Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
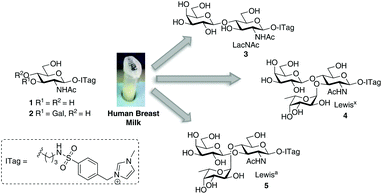
Imidazolium-labeled glycosides as probes to harness glycosyltransferase activity in human breast milk†
I.
Sittel
and
M. C.
Galan
 *
*
School of Chemistry, University of Bristol, Cantock's Close, Bristol BS8 1TS, UK. E-mail: m.c.galan@bristol.ac.uk; Fax: (+)44 (0)117 925 1295
First published on 6th April 2017
Abstract
Imidazolium-labeled (ITag-) glycosides are used to harness the glycosyltransferase activity directly from human breast milk. The covalently attached ionic labels provide a bifunctional chemical handle that is used to monitor reaction progress by MS, as well as aid in product purification from complex mixtures. The technology is exemplified in the synthesis of biologically relevant oligosaccharide analogs, LacNAc-ITag, ITag-Lewisx and ITag-Lewisa, in a matter of days from human breast milk without having to isolate specific enzymes.
The growing appreciation of oligosaccharides and glycoconjugates as key mediators in many important biological processes from cell signaling, embryogenesis, neural development to pathogen recognition, inflammation, innate immune responses and cancer, has led to their use as probes for biological research and as key compounds for drug and vaccine development.1 Despite the many recent advances in the field, progress in this area is still impeded by a lack of general and efficient methods for the routine preparation of complex oligosaccharides. Traditional chemical approaches rely on lengthy and time consuming chemical steps to achieve good yields and stereocontrol.2 On the other hand, the use of carbohydrate processing enzymes such as glycosyltransferases, to construct oligosaccharides offers several advantages over chemical approaches, since these enzymes are able to form glycosidic bonds with exquisite regio- and stereocontrol by catalyzing the transfer of an activated sugar donor to growing oligosaccharide chains without the need for protecting groups.2d,3 However, access to these enzymes is limited as the proteins can be challenging to express, expensive to employ on scale and only a handful are currently commercially available.4
Human breast milk (HBM) is an extremely complex and highly variable biofluid comprised of carbohydrates, proteins, lipids and enzymes, which has evolved over millennia to nourish infants and protect them from disease, whilst their own immune system matures.5 HBM contains more than 200 different unbound human milk oligosaccharides (HMOs),5b,6 consisting of five monosaccharide building blocks: galactose (Gal), glucose (Glc), N-acetylglucosamine (GlcNAc), fucose (Fuc) and sialic acid (Neu5Ac). All HMOs carry lactose (Gal-β-1,4-Glc) at the reducing end, which can be elongated through a β-1,3- or β-1,6-linkage by two different disaccharides, either Gal-β-1,3-GlcNAc (lacto-N-biose, type 1 chain) or Gal-β-1,4-GlcNAc N-acetyllactosamine (LacNAc, type 2 chain). Further chain elongation can take place by the incorporation of Fuc and Neu5Ac units.7 Our understanding of the biosynthesis of these oligosaccharides is limited, since most of the specific glycosyltransferases that are responsible for the formation of HMO structures with specific glycosidic linkages have not been identified.7b,8 In addition, HMO-specific glycosyltransferase expression levels are determined by the blood group, diet and environmental and genetic factors as well as lactational period of the donor.7a,9
Ionic liquid-based labels have emerged as a new class of chemical labels that can be employed for the immobilization and mass spectrometry (MS) detection of reagents from complex reaction mixtures, due to their unique physical and chemical properties.10 Our group10c,11 and others12 have reported the synthesis of inexpensive and versatile imidazolium-based tags (ITags) and their application to chromatography-free chemical oligosaccharide synthesis. Furthermore, in our work, we demonstrated that the cationic labels are also compatible with enzymatic transformations and that ITag-glycoside substrates could be employed for the qualitative and quantitative reaction monitoring, using MS techniques, of bovine milk β-1,4-galactosyltransferase (β-1,4-GalT),13 human α-1,3-fucosyltransferase (α-1,3-FucT VI)14 and α-2,3-sialyltransferase (α-2,3-ST) from pasteurella multocida, as well as α-2,6-sialyltransferase (α-2,6-ST)15 from photobacterium damsela. More recently, ITag-labeled mannosamine was also employed for the non-covalent labeling of live cells,16 demonstrating the versatility of the probes.
Encouraged by the simplicity and practicality of the approach, we hypothesized that ITag-glycan probes would be ideally suited to harness the biosynthetic machinery (e.g. glycosyltransferases) present in HBM, and thus gain expedient access to biologically important oligosaccharides, without the need to isolate or express specific enzymes (Fig. 1).
Initial experiments began by assessing our ability to monitor mixtures of ITag-glycoside products that had been generated in vitro in a multi-enzyme environment. To that end, methylimidazolium N-benzenesulfonyl-linked GlcNAc (GlcNAc-ITag) 1, which was prepared in 4 steps and 59% overall yield from azidopropyl GlcNAc 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 (see ESI† for details), was incubated in 3-morpholinopropane-1-sulfonic acid (MOPS) buffer (pH 7.8) at 37 °C with three different recombinant enzymes: β-1,4-GalT, which catalyzes the transfer of galactose from uridine 5′-diphosphogalactose (UDP-Gal) to C4 of GlcNAc to yield ITag-LacNAc 3; α-2,3-ST, which in the presence of cytidine 5′-monophospho-N-acetylneuraminic acid (CMP-Neu5Ac) will add Neu5Ac at C3′ of the LacNAc disaccharide to yield 3′-Sialyl-LacNAc-ITag 9 and α-1,3-FucT VI, which can fucosylate 3 at C3 of the GlcNAc unit in the presence of guanosine 5′-diphospho-β-L-fucose (GDP-Fuc) to form the ITag-LewisX trisaccharide [Fuc-α-1,3-(Gal-β-1,4-)GlcNAc] 4 or generate Sialyl-LewisX-ITag 10 from 9
15 (see ESI† for details), was incubated in 3-morpholinopropane-1-sulfonic acid (MOPS) buffer (pH 7.8) at 37 °C with three different recombinant enzymes: β-1,4-GalT, which catalyzes the transfer of galactose from uridine 5′-diphosphogalactose (UDP-Gal) to C4 of GlcNAc to yield ITag-LacNAc 3; α-2,3-ST, which in the presence of cytidine 5′-monophospho-N-acetylneuraminic acid (CMP-Neu5Ac) will add Neu5Ac at C3′ of the LacNAc disaccharide to yield 3′-Sialyl-LacNAc-ITag 9 and α-1,3-FucT VI, which can fucosylate 3 at C3 of the GlcNAc unit in the presence of guanosine 5′-diphospho-β-L-fucose (GDP-Fuc) to form the ITag-LewisX trisaccharide [Fuc-α-1,3-(Gal-β-1,4-)GlcNAc] 4 or generate Sialyl-LewisX-ITag 10 from 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17 (Scheme 1). Incubations were carried out in the presence of an excess of an equimolar mixture of UDP-Gal, CMP-Neu5Ac and GDP-Fuc, which was added portion-wise over 3 days. LC-MS was used to analyze and monitor reaction progress. Four different ITagged species were identified in the mixture: starting material 1 ([M+] 513), LacNAc-ITag 3 ([M+] 675) and trisaccharides 4 ([M+] 821) and 9 ([M+] 966) in a ratio of 12
17 (Scheme 1). Incubations were carried out in the presence of an excess of an equimolar mixture of UDP-Gal, CMP-Neu5Ac and GDP-Fuc, which was added portion-wise over 3 days. LC-MS was used to analyze and monitor reaction progress. Four different ITagged species were identified in the mixture: starting material 1 ([M+] 513), LacNAc-ITag 3 ([M+] 675) and trisaccharides 4 ([M+] 821) and 9 ([M+] 966) in a ratio of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42
42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 44 for 1
44 for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 as determined by LC-MS. Comparison of retention times and MS data to that of isolated ITag-materials,13–15 confirmed the structure identity of the individual ITag-glycosides.
9 as determined by LC-MS. Comparison of retention times and MS data to that of isolated ITag-materials,13–15 confirmed the structure identity of the individual ITag-glycosides.
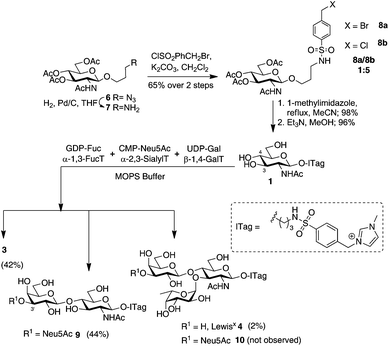 | ||
| Scheme 1 One-pot in vitro chemo-enzymatic synthesis of ITagged human milk oligosaccharides 3, 4 and 9. See ESI Fig. S1† for traces. | ||
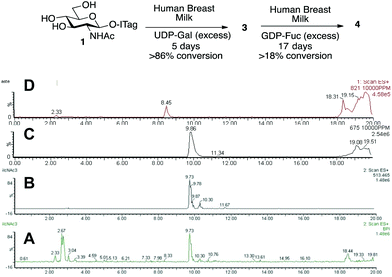
Having demonstrated that we are able to detect ITag-glycosylation products from complex enzymatic mixtures, even at low concentrations (μM), GlcNAc-ITag 1, which is a common motif in both type 1 and type 2 HMO chains,7a was used as a decoy to hijack glycosyltransferase activity in HBM. β-Linked GlcNAc units are further elongated in HMOs with a β-1,3- or β-1,4-linked galactose unit. Thus 1 was incubated at 37 °C with unpasteurized HBM (1 mg mL−1) in the presence of excess UDP-Gal (2.4 equiv.), which was added portion-wise over 5 days, in order to bias the production of galactosylated species (Scheme 2). It is important to highlight that no reaction was observed without addition of the sugar nucleotide (Fig. S3 in ESI†). To our delight, LC-MS analysis of the milk sample showed >86% conversion of 1 to a galactose-containing ITag-disaccharide (M+ 675). Importantly, ITag-oligosaccharide products display significantly different retention times in reverse-phase HPLC from other components in HBM (including unlabeled HMOs) to allow their efficient isolation. NMR analysis of the purified ITag-disaccharide confirmed the structure as β-1,4-linked LacNAc-ITag 3.18 No β-1,3-linked galactose containing disaccharide was detected in the mixture, even though it is a common fragment found among HMOs.19 This could be attributed to a number of factors e.g. low concentrations of the specific enzyme in the milk sample, low activity of the enzyme towards our ITag-substrate or an increased hydrolase activity towards the β-1,3-linked lacto-N-biose-ITag dimer.20
HMO fucosylation is a common modification, which is mostly dependent on the Lewis blood group of each individual donor. Milk donors can be divided into four different groups depending on their Lewis and secretor gene activity.21 The best known examples of fucosyltransferases found in HBM are α-1,2-fucosyltransferase (FUTII)22 and α-1,3/4-fucosyltransferase (FUTIII),23 which are responsible for the formation of α-1,2- and α-1,3/4-linked fucosides, respectively, and Lewis gene-independent α-1,3-fucosyltransferases.24 In order to target HBM fucosyltransferase activity, previously isolated ITag-LacNAc 3, which has been identified as a substrate for this class of enzymes, was incubated with excess GDP-Fuc in HBM as before (see ESI† for full details). Excitingly, a new monofucosylated ITag-trisaccharide (M+ 821) structure was observed by LC-MS analysis, which, after 18 days of incubation and 18% conversion, was isolated. Following NMR characterization, this structure could be assigned to α-1,3-linked ITag-Lewisx derivative 4 (Scheme 2). No other fucosylated species were isolated, which is in agreement with the fucosylation pattern found in HBM for LacNAc dimers9b,25 and the known specificity for HBM purified FUTIII.26 In addition to fucosyltransferase activity, α-fucosidase activity has also been identified in HBM27 and that might be accountable for the lower conversions observed.
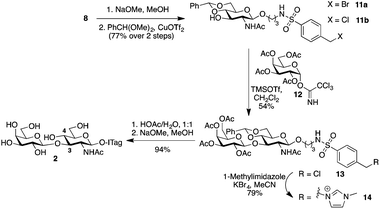
Type 1, β-1,3-linked disaccharide lacto-N-biose, is also found fucosylated in HBM.28 Possible sites for fucosylation include the common α-1,2-linkage to the terminal Gal residue of lacto-N-biose, as well as at C4 to form an α-1,4-linkage to the sub-terminal GlcNAc unit; the latter is catalyzed by the same enzyme that transfers a fucose unit to C3 of LacNAc to form Lewisx.22b Since we did not observe any β-1,3-linked disaccharides in our initial milk incubations with 1, we decided to synthetically prepare ITag-lacto-N-biose 2 (Scheme 3) and explore whether fucosyltransferases activity in the milk could be exploited in this instance. To that end, 4-(halomethyl)-benzenesulfonamide-conjugate 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14 was deacetylated using sodium methoxide in methanol. 4,6-O-Benzylidene acetal protection, using benzaldehyde dimethylacetal and copper triflate as the catalyst,29 produced glycoside acceptor 11 in 77% yield over the two steps, with a 1
14 was deacetylated using sodium methoxide in methanol. 4,6-O-Benzylidene acetal protection, using benzaldehyde dimethylacetal and copper triflate as the catalyst,29 produced glycoside acceptor 11 in 77% yield over the two steps, with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio of 11a
2 ratio of 11a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11b, ready to be glycosylated at C3. Chemical glycosylation of 11b with galactose trichloroacetimidate 12 afforded protected lacto-N-bioside 13 with complete β-stereocontrol in a 54% yield. Disaccharide 13 was subsequently ITagged by SN2 halide displacement with 1-methylimidazole to give 14 in 79% yield. Finally, global deprotection was carried out in two sequential steps: firstly acetal hydrolysis was performed with acetic acid/water, which was followed by acetate removal in the presence of sodium methoxide in methanol to reveal fully unprotected ITag-lacto-N-biose 2 in 94% yield.
11b, ready to be glycosylated at C3. Chemical glycosylation of 11b with galactose trichloroacetimidate 12 afforded protected lacto-N-bioside 13 with complete β-stereocontrol in a 54% yield. Disaccharide 13 was subsequently ITagged by SN2 halide displacement with 1-methylimidazole to give 14 in 79% yield. Finally, global deprotection was carried out in two sequential steps: firstly acetal hydrolysis was performed with acetic acid/water, which was followed by acetate removal in the presence of sodium methoxide in methanol to reveal fully unprotected ITag-lacto-N-biose 2 in 94% yield.
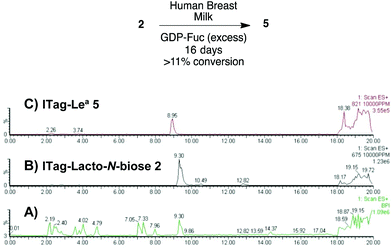
Milk incubation of 2 in the presence of an excess of GDP-Fuc (3 equiv.) in HBM was carried out as before and the reaction was monitored by LC-MS (Scheme 4). Delightfully, a new trisaccharide species was detected in the LC-MS trace, with a molecular weight consistent with a fucosylated lacto-N-biose moiety (M+ 821). A maximum conversion of 11% was observed after 16 days of incubation. After HPLC isolation and NMR structural characterization, we were able to assign the new trisaccharide to ITag-Lewisa5 (Fuc-α-1,4-[Gal-β-1,3]-GlcNAc). It is likely that the enzyme responsible for fucosylation of both disaccharides 2 and 3 is α-1,3/4-fucosyltransferase (FUTIII), which has been previously isolated from HBM.28,30
General procedure for HBM incubations
Acceptor (1, 2 or 3) and donor nucleotide (UDP-Gal or GDP-Fuc) were dissolved in HBM, which had been allowed to warm to 37 °C, and the mixture was incubated at 37 °C until no further conversion was observed. Donor nucleotide and fresh milk were added throughout the incubation period and in the case of fucosylated targets, the sample was freeze dried when no further conversion was observed and the reaction re-started by addition of fresh milk and GDP-Fucose to the dried sample (see specific details in ESI†). The reaction was then quenched by cooling on ice and the mixture filtered with ISO-Disc Filters (N 4-2, Nylon 4 mm × 0.2 μm) prior to analysis.In conclusion, although metabolic oligosaccharide pathways present a number of significant challenges, we have shown that imidazolium-labeled (ITag-) glycosides can be used to harness glycosyltransferase activity directly from human breast milk, without having to isolate the specific enzymes. We demonstrate that ITags covalently attached to glycoside substrates provide a bifunctional chemical handle that can be used to monitor reaction progress by MS, as well as aid in the purification of the products from complex mixtures. Using this technology, we were able to access a series of biologically important oligosaccharides, LacNAc-ITag, ITag-Lewisx and ITag-Lewisa, in a matter of days from a readily available source, human breast milk. We believe that these cationic probes offer a practical and novel technology to harness enzyme activity in complex biosynthetic pathways. We believe that the strategy showcased here will find applications in and beyond the field of carbohydrates.
This research was supported by EPSRC CAF EP/L001926/1 and ERC-COG: 648239. We thank Anthony P. Corfield, Matthew J. Watt and Chloe C. Medina for fruitful discussions and Mrs Marion Copeland from The Precious Milk Bank (Southmead Hospital Bristol) for donation of milk samples.
Notes and references
- (a) E. K. McCranie and B. O. Bachmann, Nat. Prod. Rep., 2014, 31, 1026 RSC; (b) J. Bauer and H. M. I. Osborn, Future Med. Chem., 2015, 7, 2285 CrossRef CAS PubMed; (c) A. Varki, Glycobiology, 2017, 27, 3 CrossRef PubMed.
- (a) M. C. Galan, D. Benito-Alifonso and G. M. Watt, Org. Biomol. Chem., 2011, 9, 3598 RSC; (b) J. P. Yasomanee and A. V. Demchenko, Trends Glycosci. Glycotechnol., 2013, 25, 13 CrossRef CAS; (c) A. Fernandez-Tejada, F. J. Canada and J. Jimenez-Barbero, ChemMedChem, 2015, 10, 1291 CrossRef CAS PubMed; (d) L. Krasnova and C. H. Wong, Annu. Rev. Biochem., 2016, 85, 599 CrossRef CAS PubMed; (e) A. P. Demchenko, Handbook of Chemical Glycosylation: Advances in Stereoselectivity and Therapeutic Relevance, Wiley, 2008 Search PubMed.
- (a) T. Murata and T. Usui, Trends Glycosci. Glycotechnol., 2000, 12, 161 CrossRef CAS; (b) A. Trincone and A. Giordano, Curr. Org. Chem., 2006, 10, 1163 CrossRef CAS; (c) M. M. Palcic, Curr. Opin. Chem. Biol., 2011, 15, 226 CrossRef CAS PubMed.
- (a) R. M. Schmaltz, S. R. Hanson and C. H. Wong, Chem. Rev., 2011, 111, 4259 CrossRef CAS PubMed; (b) P. M. Danby and S. G. Withers, ACS Chem. Biol., 2016, 11, 1784 CrossRef CAS PubMed; (c) M. C. Galan, A. P. Venot, J. Glushka, A. Imberty and G. J. Boons, J. Am. Chem. Soc., 2002, 124, 5964 CrossRef CAS PubMed; (d) M. C. Galan, A. P. Venot and G. J. Boons, Biochemistry, 2003, 42, 8522 CrossRef CAS PubMed; (e) M. C. Galan, A. P. Venot, J. Glushka, A. Imberty and G. J. Boons, Org. Biomol. Chem., 2003, 1, 3891 RSC; (f) M. C. Galan, C. S. Dodson, A. P. Venot and G. J. Boons, Bioorg. Med. Chem. Lett., 2004, 14, 2205 CrossRef CAS PubMed; (g) M. C. Galan, A. P. Venot, R. S. Phillips and G. J. Boons, Org. Biomol. Chem., 2004, 2, 1376 RSC; (h) C. A. G. M. Weijers, M. C. R. Franssen and G. M. Visser, Biotechnol. Adv., 2008, 26, 436 CrossRef CAS PubMed.
- (a) N. J. Andreas, B. Kampmann and K. M. Le-Doare, Early Hum. Dev., 2015, 91, 629 CrossRef CAS PubMed; (b) L. Bode, Early Hum. Dev., 2015, 91, 619 CrossRef CAS PubMed; (c) T. W. Hale and P. Hartmann, Textbook of Human Lactation, Hale Publishing, 2007 Search PubMed.
- Y. Yu, S. Mishra, X. Song, Y. Lasanajak, K. Bradley, M. Tappert, G. Air, D. Steinhauer, S. Halder, S. Cotmore, P. Tattersall, M. Agbandje-McKenna, R. Cummings and D. Smith, J. Biol. Chem., 2012, 287, 44784 CrossRef CAS PubMed.
- (a) L. Bode, Glycobiology, 2012, 22, 1147 CrossRef CAS PubMed; (b) X. Chen, Adv. Carbohydr. Chem. Biochem., 2015, 72, 113 CrossRef PubMed.
- D. S. Newburg, Curr. Med. Chem., 1999, 6, 117 CAS.
- (a) G. Y. Wiederschain and D. S. Newburg, J. Nutr. Biochem., 2001, 12, 559 CrossRef CAS PubMed; (b) S. Thurl, M. Munzert, J. Henker, G. Boehm, B. Muller-Werner, J. Jelinek and B. Stahl, Br. J. Nutr., 2010, 104, 1261 CrossRef CAS PubMed.
- (a) J. P. Hallett and T. Welton, Chem. Rev., 2011, 111, 3508 CrossRef CAS PubMed; (b) M. M. A. Pereira, Curr. Org. Chem., 2012, 16, 1680 CrossRef CAS; (c) M. C. Galan, R. A. Jones and A. T. Tran, Carbohydr. Res., 2013, 375, 35 CrossRef CAS PubMed; (d) T. Nokami, J. Synth. Org. Chem., Jpn., 2014, 72, 797 CrossRef CAS.
- A. T. Tran, R. Burden, D. T. Racys and M. C. Galan, Chem. Commun., 2011, 47, 4526 RSC.
- (a) L. Gillard, A. T. Tran, F. D. Boyer and J. M. Beau, Eur. J. Org. Chem., 2016, 1103 CrossRef CAS; (b) L. Ma, S. Sun, X. B. Meng, Q. Li, S. C. Li and Z. J. Li, J. Org. Chem., 2011, 76, 5652 CrossRef PubMed; (c) J. Y. Huang, M. Lei and Y. G. Wang, Tetrahedron Lett., 2006, 47, 3047 CrossRef CAS; (d) X. He and T. H. Chan, Tetrahedron, 2006, 62, 3389 CrossRef CAS.
- M. C. Galan, A. T. Tran and C. Bernard, Chem. Commun., 2010, 46, 8968 RSC.
- M. C. Galan, A. T. Tran, K. Bromfield, S. Rabbani and B. Ernst, Org. Biomol. Chem., 2012, 10, 7091 CAS.
- I. Sittel and M. C. Galan, Bioorg. Med. Chem. Lett., 2015, 25, 4329 CrossRef CAS PubMed.
- D. Benito-Alifonso, S. Tremel, J. C. Sadler, M. Berry and M. C. Galan, Chem. Commun., 2016, 52, 4906 RSC.
- M. M. Palcic and O. Hindsgaul, Trends Glycosci. Glycotechnol., 1996, 8, 37 CrossRef CAS.
- Although no significant galactosidase activity was observed, conversion levels seemed to stagnate without portion-wise addition of UDP-Galactose, likely due to donor hydrolysis.
- P. Chaturvedi, C. Warren, M. Altaye, A. Morrow, G. Ruiz-Palacios, L. Pickering and D. Newburg, Glycobiology, 2001, 11, 365 CrossRef CAS PubMed.
- (a) G. Y. Wiederschain and D. S. Newburg, Bioactive Components of Human Milk, 2001, vol. 501, p. 573 Search PubMed; (b) Note that fresh milk was added during the incubation to keep enzyme levels up.
- S. Thurl, J. Henker, M. Siegel, K. Tovar and G. Sawatzki, Glycoconjugate J., 1997, 14, 795 CrossRef CAS PubMed.
- (a) L. Shen, E. F. Grollman and V. Ginsburg, Proc. Natl. Acad. Sci. U. S. A., 1968, 59, 224 CrossRef CAS PubMed; (b) T. Kumazaki and A. Yoshida, Proc. Natl. Acad. Sci. U. S. A., 1984, 81, 4193 CrossRef CAS PubMed.
- E. F. Grollman, A. Kobata and V. Ginsburg, J. Clin. Invest., 1969, 48, 1489 CrossRef CAS PubMed.
- S. Thurl, J. Henker, M. Siegel, K. Tovar and G. Sawatzki, Glycoconjugate J., 1997, 14, 795 CrossRef CAS PubMed.
- (a) S. Thurl, M. Munzert, J. Henker, G. Boehm, B. Mueller-Werner, J. Jelinek and B. Stahl, Br. J. Nutr., 2010, 104, 1261 CrossRef CAS PubMed; (b) S. S. van Leeuwen, R. J. W. Schoemaker, G. J. Gerwig, E. J. M. van Leusen-van Kan, L. Dijkhuizen and J. P. Kamerling, Glycobiology, 2014, 24, 728 CrossRef CAS PubMed.
- K. Stangier, M. M. Palcic, D. R. Bundle, O. Hindsgaul and J. Thiem, Carbohydr. Res., 1997, 305, 511 CrossRef CAS PubMed.
- G. Y. Wiederschain and D. S. Newburg, Biochem. Mol. Med., 1996, 58, 211 CrossRef CAS PubMed.
- P. H. Johnson, A. S. R. Donald, J. Feeney and W. M. Watkins, Glycoconjugate J., 1992, 9, 251 CrossRef CAS PubMed.
- A. Tran, R. Jones, J. Pastor, J. Boisson, N. Smith and M. Galan, Adv. Synth. Catal., 2011, 353, 2593 CrossRef CAS.
- M. M. Palcic, A. P. Venot, R. M. Ratcliffe and O. Hindsgaul, Carbohydr. Res., 1989, 190, 1 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Full experimental and characterization data for all compounds, including NMR spectra and LC-MS traces. See DOI: 10.1039/c7ob00550d |
| This journal is © The Royal Society of Chemistry 2017 |