this step are coded by the genes BcBOT1 and BcBOT3. Molecular models reveal a structural similarity between the presilphiperfolan-8α-ol substrate and a cameroonan-7α-ol in which a presilphiperfolanol 11α-hydroxyl group and a cameroonan-7α-hydroxyl group would also be juxtaposed.
Compound 21 was the most polar metabolite. The 1H and 13C NMR spectra, (Table S6†) contained signals assigned to two tertiary methyl groups, five methylenes, three methines and three quaternary carbon atoms and two carbonyl groups at δC 180.6 and 175.7. These were assigned to a carboxylic acid and an ester function. The characteristic cameroonan-7α-ol, H-7, singlet was shifted to δH 4.27 and the presence of IR absorption at 1810 cm−1 was indicative of the presence of a β-lactone. There were HMBC correlations between H-12 and C-3, C-4, C-5 and C-8 and between H-13 and C-5, C-6, C-7 and C-14 thus locating the carboxyl group at C-15. Compound 21 was therefore assigned the structure of 7-α-hydroxycameroonan-14,15-dioic acid 14,7-lactone.
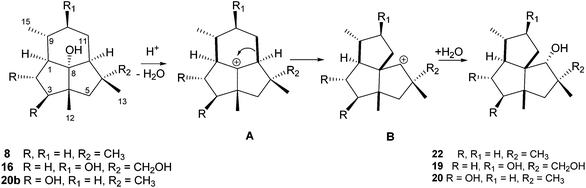
Biosynthetically these cameroonane metabolites may be formed by the rearrangement of presilphiperfolan-8α-ol which has laboratory precedent.22 The loss of the 8α-hydroxyl group may generate a C-8 carbocation (A) which may be followed by the rearrangement of the C-11:C-7 bond to C-8 to yield a C-7 carbocation (B), Scheme 3. This is then discharged by a hydroxyl group to form the cameroonan-7α-ol (22). The geometrical requirements for this rearrangement have been examined. An X-ray crystal structure of the p-nitrobenzoate of presilphiperfolan-8α-ol revealed that there is a trans-relationship between the departing 8α-hydroxyl group and the migrating C-11:C-7 bond in a chair six-membered ring. It is likely that this rearrangement leads to the formation of the cameroonanes in B. cinerea.22
In the same line compound 19, Scheme 3, should be formed from the isolated compound 16, while compound 20 must arise from the rearrangement of the hypothetical precursor 20b.
Compounds with the cameroonane carbon skeleton have not been detected previously as the metabolites of B. cinerea. Fermentation of the intact B. cinerea for 15 and 17 days led to the isolation of botryanes and the presilphiperfolane 20c, Scheme 2, the precursor of botrydial (1) by oxidative cleavage of the C10–C15 bond.19 The presence of the C-11 hydroxyl group (C-4 in botryane numbering, see 8) which is introduced by the enzyme system mediated by BcBOT4, appears to inhibit the rearrangement. It could be that transannular hydrogen bonding to the C-8 hydroxyl group and interactions between the C-11 hydroxyl group and the methyl groups at C-4, lead to a distortion of the chair form of the six-membered ring and so reduces the facility with which the ring-contraction occurs.
Experimental
General experimental procedures
Purification by semipreparative and analytical HPLC was performed with a Hitachi/Merck L-6270 apparatus equipped with a differential refractometer detector (RI-7490). A LiChrospher® Si 60 (5 μm) LiChroCart® (250 mm × 4 mm) column and a LiChrospher® Si 60 (10 μm) LiChroCart® (250 mm × 10 mm) were used in isolation experiments. Silica gel (Merck) was used for column chromatography. TLC was performed on Merck Kieselgel 60 F254, 0.25 mm thick. Melting points were measured with a Reichert-Jung Kofler block and are uncorrected. Optical rotations were determined with a digital polarimeter. Infrared spectra were recorded on a FT-IR spectrophotometer and reported as wave number (cm−1). 1H and 13C NMR measurements were recorded on Agilent 500 and 600 MHz spectrometers with SiMe4 as the internal reference. Chemical shifts were referenced to CDCl3 (δH 7.25, δC 77.0). NMR assignments were made using a combination of 1D and 2D techniques. Multiplicities are described using the following abbreviations: s = singlet, d = doublet, t = triplet, q = quarter; quint = quintuplet; sext = sextuplet; m = multiplet, br = broad. High-Resolution Mass Spectroscopy (HRMS) was performed either with a double-focusing magnetic sector mass spectrometer in a QTOF mass spectrometer in positive or negative ion ESI or APCI modes (APGC+ for samples analysed by GC chromatography).Fungal material
For the isolation of metabolites, the null mutant ΔBcbot4 of Botrytis cinerea was cultured in 200 mm Petri dishes containing 100 mL of malt agar medium (6 L) for 17 days at 23 °C under white light. After the growth time, the media were extracted with ethyl acetate. The extracts were dried over sodium sulfate, and the solvent was evaporated to give 6.7 g of extract.
Gene deletion of the cytochrome P450-encoding gene bcbot4
The B. cinerea gene bcbot4 (GenBank: AY277723.2) was deleted by homologous recombination. The 5′ genomic non-coding regions of bcbot4 (1442 pb) was amplified by PCR. In the same way, we obtained the 3′ regions of bcbot4 (866 pb) and the nourseothricin resistance gene (nat). For each gene, a Knock-Out (KO) cassette consisting of the 5′ region of bcbot, the nat resistance gene and the 3′ region bcbot was generated by double-joint PCR fusion24 as previously described for B. cinerea.25 All the PCR primers have been reported.19Protoplasts from the B05.10 strain were prepared and transformed as previously described.19,26 Additionally, the genetic purity of the KO transformants was checked by the absence of the bcbot4 WT allele. Information about the B. cinerea KO mutant used in this study and other published mutants is available at http://botbioger.versailles.inra.fr/botmut/.
Isolation of the metabolites
Fractionation of the extract was carried out on a silica column to give six fractions using 100% hexane (5.3 mg), hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (70
ethyl acetate (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) (1.7 g), hexane
30) (1.7 g), hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (40
ethyl acetate (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60) (2.1 g), hexane
60) (2.1 g), hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (10
ethyl acetate (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90) (1.25 g), ethyl acetate
90) (1.25 g), ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (70
methanol (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) (1.3 g) and 100% methanol (303.8 mg).
30) (1.3 g) and 100% methanol (303.8 mg).
In addition to the known compounds 2–15,19 which were obtained from the fractions of medium polarity, the sesquiterpenes presilphiperfolan-8α,10β,14-triol (16) (93 mg), 8α,14-dihydroxypresilphiperfolan-10-one (17) (1.5 mg), 8α,10β,14-trihydroxypresilphiperfol-1-ene (18) (1 mg), cameroonan-7α,10β,14-triol (19) (102 mg) and 7α-hydroxycameroonane-14,15-dioic acid 14-7-lactone (21) (6.71 mg) were isolated and characterized. Compounds 16 (2.03 mg), 19 (1.27 mg) and 20 (1.54 mg) were acetylated. For acetylation, compounds were dissolved in the minimum quantity of dry pyridine (0.5 mL), and acetic anhydride (30 mg) was added to the solution. The mixture was kept at room temperature with stirring for 24 h. The solvent was evaporated to give derivatives 16a (2.20 mg), 19a (1.32 mg) and 20a (1.49 mg).
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3CO), 2.05 (s, 3H, CH3CO), 1.98 (1H, m, H-2α), 1.94 (1H, d, J = 12.1 Hz, H-5α), 1.90 (1H, m, H-3), 1.88 (1H, m, H-9), 1.88 (1H, m, superimposed, H-11), 1.68 (1H, td, J = 11.0, 6.2 Hz, H-1), 1.58 (1H, m, H-11), 1.46 (1H, ddd, J = 11.2, 9.1, 3.9 Hz, H-7), 1.34 (s, 3H, H-12), 1.24 (1H, m, H-3′), 1.21 (s, 3H, H-13), 1.14, (1H, d, J = 12.1 Hz, H-5β), 0.89 (d, J = 6.3, 3H, H-15). 13C NMR (125 MHz, CDCl3) δ, 170.9 (C, CH3CO), 170.7 (C, CH3CO), 94.44, (C, C-8), 77.11 (CH, C-10), 74.7 (CH2, C-14), 56.13 (C, C-4), 51.64 (C, C-6), 46.9 (CH, C-7), 44.6 (CH, C-9), 43.61 (CH2, C-5), 41.0 (CH, C-1), 33.23 (CH2, C-2), 33.07 (CH2, C-3), 32.4 (CH2, C-11), 28.32 (CH3, C-12), 22.53 (CH3, C-13), 21.2 (
3CO), 2.05 (s, 3H, CH3CO), 1.98 (1H, m, H-2α), 1.94 (1H, d, J = 12.1 Hz, H-5α), 1.90 (1H, m, H-3), 1.88 (1H, m, H-9), 1.88 (1H, m, superimposed, H-11), 1.68 (1H, td, J = 11.0, 6.2 Hz, H-1), 1.58 (1H, m, H-11), 1.46 (1H, ddd, J = 11.2, 9.1, 3.9 Hz, H-7), 1.34 (s, 3H, H-12), 1.24 (1H, m, H-3′), 1.21 (s, 3H, H-13), 1.14, (1H, d, J = 12.1 Hz, H-5β), 0.89 (d, J = 6.3, 3H, H-15). 13C NMR (125 MHz, CDCl3) δ, 170.9 (C, CH3CO), 170.7 (C, CH3CO), 94.44, (C, C-8), 77.11 (CH, C-10), 74.7 (CH2, C-14), 56.13 (C, C-4), 51.64 (C, C-6), 46.9 (CH, C-7), 44.6 (CH, C-9), 43.61 (CH2, C-5), 41.0 (CH, C-1), 33.23 (CH2, C-2), 33.07 (CH2, C-3), 32.4 (CH2, C-11), 28.32 (CH3, C-12), 22.53 (CH3, C-13), 21.2 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3CO), 21.0 (CH3CO), 17.20 (CH3, C-15). HRMS (APGC+): calcd for C19H31O5 [M + H]+ 339.2171, found 339.2162.
H3CO), 21.0 (CH3CO), 17.20 (CH3, C-15). HRMS (APGC+): calcd for C19H31O5 [M + H]+ 339.2171, found 339.2162.
The diffraction frames were integrated using the program SAINT, and the integrated intensities were corrected for Lorentz_polarization effects and absorption with SADABS. The structure was solved by direct methods and refined to all 3237 unique Fo2 by full matrix least-squares calculations using SHELXL-97. All the hydrogen atoms, except the hydrogens of the –OH groups, were placed at idealized positions and refined as rigid atoms. The positions of the OH hydrogens are found in the diffraction map and refined as rigid atoms. Final R1 indices [2699 with I > 2σ(I)] 0.0465, wR2 = 0.1074. R indices (all data) R1 = 0.0578, wR2 = 0.1150. S[F2] 1.050 for 173 refined parameters. Crystal data: C15H26O3, M = 254.36, orthorhombic, a = 9.9836(8) Å, b = 11.1199(9) Å, c = 12.6582(11) Å, α = β = γ = 90°, V = 1405.3(2) Å3, dimensions 0.50 × 0.28 × 0.10 mm, space group: P212121, Z = 4, F(000) = 560, μ = 0.081 mm−1. Crystallographic data of presilphiperfolan-8α,10β,14-triol have been deposited at the Cambridge Crystallographic Data Centre under the reference number CCDC 1547406.†
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3–CO), 2.04 (3H, s, C
3–CO), 2.04 (3H, s, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3–CO), 2.01 (3H, s, C
3–CO), 2.01 (3H, s, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3–CO), 1.90 (1H, m, H-2), 1.89 (1H, dd 12.3, 6.0 Hz, H-11) 1.80 (1H, d, J = 14.4 Hz, H-5), 1.64 (1H, dd, J = 12.3, 10.4 Hz, H-11′), 1.54 (2H, m, H-3, H-3′), 1.49 (1H, m, H-9) 1.37 (1H, m, H-2′) 1.36 (1H, d, J = 14.4 Hz, H-5′), 1.20 (3H, s, H-13), 1.08 (3H, s, H-12), 0.98 (3H, d, J = 6.5 Hz, H-15). 13C NMR (125 MHz, CDCl3) δ, 171.2 (C, CH3
3–CO), 1.90 (1H, m, H-2), 1.89 (1H, dd 12.3, 6.0 Hz, H-11) 1.80 (1H, d, J = 14.4 Hz, H-5), 1.64 (1H, dd, J = 12.3, 10.4 Hz, H-11′), 1.54 (2H, m, H-3, H-3′), 1.49 (1H, m, H-9) 1.37 (1H, m, H-2′) 1.36 (1H, d, J = 14.4 Hz, H-5′), 1.20 (3H, s, H-13), 1.08 (3H, s, H-12), 0.98 (3H, d, J = 6.5 Hz, H-15). 13C NMR (125 MHz, CDCl3) δ, 171.2 (C, CH3![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) O), 170.75(C, CH3
O), 170.75(C, CH3![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) O), 170.4 (C, CH3
O), 170.4 (C, CH3![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) O), 89.12 (CH, C-7), 78.7 (CH, C-10), 68.93 (CH2, C-14), 62.9 (C, C-8), 47.8 (CH2, C-5), 49.25 (C, C-4), 49.15 (CH, C-1), 46.80 (CH, C-9), 42.35 (C, C-6), 40.1 (CH2, C-11), 38.91 (CH2, C-3), 28.85 (CH2, C-2), 26.73 (CH3, C-13), 24.93(CH3, C-12), 21.13 (C
O), 89.12 (CH, C-7), 78.7 (CH, C-10), 68.93 (CH2, C-14), 62.9 (C, C-8), 47.8 (CH2, C-5), 49.25 (C, C-4), 49.15 (CH, C-1), 46.80 (CH, C-9), 42.35 (C, C-6), 40.1 (CH2, C-11), 38.91 (CH2, C-3), 28.85 (CH2, C-2), 26.73 (CH3, C-13), 24.93(CH3, C-12), 21.13 (C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3CO), 21.04 (C
3CO), 21.04 (C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3CO), 20.89 (C
3CO), 20.89 (C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3CO), 16.70 (CH3, C-15). HRMS (APGC+): calcd for C21H33O6 [M + H]+ 381.2277, found 381.2256.
3CO), 16.70 (CH3, C-15). HRMS (APGC+): calcd for C21H33O6 [M + H]+ 381.2277, found 381.2256.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3–CO), 2.08 (1H, m, H-1), 2.05 (3H, s, C
3–CO), 2.08 (1H, m, H-1), 2.05 (3H, s, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3–CO), 2.05 (3H, s, C
3–CO), 2.05 (3H, s, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3–CO), 1.99 (1H, m superimposed, H-9), 1.95 (1H, m, superimposed, H-10α), 1.80 (1H, dddd, J = 12.7, 6.5, 4.3 Hz, H-10β), 1.64 (1H, m, H-11), 1.39 (1H, d, J = 13.9 Hz, H-5β), 1.10 (3H, s, H-14), 1.22 (1H, m, H-11′), 1.02 (3H, s, H-12), 1.0 (3H, s, H-13), 0.96 (3H, d, J = 6.8 Hz, H-15). 13C NMR (125 MHz, CDCl3) δ 170.4(CH3
3–CO), 1.99 (1H, m superimposed, H-9), 1.95 (1H, m, superimposed, H-10α), 1.80 (1H, dddd, J = 12.7, 6.5, 4.3 Hz, H-10β), 1.64 (1H, m, H-11), 1.39 (1H, d, J = 13.9 Hz, H-5β), 1.10 (3H, s, H-14), 1.22 (1H, m, H-11′), 1.02 (3H, s, H-12), 1.0 (3H, s, H-13), 0.96 (3H, d, J = 6.8 Hz, H-15). 13C NMR (125 MHz, CDCl3) δ 170.4(CH3![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) O), 170.4(CH3
O), 170.4(CH3![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) O), 170.3(CH3
O), 170.3(CH3![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) O), 89.9 (CH, C-7), 85.3 (CH, C-3), 83.2 (CH, C-2), 65.7 (C, C-8), 55.9 (CH, C-1), 52.4 (CH2, C-5), 50.6 (C, C-4), 42.1 (C, C-6), 40.3 (CH, C-9), 35.6 (CH2, C-10), 35.1, 30.3 (CH3, C-14), 25.0 (CH3, C-12), 21.1 (C
O), 89.9 (CH, C-7), 85.3 (CH, C-3), 83.2 (CH, C-2), 65.7 (C, C-8), 55.9 (CH, C-1), 52.4 (CH2, C-5), 50.6 (C, C-4), 42.1 (C, C-6), 40.3 (CH, C-9), 35.6 (CH2, C-10), 35.1, 30.3 (CH3, C-14), 25.0 (CH3, C-12), 21.1 (C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3CO), 21.2 (C
3CO), 21.2 (C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3CO), 20.9 (C
3CO), 20.9 (C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3CO), 20.5 (CH3, C-15), 19.8 (CH3, C-13). HRMS (APGC+): calcd for C21H33O6 [M + H]+ 381.2277, found 381.2264.
3CO), 20.5 (CH3, C-15), 19.8 (CH3, C-13). HRMS (APGC+): calcd for C21H33O6 [M + H]+ 381.2277, found 381.2264.
Acknowledgements
This research was supported by a grant from Ministerio de Economía y Competitividad (AGL2015-65684-C2-1-R) (MINECO-FEDER). The use of NMR facilities at the Servicio Centralizado de Ciencia y Tecnología (SCCYT) of the University of Cádiz is acknowledged.Notes and references
- Y. Elad, B. Williamson, P. Tudzynski and N. Delen, Botrytis: Biology, pathology and control, 2007 Search PubMed.
- Botrytis-the fungus, the pathogen and its management in agricultural systems, ed. S. Fillinger and Y. Elad, Springer International Publishing, 2016 Search PubMed.
- E. M. Govrin and A. Levine, J. Nat., 2000, 10, 751–757 CAS.
- I. Kars and J. Van Kan, in Botrytis: biology, pathology and control, ed. Y. Elad, B. Williamson, P. Tudzynski and N. Delen, Kluwer Academic Publisher, 2007, pp. 99–118 Search PubMed.
- I. G. Collado and M. Viaud, in Botrytis-the fungus, the pathogen and its management in agricultural systems, ed. S. Fillinger and Y. Elad, Springer International Publishing, 2016, pp. 291–313 Search PubMed.
- J. L. Reino, R. Durán-Patrón, I. Segura, R. Hernández-Galán, H. H. Riese and I. G. Collado, J. Nat. Prod., 2003, 66, 344–349 CrossRef CAS PubMed.
- N. Deighton, I. Muckenschnabel, A. J. Colmenares, I. G. Collado and B. Williamson, Phytochemistry, 2001, 57, 689–692 CrossRef CAS PubMed.
- A. J. Colmenares, J. Aleu, R. Durán-Patrón, I. G. Collado and R. Hernández-Galán, J. Chem. Ecol., 2002, 28, 997–1005 CrossRef CAS PubMed.
- I. G. Collado, A. J. M. Sánchez and J. R. Hanson, Nat. Prod. Rep., 2007, 24, 674–686 RSC.
- H. Tani, H. Koshino, E. Sakuno and H. Nakajima, J. Nat. Prod., 2005, 68, 1768–1772 CrossRef CAS PubMed.
- H. Tani, H. Koshino, E. Sakuno, H. G. Cutler and H. Nakajima, J. Nat. Prod., 2006, 69, 722–725 CrossRef CAS PubMed.
- H. G. Cutler, S. R. Parker, S. a. Ross, F. G. Crumley and P. R. Schreiner, Biosci., Biotechnol., Biochem., 1996, 60, 656–658 CrossRef CAS PubMed.
- F. R. Rossi, A. Gárriz, M. Marina, F. M. Romero, M. E. Gonzalez, I. G. Collado and F. L. Pieckenstain, Mol. Plant-Microbe Interact., 2011, 24, 888–896 CrossRef CAS PubMed.
- V. Siewers, M. Viaud, D. Jimenez-Teja, I. G. Collado, C. S. Gronover, J.-M. Pradier, B. Tudzynski and P. Tudzynski, Mol. Plant-Microbe Interact., 2005, 18, 602–612 CrossRef CAS PubMed.
- C. Pinedo, C. M. Wang, J. M. Pradier, B. Dalmais, M. Choquer, P. Le Pêcheur, G. Morgant, I. G. Collado, D. E. Cane and M. Viaud, ACS Chem. Biol., 2008, 3, 791–801 CrossRef CAS PubMed.
- S. Fillinger, J. Amselem, F. Artiguenave, A. Billaut, M. Choquer, A. Couloux, C. Cuomo, M. B. Dickman, E. Fournier, A. Gioti, C. Giraud and C. Kodira, Macromol. Grape Wines, 2007, 125–133 Search PubMed.
- J. Amselem, C. A. Cuomo, J. A. L. van Kan, M. Viaud, E. P. Benito, A. Couloux, P. M. Coutinho, R. P. de Vries, P. S. Dyer, S. Fillinger, E. Fournier, L. Gout, M. Hahn, L. Kohn, N. Lapalu, K. M. Plummer, J. M. Pradier, E. Quévillon, A. Sharon, A. Simon, A. Have, B. Tudzynski, P. Tudzynski, P. Wincker, M. Andrew, V. Anthouard, R. E. Beever, R. Beffa, I. Benoit, O. Bouzid, B. Brault, Z. Chen, M. Choquer, J. Collémare, P. Cotton, E. G. Danchin, C. Da Silva, A. Gautier, C. Giraud, T. Giraud, C. Gonzalez, S. Grossetete, U. Güldener, B. Henrissat, B. J. Howlett, C. Kodira, M. Kretschmer, A. Lappartient, M. Leroch, C. Levis, E. Mauceli, C. Neuvéglise, B. Oeser, M. Pearson, J. Poulain, N. Poussereau, H. Quesneville, C. Rascle, J. Schumacher, B. Ségurens, A. Sexton, E. Silva, C. Sirven, D. M. Soanes, N. J. Talbot, M. Templeton, C. Yandava, O. Yarden, Q. Zeng, J. A. Rollins, M. H. Lebrun and M. Dickman, PLoS Genet., 2011, 7, e1002230 CAS.
- A. Porquier, G. Morgant, J. Moraga, B. Dalmais, I. Luyten, A. Simon, J. M. Pradier, J. Amselem, I. G. Collado and M. Viaud, Fungal Genet. Biol., 2016, 96, 33–46 CrossRef CAS PubMed.
- J. Moraga, B. Dalmais, I. Izquierdo-Bueno, J. Aleu, J. R. Hanson, R. Hernández-Galán, M. Viaud and I. G. Collado, ACS Chem. Biol., 2016, 11, 2838–2846 CrossRef CAS PubMed.
- R. Duran-Patron, J. M. Cantoral, R. Hernandez-Galan, J. R. Hanson and I. G. Collado, J. Chem. Res., 2004, 441–443 CrossRef CAS.
- P. Weyerstahl, P. Weyerstahl, H. Marschall, H. Marschall, I. Seelmann, I. Seelmann, J. Jakupovic and J. Jakupovic, Eur. J. Org. Chem., 1998, 1205–1212 CrossRef CAS.
- R. M. Coates, Z. Ho, M. Klobus and S. R. Wilson, J. Am. Chem. Soc., 1996, 118, 9249–9254 CrossRef CAS.
- T. Quidde, A. E. Osbourn and P. Tudzynski, Physiol. Mol. Plant Pathol., 1998, 52, 151–165 CrossRef CAS.
- J. H. Yu, Z. Hamari, K. H. Han, J. A. Seo, Y. Reyes-Domínguez and C. Scazzocchio, Fungal Genet. Biol., 2004, 41, 973–981 CrossRef CAS PubMed.
- Z. Antal, C. Rascle, A. Cimerman, M. Viaud, G. Billon-Grand, M. Choquer and C. Bruel, PLoS One, 2012, 7, e48134 CAS.
- A. Simon, B. Dalmais, G. Morgant and M. Viaud, Fungal Genet. Biol., 2013, 52, 9–19 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Details of spectroscopic and characterization data, and reproduction of 1H- and 13C-NMR spectra. CCDC 1547406. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c7ob01088e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |