Gold nanoparticle-based colorimetric biosensors
H.
Aldewachi†
 *ad,
T.
Chalati†
*ad,
T.
Chalati†
 *ab,
M. N.
Woodroofe
a,
N.
Bricklebank
a,
B.
Sharrack
c and
P.
Gardiner
*ab,
M. N.
Woodroofe
a,
N.
Bricklebank
a,
B.
Sharrack
c and
P.
Gardiner
 a
a
aBiomolecular Sciences Research Centre, Sheffield Hallam University, Sheffield, UK. E-mail: Aldewachi@gmail.com
bSchool of Pharmacy, University of Lincoln, Lincoln, UK. E-mail: tchalati@lincoln.ac.uk
cDepartment of Neurology, University of Sheffield, Sheffield, UK
dPharmacy College, University of Mosul, Mosul, Iraq
First published on 28th November 2017
Abstract
Gold nanoparticles (AuNPs) provide excellent platforms for the development of colorimetric biosensors as they can be easily functionalised, displaying different colours depending on their size, shape and state of aggregation. In the last decade, a variety of biosensors have been developed to exploit the extent of colour changes as nano-particles (NPs) either aggregate or disperse, in the presence of analytes. Of critical importance to the design of these methods is that the behaviour of the systems has to be reproducible and predictable. Much has been accomplished in understanding the interactions between a variety of substrates and AuNPs, and how these interactions can be harnessed as colorimetric reporters in biosensors. However, despite these developments, only a few biosensors have been used in practice for the detection of analytes in biological samples. The transition from proof of concept to market biosensors requires extensive long-term reliability and shelf life testing, and modification of protocols and design features to make them safe and easy to use by the population at large. Developments in the next decade will see the adoption of user friendly biosensors for point-of-care and medical diagnosis as innovations are brought to improve the analytical performances and usability of the current designs. This review discusses the mechanisms, strategies, recent advances and perspectives for the use of AuNPs as colorimetric biosensors.
1. Introduction
Gold nanoparticles (AuNPs) (derived from the Greek word nanus, meaning dwarf) are currently used in a variety of biomedical applications, due to their size-dependent chemical, electronic and optical properties. The pertinent physical properties include a high surface-to-volume ratio, biocompatibility, in addition, the dimensions of the metal NPs are comparable to those of biomolecules such as proteins (enzymes, antibodies) or DNA, whose dimensions are in the range of 2–20 nm thus imparting a structural compatibility of these two classes of material.1,2 Moreover, the multitude of available shapes of AuNPs allow easy surface functionalisation with probes and other compounds of interest, thus making them amenable to a variety of detection modalities and techniques as shown in Fig. 1.3–5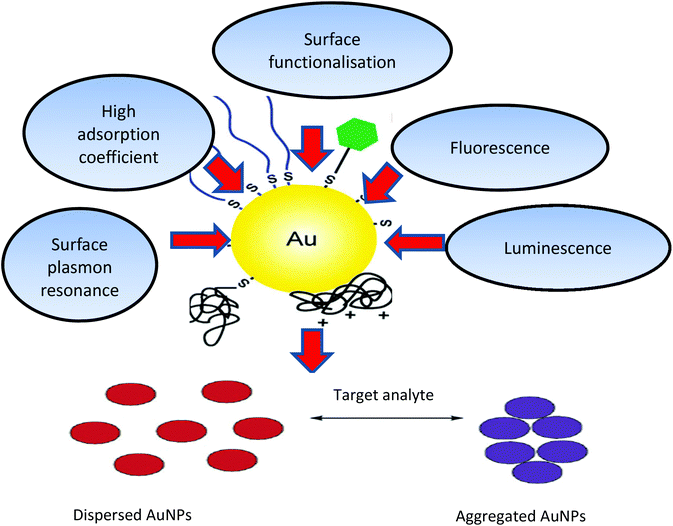 | ||
| Fig. 1 Physical and chemical properties of AuNPs and schematic illustration of AuNPs (aggregation/dispersion) colorimetric based detection systems. The unique properties of AuNPs, such as their high absorption coefficient, scattering flux, luminescence and conductivity, as well as their ability to enhance electromagnetic fields, quench (or enhance) fluorescence and catalyse reactions, provide numerous possibilities to exploit these particles for sensing and quantification purposes.10,186,187 | ||
Modern scientific evaluation of colloidal Au began with Michael Faraday's work in the 1850s, which recognised that the colour observed was due to the size of the Au particles.6 Faraday's rationale in investigating colloidal phenomena followed his interests in the interaction between light and matter. Technological advances in the development of molecular beam techniques have made it possible to study and determine the morphological, physical and chemical properties of AuNPs.7,8
The chronology of the historical events that led to the development of nanotechnology is listed in (Table 1).
| Period | Inventors | Achievement | Ref. |
|---|---|---|---|
| 4th Century A. D | Romans | Lycurgus Cup | 40 |
| 17th Century | Andreas Cassius | “Purple of Cassius” for glass staining | 41 |
| 1857 | Faraday | Scientific evaluation of AuNPs colour change | 6 |
| 1902 | Zsigmondy | Invention of ultra-microscope | 42 |
| 1908 | Mie | Mie theory – Fundamental understanding of the NPs interactions with electromagnetic radiation | 43 |
| 1951–1973 | Frens–Turkevich | Synthesis of AuNP preparations | 44 and 45 |
| 1960 | Feynman | Mention of NPs in a famous lecture in which he propounded that “There's plenty of room at the bottom” meaning at the nanoscale. | 46 |
| 1971 | Faulk and Taylor | First biomedical use in immunochemical staining | 47 |
Colloidal Au has been used since the last century for the study of cerebrospinal fluid by using the Lange reaction in which colloidal gold reaction was used to detect infection of the central nervous system for the early diagnosis and prognosis of obscure neurological conditions. Briefly, a series of spinal fluid samples diluted in aqueous saline solution were treated with a colloidal gold solution and the reaction was allowed to stand for 24 hours to get a final reading (Fig. 2). Depending on the pathological conditions investigated, a certain pattern of colour change of the colloidal auric solution occurs. This application may have been the first recorded practical application of extremely small gold particles for biosensing.9
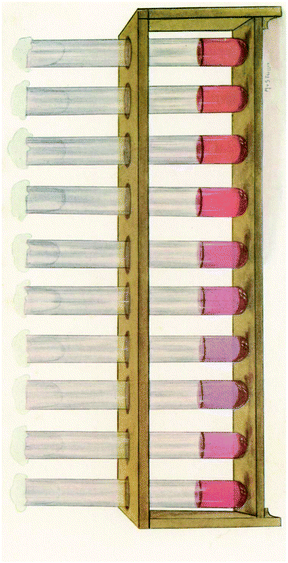 | ||
| Fig. 2 The reaction of Lange gold chloride test on the cerebrospinal fluid in congenital syphilis. This figure has been reproduced from ref. 9. | ||
The use of AuNPs for colorimetric sensing exploits the interparticle distance dependent localised surface plasmon resonance (LSPR) property.10–12 Biological processes and biomolecular-interactions can be monitored because they can be used to control the dispersion and aggregation of the NPs. Depending on the size of the AuNPs, controlled aggregation of the particles can result in colour changes,13–15 from pink to violet to pale blue. This phenomenon has been used in home pregnancy tests and in analyses for specific gene sequences16,17 and for colorimetric detection of a variety of analytes.18–20 It is the anticipated colour changes during aggregation (or dispersion of aggregates) that provide a platform for colorimetric detection using AuNPs as signal transducers.21,22 The aggregated AuNPs not only give different colours from dispersed AuNPs but also different surface enhancement abilities of Raman scattering.23
Typically, NPs biosensors consist of immobilised ligand or a biological substrate which undergo transformation in the presence of an analyte.24 The analyte induces a modification in the substrate composition or conformation, thus imparting a change to the environment of the NPs, resulting in changes in the aggregation status of NPs leading to detectable colour changes.25–27 In order to develop efficient and reliable sensors, it is essential to improve both the recognition and transduction processes through the development of novel materials. Nanomaterials can be fabricated to produce platforms with improved signal to noise (S/N) ratios by miniaturisation of the sensor elements.28
Traditionally, analyte detection and quantification involve recording the absorbance or fluorescence emitted from substrates modified with chromogenic or fluorogenic reporter molecules. Fluorescence resonance energy transfer (FRET) substrates or indirect sensor systems depend on sensing the reactions of unmodified substrates to produce a detectable spectroscopic signal. Although these approaches, in general, are adequate for many applications (commercial assays kits are available), there is a need to improve sensitivity for a variety of other applications.29–33
NPs-based assays have now made it possible to measure minute changes in enzyme activity with high accuracy and precision.34 A variety of NPs-based sensing systems have been developed with varying degrees of success and reliability. These sensing systems are valuable tools for the measurement of enzyme activity in real time and can be used for high-throughput screening of enzyme activity and in drug discovery.35 This review includes a brief introduction to the physical phenomenon (i.e., colours) associated with AuNPs and their aggregation, discussion of the inter-particle forces of AuNPs and factors controlling AuNPs colloidal stability and aggregation, along with general strategies for the stabilisation or aggregation of the colloidal particles. The focus is on the mechanisms and strategies of biosensing assays, the common challenges, current trends in the field, real sample analysis, and exploitation of the anisotropic properties of AuNPs as colorimetric probes.36–38
2. Localised surface plasmon resonance (LSPR) of AuNPs
The fundamental concept underpinning light absorption by metal NPs is the coherent oscillation of the conduction band electrons induced by the interacting electromagnetic field.26 In the case of AuNPs, the oscillations involve the 6s electrons in the conduction band. The boundary and surface effects become more dominant when the particle dimensions become less than 100 nm. The optical properties of small metal NPs are dominated by the collective oscillation of the conduction electrons.39 When the incident photon frequency is resonant with the collective oscillation of the conduction band electrons, the radiation is absorbed to give an absorption band. This phenomenon is known as localized surface plasmon resonance (LSPR).Unique LSPR properties and the colours associated with the aggregation and dispersion of the colloidal particles make AuNPs ideal colorimetric reporters for biological analysis.
It has been found that the electromagnetic coupling of NPs becomes effective when the inter-particle distances are less than 2.5 times their individual diameters.48,49 Aggregation can induce the coupling of the AuNPs plasmon modes, producing a red shift and broadening of the SPR band, associated with the longitudinal resonance in the optical spectrum.8 As a result, aggregation usually changes the original red colour of the AuNPs dispersion into purple or blue.50 The strong enhancement of the localized electric field within the interparticle spacing, broadens and red shifts the SPR spectra. Interparticle plasmon coupling is rather complex and dependent on a number of factors such as aggregate morphology and NPs density.19
Aggregation or dispersion of NPs can be initiated by changes in the external environment. These stimuli can cause the AuNPs to either aggregate or disperse, accompanied by a shift in the absorption spectrum of the AuNPs, and consequently the colour of the colloidal solution changes. The extent of aggregation/re-dispersion is proportional to the absorption peak shift, and thus the extent of signal change is quantifiable offering a direct measure of the effect of the inducing agent e.g., an active enzyme.51 For AuNPs in the size range of 13–300 nm, progressively increased aggregation is characterized by a gradual decrease of the plasmon peak at 520 nm and the appearance of a new peak between ∼600–700 nm with colour change of the resultant solution ranging from red to purple or blue depending on the degree of aggregation (Fig. 3A) whereas the band at 520 nm is the SPR of the spherical AuNPs, however with aggregation the new band appear 600–700 nm appears (see Fig. 3B). In some cases, the LSPR spectrum may be featureless and the solution could turn colourless, showing extreme aggregation, when the plasmon band shifts into the infra-red (IR) region where most visible light is scattered.
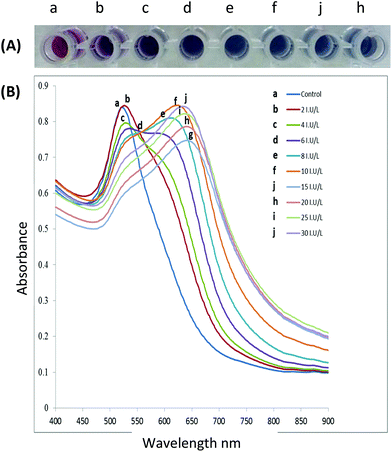 | ||
| Fig. 3 (A) The gradual colour change as functionalised AuNPs are exposed to increasing enzyme activity resulting in increased aggregation, (B) spectral changes of AuNPs solutions after incubation with increasing enzyme activity (incubation of peptide capped AuNPs with the target enzyme for 10 minutes at 37 °C). This figure has been adapted from ref. 100 with permission from Elsevier. | ||
The sensitivity of a colorimetric assay is dependent on the molar extinction coefficients of the NPs plasmon bands, which are in turn determined by the material composition and particle size.21,52 Larger NPs offer higher sensitivity as their surface plasmons have higher molar extinction coefficients. It has been found that the molar extinction coefficients triple in magnitude when the AuNPs size increases from 4 to 35 nm.53–55 The ratio of maximum absorbance at the longer and original wavelength (e.g., A600/A520 for AuNPs) is often used to quantify the extent of aggregation and it is known as an aggregation parameter.56,57
3. Colloidal AuNPs stabilization
A major consideration in the use of AuNPs in a variety of biomedical applications is that the stability of colloidal solutions and changes in aggregation should be controllable and predictable. Stability of the gold solutions can be achieved by the introduction of molecules that interact with AuNPs through bonding and/or electrostatic interactions.58,59For this purpose, various biological probes have been used for the stabilization and/or functionalisation of colloidal AuNPs; these include nucleic acids, enzymes, receptors, lectins, antibodies and superantigens.60–64
3.1 DLVO and non-DLVO forces
The stabilisation phenomenon of Au particles in a well-defined and controlled assembly has been generally explained by the Derjaguin–Landau–Verwey–Overbeck (DLVO) theory,65,66 usually expressed in terms of the balance between attractive van der Waals forces and the repulsive electrostatic interactions, to describe the colloidal stability of aqueous dispersions.66 The DLVO theory accounts for the potential energy variations that occur when two particles approach each other with the resultant net attraction and repulsion forces as a function of interparticle distance.65–67According to this theory, the net interaction potential G between two AuNPs is determined by the van der Waals attraction (GvdW) and electrostatic repulsion (Gelec) values,
| ΔG = ΔGvan der Waals + ΔGelectrostatic | (1) |
Although there is considerable body of research information in support of the DLVO model, it has long been understood that the classical DLVO theory does not completely explain the results across different experimental settings. For example, the prediction of the model is contradicted when two particles or surfaces approach closer than a few nanometres.68 In this instance, non-DLVO forces such as different hydrophobic, steric and solvation forces come into play. As a result, the traditional DLVO theory has thus been extended to include the latter as described in the following equation:
| ΔG = ΔGvan der Waals attraction + ΔGelectrostatic repulsion + ΔGnon-DLVO forces | (2) |
3.2 Stabilization strategies
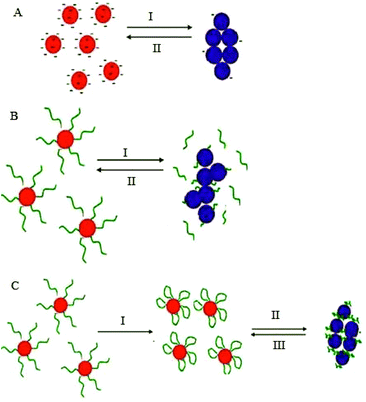
Stabilization of colloidal Au is achieved either electrostatically or sterically by the use of various surface ligands or by a combination of both electrostatic and steric repulsion forces (Fig. 4A–C).58,69–71 In aqueous solutions, zero charged bare AuNPs tend to aggregate because of van der Waals attractive forces. The fundamental importance in the design of AuNPs-based colorimetric sensing platforms is to achieve colloidal stabilization by introducing repulsive forces between the particles in order to prevent colloidal aggregation in the absence of the target analyte. AuNPs stabilization or aggregation depends on the net potential between interparticle attractive and repulsive forces.58,69–73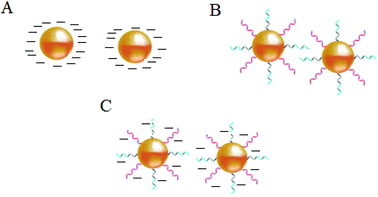 | ||
| Fig. 4 Schematic illustration of NPs colloidal stabilisation: (A) electrostatic, (B) steric, and (C) electrosteric mechanisms. | ||
Further control of AuNPs stability is usually achieved through the introduction of colloidal stabilizers by using chemical coupling methods (e.g., Au–thiol, Au–amine), electrostatic and physical adsorption, etc. Common colloidal stabilizers include charged small molecules, polymers and polyelectrolytes. In the case of electrostatic stabilization (Fig. 4A), each particle carries a “similar” electrical charge, and together with the counter ions in the medium, they form a repulsive electric double layer that stabilizes the colloid against van der Waals attractive forces.74 Since the thickness of the electrical double layer is governed by the bulk ionic strength of the liquid medium, electrostatic stabilization is highly sensitive to salt concentration.72,73
With steric stabilization (Fig. 4B), ligands adsorbed or bound to the particle surfaces form a physical barrier that prevents the particles from crowding. The strength of this stabilization effect is not sensitive to change in ionic strength but determined by molecular size and capping density. Both electrostatic and steric stabilization are known as the electrosteric effect (Fig. 4C). Biomolecules, for example, drugs, peptides, nucleic acids, and proteins that are highly charged and have polymeric properties can be effective stabilizers.69–71
The common approach for the use of AuNPs in colorimetric assays involves the preparation of a stable colloidal solution, whose stability is disrupted upon the addition of the analyte with resultant colour change.75 The mechanisms involved in interactions between the colloidal particles leading to aggregation or dispersion of the particles are given below:
(i) formation of interparticle bond crosslinking (CL), (ii) removal of colloidal stabilisation leading to non-crosslinking (NCL) aggregation, (iii) there is also another colorimetric sensing strategy that does not rely on changes in NPs stability, but on morphology changes induced by a chemical reaction on the surface of the metal NP (AuNPs based plasmonic ELISA sensors).
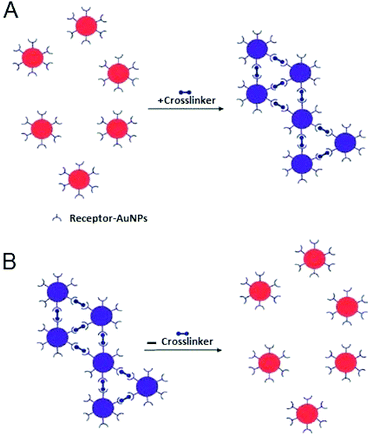
4. Interparticle crosslinking (CL) aggregation
Interparticle CL aggregation is a mechanism in which AuNPs are brought together through the formation of linkages between the individual particles. This occurs either by using crosslinker molecules that have two binding sites linking two AuNPs to each other (i.e., peptides with thiol and/or guanidine anchors) (Fig. 5) or by the direct interaction (without crosslinkers) such as antigen–antibody interactions and DNA hybridisation.76 In either case, specific binding forces (H-bonding, electrostatic interaction, metal–ligand coordination, etc.) associated with biological recognition events (DNA hybridisation, ligand–DNA binding, DNA cleavage, etc.) overcome the interparticle repulsive forces (electrostatic and/or steric repulsion) with resultant typical aggregation.11Table 2 summarises typical applications of AuNPs-based colorimetric biosensing assays that rely on interparticle CL aggregation mechanism. The first platform for the detection of target analytes relies on the presence of crosslinkers that bind to sites on the AuNPs surface (Fig. 5). This platform (Fig. 5A) has also been utilised for the detection of a variety of biological molecules.77–81 In contrast, already crosslinked AuNPs aggregates can also be harnessed to detect analytes that cleave the crosslinkers and this leads to redispersion of the AuNPs aggregates (Fig. 5B). A reverse colour change (blue-to-red) occurs in this case.| Target | Cross linker | Detection limit | Ref. |
|---|---|---|---|
| Kinase | γ-Biotin-ATP | n/a | 89 |
| Thrombin, lethal factor | Two-cysteine-containing peptide | 5 nM | 142 |
| 25 nM | |||
| Factor XIII | ε-(γ-Glutamyl) lysine bonds | 0.01 U mL−1 | 143 |
| Histone methyl transferase Acetyltransferae | IgG antibody | 0.2 nM | 144 |
| 0.5 nM | |||
| Thermolysin | Acetylated Cys residues at both peptides termini | 90 zg mL−1(<380 molecule) | 56 |
| 10 ag mL−1 | |||
| MMP 7 | Peptide substrates with hexahistidine at both ends | 3 nM | 145 |
| Phospho lipase A2 | Synthetic polypeptide JR2KC2 | 700 pM | 146 |
| Thrombin | AuNP-conjugated polyelectrolyte | 10 nM | 147 |
| Horse radish peroxidase | Ferrocene dimer | 1.75 ag mL−1 | 112 |
| Telomerase | Cysteine | 27 cells per μL | 148 |
| β-Lactamase | Dithiol-modified 1,2-bis (2-amino ethoxy)ethane | n/a | 87 |
| LSD1 nuclease | Biotin–avidin | 130 pM | 62 |
| Polyanionic ssDNA | 95 | ||
| ALP | Zn | n/a | 149 |
| Pyrophosphatase | |||
| Apyrase | |||
| Trypsin/MMP-2 | Polypeptide | 5 nM | 99 |
| DPP-IV | Tripeptide | 70 U L−1 | 150 |
| human RecQ4 Helicase | DNA base pair | n/a | 151 |
| Lysine | Cadaverine | 2.5 μM | 152 |
| Decarboxylase | |||
| Histidine | Histamine | 0.9 μM | |
| Decarboxylase |
Liu and his colleagues pioneered this sensing strategy.82–84 Laromaine et al. (2007)56 applied a similar strategy for the detection of protease activity. In their assay, the particles were modified with a short protease substrate terminated with an Fmoc-group that causes the particles to aggregate. When cleaved by the target protease the Fmoc group was removed and the particles were re-dispersed, enabling detection of thermolysin down to 90 zg mL−1.56
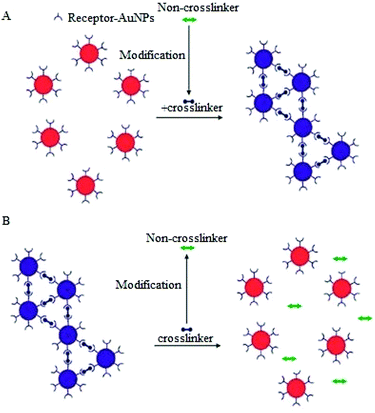
A variation of the CL aggregation method relies on biological transformation of molecules that are not crosslinkers into crosslinkers (or vice versa) or modification of ligand bound AuNPs to induce either aggregation or dispersion of the AuNPs (Fig. 6). Typical examples of the use of this modification of crosslinkers include protease detection assays, in which a protease degrades a peptide linker followed by dispersion of the AuNPs aggregates.85
A similar strategy has also been applied to the detection of kinase, beta-lactamase and phosphatase.86–88 Detection of kinase activity is a classic example of assays that involve the modification of receptor functionalised AuNPs.89 In this approach different forms of functionalised AuNPs are utilized, one is avidin modified AuNPs and the second is the pentapeptide (CALNN) capped AuNPs decorated with γ-biotin-ATP, which when treated with a phosphatase enzyme results in the binding of biotinylated substrate to the avidin modified NPs.89
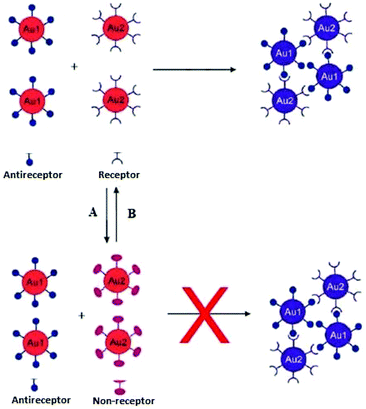
Interparticle CL aggregation can also take place without crosslinkers (Fig. 7), especially when the AuNPs are functionalised with two different types of ligands that are complementary to each other. For example, DNA base-pair recognition was exploited for the coupling of DNA-modified NPs to each other by hybridisation with complementary linker strands. This approach has been used as probes for the detection of endonuclease (DNase I) activity and its inhibitors.90 Double-stranded (ds) DNA strands break down into single-stand (ss) DNA, resulting in the separation of AuNPs followed by a bluish-purple to red colour change. Alternatively, the click chemistry reactions-based covalent bonding, especially Cu(I)-catalysed 1,3-dipolar cycloaddition of azides and alkynes (CuAAC), has been widely used in the design of CL colorimetric nanosensors. In this strategy, AuNPs were typically modified with azide and alkyne groups by the ligand exchange reaction, and thus the existence of Cu(I) would trigger the CuAAC reaction and then crosslink the azide-AuNPs and alkyne-AuNPs to cause their aggregation. This aggregation of NPs leads to the color change of AuNPs from red to blue, and the status of aggregation can be correlated to the concentration of Cu(I).91 (Table 3) displays different sensing platforms that use the CL aggregation mechanism without crosslinkers and some representative target analytes.
In summary, AuNPs-based colorimetric bioassays, which utilize interparticle CL aggregation mechanism, rely on interparticle forces and sometimes chemical interactions. This aggregation mechanism is mainly employed for targets that can be directly or indirectly modified such that they form interparticle bonds. This type of assay is mainly limited to cases in which the analytes and/or ligands have at least two binding sites. AuNPs aggregation by interparticle CL is sometimes a time-consuming process requiring several hours before aggregation and colour change are complete. Aggregation is mainly driven by random collisions between NPs with relatively slow Brownian motion.60 In addition, this interaction is hampered, if the probes have poorly accessible substrate ligands sites, thus resulting in long detection times because of slow enzyme kinetics.82–84
5. Non-crosslinking (NCL) aggregation
To overcome the difficulties associated with the CL based AuNPs aggregation, an alternative approach to control AuNPs aggregation by harnessing the ability of AuNPs to interact without actual bond formation may be desirable. This strategy eliminates the CL requirement for multiple binding sites in order to link the AuNPs thus making it possible to use a facile experimental design. In such a system, AuNPs aggregation is induced by the stronger interaction between ligands and target analytes, accompanied with the release of the ligand or part of it from the surface of NPs, without the formation of interparticle bonds, allowing real-time measurement of enzyme activity in contrast to the relatively slow process induced by inter-particle CL. Sato and his colleagues pioneered the use of this strategy for the detection of polynucleotide polymorphisms.52,60,92 When a target polynucleotide is complementary to the ligand in sequence and chain length, it will hybridise to the DNA onto the AuNPs, this in turn alters NPs resistance to salt-induced aggregation.22 Examples of NCL aggregation mechanism and some representative analytes are listed in Table 4.| Target | Detection limit | Sample type | Ref. |
|---|---|---|---|
| λ exonuclease | n/a | n/a | 94 |
| MMP-7 | 5 nM (0.1 mg mL−1). | n/a | 158 |
| Lysozyme | 80 pg mL−1 | Serum | 130 |
| Thrombin | 0.04 pM | Plasma | 133 |
| Kinase (PKA, PKCα, and MAPKAPK-2) and its inhibitors | n/a | Cell lysate* B16 melanoma | 98 and 159 |
| c-Src tyrosine kinase (gold nanorods) | n/a | n/a | 160 |
| Lipase | 2.8 × 10 mg mL−1 | Fermentation broth of Bacillus subtilis | 161 |
| Trypsin | 1.25 × 10−2 | n/a | 162 |
| MMP-2 | 20 ng mL−1 | ||
| MMP-1, MMP-2 and MMP-7 | n/a | Plasma | 163 |
| ALP | 0.01 U mL−1 | n/a | 164 |
| ALP | 10 units per L | Serum | 165 |
| Thrombin | 10 nM | n/a | 81 |
| Cathepsin B | 5 nM | n/a | 166 |
| Pyrophosphatase | 0.010 U | n/a | 167 |
| Acetyl choline esterase | 1.0 mU mL−1 | CSF | 155 |
| Immunoglobulin A1 protease | n/a | n/a | 168 |
| Choline Esterase (gold nanorods) | 0.018 μU mL−1 | Serum | 169 |
| β-Galactosidase | 9.2 nM | n/a | 170 |
| β-Glucosidase | 22.3 nM | ||
| Penicillin G acylase | 0.007 mg mL−1 | n/a | 171 |
| Telomerase | 1 HeLa cell per μL | n/a | 172 |
| S-Adenosyl homocysteine hydrolase | 100 u L−1 (∼6 nM) | n/a | 173 |
| Caspase-3 | 0.005 mg | Jurkat T lymphocyte cells | 174 |
| Telomerase | 1000 HeLa cells | n/a | 175 |
| Adenosine deaminase | 1.526 U L−1 | n/a | 176 |
| ALP | 16 nM | n/a | 96 |
| DNA adenine methylation methyltransferase | n/a | 0.3 units per mL | 177 |
| Adenosine deaminase | n/a | n/a | 178 |
| Heparinase III | Serum & plasma | n/a | 179 |
| DNAse I | n/a | n/a | 180 |
| Telomerase | 29 HL-60 cells per mL (0.04 mU mL−1) | n/a | 181 |
| Gelatinase | n/a | n/a | 183 |
Liu et al. designed a highly sensitive and selective rhodamine B-functionalised gold nanoparticle (RB-AuNP)-based assay with dual readouts (colorimetric and fluorometric) for monitoring the level of AChE and inhibitor screening. AChE is able to hydrolyze its substrate acetylthiocholine to generate thiocholine, which is comprised of a thiol moiety and a positively charged group. The thiol group shows high affinity toward AuNP to induce a partial replacement of the fluorescent ligands such as rhodamine B from AuNP surfaces. At the same time, the positively charged tail groups interact with the negatively charged-ligand residues on AuNP surfaces, causing aggregation of AuNPs and the change of solution colour.93
5.1. Modification of electrostatic surface stabilisation
Electrostatic stabilisation of AuNPs by small charged moieties, such as citrate-capped AuNPs, undergo aggregation when surface charges are screened by the addition of salt, making it possible for the charged ligands to be displaced by uncharged species or neutralized by oppositely-charged species (Fig. 8a). Lou et al. 2009 found that digested elements of DNA have superior ability over intact DNA to bind to unmodified colloidal AuNPs solution at high salt concentration, and to stabilise them electrostatically.94 Without the nucleases, unmodified AuNPs turn blue due to aggregation at high salt concentration, whereas the particles are stabilised in the presence of nuclease and its substrates and remain red.Recently, using the same principle, Cao and co-workers have developed a sensitive nuclease sensor in which positively charged cysteamine was used to stabilise AuNPs by electrostatic repulsion differentially in the presence and absence of thrombin.95 This platform has been validated for the detection of other enzymes or monitoring enzymatic reactions in which the reactant and product differentially affect the surface charge properties of the citrate-capped AuNPs and thus the stability of the colloidal solution.96–98
5.2. Factors affecting (electro) steric stabilisation
In NCL aggregation systems, as soon as interparticle repulsive forces are insufficient to stabilise the NPs, van der Waals attraction forces dominate with resultant aggregation.96 The loss of colloidal stabilisation, often induced by electrolytes, can be achieved by eliminating surface charges and surface-grafted (charged) polymers, and by target-induced charged ligand conformational changes.102 AuNPs aggregation that results from charged ligand conformational transformation is more complex. Surface charge characteristics e.g., charge intensity, the extent of related counter ions and entropy factors are key considerations in these systems. This necessitates comprehensive understanding of the effects of charged ligand conformations on colloidal stability and state of aggregation.
Compared to the behaviour of interparticle CL aggregation systems, the NCL aggregation mechanism has many desirable characteristics. Primarily, neither an interparticle bio recognition nor target analyte/receptor with multiple binding tags is needed. Furthermore, aggregation induced by the NCL process provides quick detection, often offering relatively short assay times, in the order of a few minutes.103 Once the inter-particle repulsive forces are significantly weakened, the inter-particle attractive van der Waals forces predominate leading to rapid aggregation.60,104,105 High ionic strength solutions are frequently utilised to adjust AuNPs stability and aggregation in NCL systems. However, it is important to determine the appropriate salt type and strength in order to obtain optimum assay performance. It should be noted that in some situations, certain salt types, and/or concentrations are undesirable or incompatible for the required signal development, therefore choosing a salt that has no effect on the biomolecular activity or a compromise between measurement performance and AuNPs stability/aggregation are essential.11
6. Seedless production of AuNPs
Another platform for the development of sensitive and simple optical methods is based on the seedless production of AuNPs. AuNPs are usually synthesized by using a reducer without the addition of NPs seeds and stabilising agent.106 Baron et al. utilised the neurotransmitters: dopamine, L-DOPA, adrenaline and noradrenaline to mediate the generation and growth of AuNPs. The plasmon absorbance of the AuNPs allowed the quantitative colorimetric detection of the neurotransmitters and was also used to detect the activity of tyrosinase.107 The production of AuNPs by the neurotransmitters enabled the measurement of the activity of the tyrosinase enzyme that catalyzes the O2-induced hydroxylation of tyrosine to L-DOPA. The latter product induces the development of AuNPs aggregation and colour change from colourless to red (Fig. 9).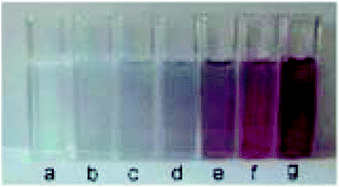 | ||
| Fig. 9 Colour changes of the AuNPs formed in the presence of different concentrations of dopamine (a) 0, (b) 2.5 × 10−6, (c) 5 × 10−6, (d) 8 × 10−6, (e) 1 × 10−5, (f) 1.5 × 10−5, and (g) 2 × 10−5 M. This figure has been adapted from ref. 107. Copyright (2005) American Chemical Society. | ||
7. High resolution multicolour nanoparticles
Gold nanoshells containing a silver alloy have also been used as nanosensors in this type of detection scheme. Gao and coworkers designed a high-resolution colorimetric protocol based on gold/silver core/shell nanorod for visual readout of alkaline phosphatase (ALP) activity by the naked-eye (Fig. 10). The method, based on the enzymatic reaction, assisted silver deposition on gold nanorods to produce colour change, which was dependent on ALP activity. Upon target incubation with the substrate, ascorbic acid 2-phosphate was hydrolysed to produce ascorbic acid, which in turn reduced silver ion to metallic silver coating on the gold nanorod, thereby resulting in a perceptible colour changes.108 Colorimetric AuNPs methods that have been developed for this application are summarized in Table 5.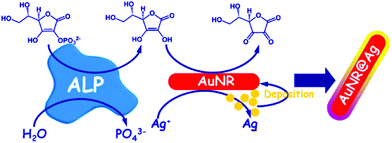 | ||
| Fig. 10 Schematic diagram of the high-resolution colorimetric assay for sensitive visual readout of phosphatase activity based on gold/silver core/shell nanorod. This figure has been adapted from ref. 107. Copyright (2014) American Chemical Society. | ||
8. Colloidal nanoparticle-based plasmonic ELISA sensors and immunoassays
A new trend has been the use of NPs with different morphology and composition as a way to tune the LSPR properties of the nanosensors. This approach is also known as single particle morphology-based methods.109–111 For example, Stevens et al. developed a new LSPR assay in which the signal is inversely proportional to the analyte concentration; that is, a lower concentration of analyte induces a larger LSPR shift. This LSPR assay is based on the growth of Ag crystals in the presence of Au nanostars, whose absorption band is in the NIR region.109 The colorimetric signal of plasmonic nanosensors can be greatly amplified by controlling the amount of gold ions on AuNPs with hydrogen peroxide generated by the enzyme glucose oxidase (Fig. 11). De la Rica and Stevens pioneered the development of this strategy and employed it for the detection of prostate specific antigen (PSA), HIV-1 capsid antigen p24, gp 120 and urease enzyme activity (Table 6).111–113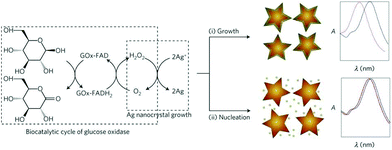 | ||
| Fig. 11 Scheme of the proposed signal-generation mechanism by means of enzyme-guided crystal growth. In the absence of the analyte, the reduction of gold ions with hydrogen peroxide occurs at a fast rate, and quasi-spherical, non-aggregated gold nanostars are blue in colour. This figure has been reproduced from ref. 109 with permission from Nature Publishing Group. | ||
| Target | Nanomaterial | Detection limit | Sample type | Ref. |
|---|---|---|---|---|
| PSA | Au nanostars | 10−18 g mL−1 | n/a | 109 |
| ALP | Gold/Silver Core/Shell Nanorod | 3.3 mU mL−1 | Serum | 108 |
| Tyrosinase | AuNPs seeds | 10 units (100 μg mL−1) | n/a | 107 |
| MMP-7 | Combined peptide–AuNP probe and nitrilotriacetic acid (NTA) modified chip | 0.097 ng ml−1 (4.8 pM) | Cell culture supernatant | 185 |
| Acetylcholine esterase | Gold nanorods | n/a | n/a | 182 |
Liu and coworkers recently reported a wash-free homogeneous plasmonic colorimetric immunoassay combined with the controlled growth of AuNPs in aqueous solutions.114 In this strategy, the inter-particle spacing in the protein–AuNP oligomers was modulated by the controlled growth on the AuNPs aggregate. Upon the gold growth, the particle size was enlarged and the inter-particle distance decreased, thereby inducing a visible optical transition. With this approach, the in vitro detection of cancer biomarkers (e.g. CEA) in serum samples from patients’ needs only 30 s by the naked eye. Because of its rapid and convenient sensing advantages, this resulted colorimetric immunoassay may allow the timely disease diagnosis.
9. Future prospects and challenges in AuNPs-based nanosensor development
Although a variety of colorimetric nanosensors have been developed in laboratory settings, several aspects related to their practical application have not yet been addressed. Transforming these sensors into point of care devices awaits further development. Key factors that have to be addressed include improved resistance to fouling of the sensors and degradation of the organic probes, in addition, coupling of the biomolecular ligands to the NPs must be carefully controlled in order to avoid denaturation and loss of reactivity.115 The production of NPs for biosensors must be reproducible with uniform size and shape. Furthermore, it is essential to be able to re-use the biosensor. Therefore, regeneration protocols have to be developed. Lack of sensitivity in some applications is a concern as perceptible colour change is difficult to measure, which restricts their application to biological sample analysis in comparison to other analytical methods such as fluorescence and chemiluminescence.51 A variety of approaches have been introduced to address some of these limitations.51,116–118Kim and colleagues have recently introduced a new approach to enhance AuNPs platform sensitivity based on centrifugation and silver enhancement. This development resulted in a threefold improvement in detection limit, but the additional steps are time consuming and add to costs.119 Moreover, an approach by Valentini et al. for the colorimetric detection of the cancer-related point mutation was demonstrated using AuNPs combined with signal enhancement on the surface of paramagnetic microparticles. This simple colorimetric assay is sensitive (with a limit of detection in the low picomolar range) and instrument-free. Its naked-eye detection, small sample volumes, and isothermal (PCR-free) amplification, make the assay suitable for point-of-care applications.120 Integration of LSPR with identification techniques (e.g., SERS) is another way allowing improvement of the AuNPs assay detection limits.36,121
In the development of colorimetric sensing assays for routine use in biological samples, it is important to take cognisance of the fact that the interferences caused by cellular components and macromolecules can affect the analytical performance of the sensor. Nevertheless, some reports of measurement of enzyme activity via AuNPs in cell lysates have shown promising results.98,122 In complex samples, such as serum, saliva or cerebrospinal fluid, AuNPs surfaces are exposed to biomolecules, cells, and tissues which can potentially impair the performance of the measurement. The occurrence of nonspecific binding and ligand exchange should be taken into account when developing biosensing applications. In addition, ligand exchange may occur in the presence of high concentrations of other thiolated molecules (e.g., intracellular glutathione).123 These interferences must be carefully studied during method validation. Potential interferences can occur when charge bearing ligands interact non-specifically with other charged biomolecules.
In order to avoid or at least minimise ligand exchange and nonspecific binding, it is essential to use a mixture of ligands, including PEGylated alkanethiols.124 The ethylene glycol group is characterised by being hydrophilic and uncharged at physiological pH imparting high stability and low nonspecific binding to metal NPs.125 The stability imparted to the NPs solution is possibly due to the flexibility of the ethylene glycol groups, which prevents aggregation. Lévy and his co-workers demonstrated that the mixed matrix method provides remarkable resistance against ligand exchange for a caspase 3 protease sensor, while not affecting its function.126 Long-term studies on the impact of these factors on analytical performance and shelf life of nanosensors are needed in order to establish their reliability.127 In addition, issues of AuNPs stability in high ionic strength solutions should also be taken into consideration and addressed. Recently, Cheng et al. have described a method for the detection of Ochratoxin in which paper-based readout is used to improve the sensitivity and accuracy of AuNP aggregation based colorimetric biosensor.128 The Integration of labelled AuNPs in microfluidics testing device is another important concern to pave the road to efficient strategies with enhanced performance and reduced complexity, e.g., aptamers were tethered to AuNPs as part of an lateral flow assay-like dry-reagent assay strip to detect thrombin.129
10. Conclusions
AuNPs enzymatic colorimetric assays are challenging conventional detection systems as they are simple methods that only require a few steps for the detection of target molecules. Furthermore, no sophisticated instrumentation is required, and they could be developed into suitable near patient testing platforms. The key objective in these assays is to control AuNPs aggregation or dispersion that is determined by inter-particle attractive and repulsive forces. A variety of factors including the type of target molecule or enzyme, the recognition ligand and the transduction method can affect the sensitivity of AuNPs-based sensors. Their unique optical properties make them suitable for high throughput and multiplexed detection techniques. The recent successes of multidisciplinary research in nanoscience have resulted in increased investment in the field of colorimetric biosensors for point of care medical devices. Advances in proteomics and genomics research for biomarkers, sensor development, imaging techniques and biomedical science will result in better control and design of nanometre-sized materials. Better understandings of the interactions that control the performance of the materials at this scale are required for optimum design and performance. The development of novel chemical tools for the study of organised assembly and modification of nanostructures will result in advancement in the design of these biosensors. Although there are many enzymes used in drug screening and biomedical applications, not all categories have attracted the same level of attention. The majority of nanosensors have been developed for hydrolysases (e.g., lysozyme)130–132 and proteases (e.g. thrombin)81,133,134 because of their well-known biochemical characteristics and facile binding to synthetic peptide substrate probes.131,135 There are, however, other groups of enzymes (e.g., epimerase, γ-glutamyltransferase, glutamine synthetase) that can be used as potential diagnostic or prognostic markers of specific diseases, yet their activity has infrequently been measured in AuNPs-based enzyme assays. A focus on these neglected enzyme groups (isomerases, transferases, and synthetases) should not only extend the spectrum using AuNPs-based enzyme assays, but also inspire the development of AuNPs-based assays for new targets.136–138Despite the benefits presented by these nano diagnostic systems, there is still a long way to go before the extensive application of colorimetric AuNPs can offer effective diagnostic tests that can be translated into the laboratory or testing near patient setting.139 There are remaining challenges and these include the following: stability issues encountered in high ionic strength samples, such as in serum and urine that induce non-specific AuNPs aggregation and hence trigger associated colour change especially bare AuNPs are deployed. The lack of integration into simple platforms that are capable of eliminating the need for sample pre-treatment, and to prevent possibility of ligand exchange and nonspecific interactions in complex matrices are major drawbacks. These limitations are currently being addressed. Nonetheless, there has been some effort in developing these technologies for point-of-care applications. For example, the Au-nanoprobe system for the characterisation of DNase I activity has been integrated into a lateral flow platform for fast and easy to use of measurements on test strips.140 This field is still in need of innovation, especially in exploiting the remarkable properties of AuNPs for biosensing applications. One such innovation is incorporating these biosensing platform into smartphones or similar portable electronic devices for mobile sensing.141
Abbreviations
| ALP | Alkaline phosphatase |
| DPP-IV | Dipeptidyl peptidase IV |
| LSD1 | Lysine-specific demethylase 1 |
| MMP | Matrix metallo proteinase |
| PSA | Prostate-specific antigen. |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to the Institute of International Education-Scholar Rescue Fund (IIE-SRF) that generously funded Dr Tamim Chalati as a post-doctoral research fellow and the Ministry of Higher Education and Scientific Research in Iraq for providing funding for a Ph.D. scholarship awarded to Dr Hasan Aldewachi.References
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607–609 CrossRef CAS PubMed.
- G. M. Whitesides, Nat. Biotechnol., 2003, 21, 1161–1165 CrossRef CAS PubMed.
- T. Zhu, K. Vasilev, M. Kreiter, S. Mittler and W. Knoll, Langmuir, 2003, 19, 9518–9525 CrossRef CAS.
- B. Harper, F. Sinche, R. Wu, M. Gowrishankar, G. Marquart, M. Mackiewicz and S. Harper, Nanomaterials, 2014, 4, 355–371 CrossRef PubMed.
- R. H. Wu, T. P. Nguyen, G. W. Marquart, T. J. Miesen, T. Mau and M. R. Mackiewicz, Molecules, 2014, 19, 6754–6775 CrossRef CAS PubMed.
- M. Faraday, Philos. Trans. R. Soc. London, 1857, 147, 145–181 CrossRef.
- P. Mulvaney, Langmuir, 1996, 12, 788–800 CrossRef CAS.
- S. Link and M. A. El-sayed, Int. Rev. Phys. Chem., 2000, 409–453 CrossRef CAS.
- C. G. Grulee and A. M. Moody, Am. J. Dis. Child., 1915, 9, 17–27 Search PubMed.
- P. Baptista, E. Pereira, P. Eaton, G. Doria, A. Miranda, I. Gomes, P. Quaresma and R. Franco, Anal. Bioanal. Chem., 2008, 391, 943–950 CrossRef CAS PubMed.
- W. Zhao, M. A. Brook and Y. Li, ChemBioChem, 2008, 9, 2363–2371 CrossRef CAS PubMed.
- C. S. Thaxton, D. G. Georganopoulou and C. A. Mirkin, Clin. Chim. Acta, 2006, 363, 120–126 CrossRef CAS PubMed.
- J. A. Creighton, C. G. Blatchford and M. G. Albrecht, J. Chem. Soc., Faraday Trans. 2, 1979, 75, 790–798 RSC.
- D. Lanqbein, Theory of Van der Walls Interactions, Springer, New York, 1974 Search PubMed.
- K. U. von Raben, R. K. Chang and B. L. Laube, Chem. Phys. Lett., 1981, 79, 465–469 CrossRef CAS.
- A. P. Alivisatos, Sci. Am., 2001, 285, 66–73 CrossRef CAS PubMed.
- R. Rojanathanes, A. Sereemaspun, N. Pimpha, V. Buasorn, P. Ekawong and V. Wiwanitkit, Taiwan J. Obstet. Gynecol., 2008, 47, 296–299 CrossRef PubMed.
- P. K. Jain, K. S. Lee, I. H. El-Sayed and M. a. El-Sayed, J. Phys. Chem. B, 2006, 110, 7238–7248 CrossRef CAS PubMed.
- S. K. Ghosh and T. Pal, Chem. Rev., 2007, 107, 4797–4862 CrossRef CAS PubMed.
- S. Basu, S. K. Ghosh, S. Kundu, S. Panigrahi, S. Praharaj, S. Pande, S. Jana and T. Pal, J. Colloid Interface Sci., 2007, 313, 724–734 CrossRef CAS PubMed.
- R. A. Reynolds, C. A. Mirkin and R. L. Letsinger, J. Am. Chem. Soc., 2000, 122, 3795–3796 CrossRef CAS.
- H. Li and L. Rothberg, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 14036–14039 CrossRef CAS PubMed.
- V. Joseph, A. Matschulat, J. Polte, S. Rolf, F. Emmerling and J. Kneipp, J. Raman Spectrosc., 2011, 42, 1736–1742 CrossRef CAS.
- Y. Chen, Y. Xianyu and X. Jiang, Acc. Chem. Res., 2017, 50, 310–319 CrossRef CAS PubMed.
- N. Dykman and L. Khlebtsov, Chem. Soc. Rev., 2012, 41, 2256–2282 RSC.
- S. Link and M. A. El-Sayed, J. Phys. Chem. B, 1999, 103, 8410–8426 CrossRef CAS.
- S. Link and M. A. El-Sayed, Annu. Rev. Phys. Chem., 2003, 54, 331–366 CrossRef CAS PubMed.
- P. E. Sheehan and L. J. Whitman, Nano Lett., 2005, 5, 803–807 CrossRef CAS PubMed.
- R. Eisenthal and M. Danson, Enzyme Assays: a Practical Approach, University Press, Oxford, 2002 Search PubMed.
- J. L. Reymond, Food Technol. Biotechnol., 2004, 42, 265–269 CAS.
- D. Wahler and J. L. Reymond, Curr. Opin. Biotechnol., 2001, 12, 535–544 CrossRef CAS PubMed.
- S. R. Corrie, J. W. Coffey, J. Islam, K. A. Markey and M. A. F. Kendall, Analyst, 2015, 140, 4350–4364 RSC.
- J.-P. Goddard and J.-L. Reymond, Trends Biotechnol., 2004, 22, 363–370 CrossRef CAS PubMed.
- E. Hutter and D. Maysinger, Trends Pharmacol. Sci., 2013, 34, 497–507 CrossRef CAS PubMed.
- J. E. Ghadiali and M. M. Stevens, Adv. Mater., 2008, 20, 4359–4363 CrossRef CAS.
- J. N. Anker, W. P. Hall, O. Lyandres, N. C. Shah, J. Zhao and R. P. Van Duyne, Nat. Mater., 2008, 7, 8–10 CrossRef PubMed.
- L. Tang and J. Li, ACS Sens., 2017, 2, 857–875 CrossRef CAS PubMed.
- P. D. Howes, R. Chandrawati and M. M. Stevens, Science, 2014, 346, 1247390–1247400 CrossRef PubMed.
- C. J. Murphy, A. M. Gole, S. E. Hunyadi, J. W. Stone, P. N. Sisco, A. Alkilany, B. E. Kinard and P. Hankins, Chem. Commun., 2008, 544–557 RSC.
- I. Freestone, N. Meeks, M. Sax and C. Higgitt, Gold Bull., 2007, 40, 270–277 CrossRef CAS.
- F. Habashi, Eur. Chem. Bull., 2016, 5, 416–419 Search PubMed.
- R. Zsigmondy, J. Am. Chem. Soc., 1909, 31, 951–952 CrossRef.
- G. Mie, Annu. Rev. Phys. Chem., 1908, 25, 329–371 Search PubMed.
- G. Frens, Nat. Phys. Sci., 1973, 241, 20–22 CrossRef CAS.
- J. Turkevich, P. C. Stevenson and J. Hillier, Discuss. Faraday Soc., 1951, 11, 55–75 RSC.
- R. P. Feynman, Eng. Sci., 1960, 23, 22–36 Search PubMed.
- W. Faulk and G. Taylor, Immunochemistry, 1971, 8, 1081–1083 CrossRef CAS PubMed.
- R. Sardar, A. M. Funston, P. Mulvaney and R. W. Murray, Langmuir, 2009, 25, 13840–13851 CrossRef CAS PubMed.
- P. K. Jain and M. A. El-sayed, Chem. Phys. Lett., 2010, 487, 153–164 CrossRef CAS.
- S. Link and M. A. El-Sayed, J. Phys. Chem. B, 1999, 103, 4212–4217 CrossRef CAS.
- D. H. Kang, J. Yu, Y. J. Choi and S. Gunasekaran, Sci. Rep., 2012, 2, 456–461 CrossRef PubMed.
- K. Sato, K. Hosokawa and M. Maeda, Nucleic Acids Res., 2005, 33, e4 CrossRef PubMed.
- J. Yguerabide and E. E. Yguerabide, Anal. Biochem., 1998, 262, 157–176 CrossRef CAS PubMed.
- J. Yguerabide and E. E. Yguerabide, Anal. Biochem., 1998, 262, 137–156 CrossRef CAS PubMed.
- X. Liu, M. Atwater, J. Wang and Q. Huo, Colloids Surf., B, 2007, 58, 3–7 CrossRef CAS PubMed.
- A. Laromaine, L. Koh, M. Murugesan, R. V. Ulijn and M. M. Stevens, J. Am. Chem. Soc., 2007, 129, 4156–4157 CrossRef CAS PubMed.
- P. V. Baptista, M. Koziol-Montewka, J. Paluch-Oles, G. Doria and R. Franco, Clin. Chem., 2006, 52, 1433–1434 CAS.
- J. C. L. De Vasconcelos, M. R. Pereira and J. L. C. Fonseca, J. Dispersion Sci. Technol., 2005, 26, 59–70 CrossRef.
- B. J. Jordan, R. Hong, B. Gider, J. Hill, T. Emrick and V. M. Rotello, Soft Matter, 2006, 2, 558 RSC.
- M. M. K. Sato and K. Hosokawa, J. Am. Chem. Soc., 2003, 125, 8102–8103 CrossRef PubMed.
- D. Aili, K. Enander, L. Baltzer and B. Liedberg, Nano Lett., 2008, 8, 2473–2478 CrossRef CAS PubMed.
- J. Y. Piao and D. S. Chung, Analyst, 2012, 137, 2669–2673 RSC.
- J. Wang, T. Duan, L. Sun, D. Liu and Z. Wang, Anal. Biochem., 2009, 392, 77–82 CrossRef CAS PubMed.
- P. L. Truong, C. Cao, S. Park, M. Kim and S. J. Sim, Lab Chip, 2011, 11, 2591–2597 RSC.
- J. Zhou, J. Ralston, R. Sedev and D. A. Beattie, J. Colloid Interface Sci., 2009, 331, 251–262 CrossRef CAS PubMed.
- T. Kim, C. H. Lee, S. W. Joo and K. Lee, J. Colloid Interface Sci., 2008, 318, 238–243 CrossRef CAS PubMed.
- C. L. De Vasconcelos, M. R. Pereira and J. L. C. Fonseca, Sens. Actuators, B, 2005, 23, 12677–12683 Search PubMed.
- L. Boinovich, Curr. Opin. Colloid Interface Sci., 2010, 15, 297–302 CrossRef CAS.
- L. H. W. Walker and S. B. Grant, Langmuir, 1996, 12, 3151–3156 CrossRef.
- W. R. J. Glomm, J. Dispersion Sci. Technol., 2005, 26, 389–414 CrossRef CAS.
- D. H. Napper, Polymeric Stabilization of Colloidal Dispersions, Academic Press, London, 1983 Search PubMed.
- R. J. Hunter, Foundations of Colloid Science, Oxford University Press, New York, 2004 Search PubMed.
- D. F. Evans and H. Wennerstrçm, the Colloidal Domain: Where Physics, Chemistry, Biology and Technology Meet, Wiley-VCH, Weinheim, 2nd edn, 1999 Search PubMed.
- A. N. Shipway, M. Lahav, R. Gabai and I. Willner, Langmuir, 2000, 16, 8789–8795 CrossRef CAS.
- D. Aili, R. Selegård, L. Baltzer, K. Enander and B. Liedberg, Small, 2009, 5, 2445–2452 CrossRef CAS PubMed.
- P. L. Truong, B. W. Kim and S. J. Sim, Lab Chip, 2012, 12, 1102 RSC.
- Y. F. Huang and H. T. Chang, Anal. Chem., 2007, 79, 4852–4859 CrossRef CAS PubMed.
- K. Quan, J. Huang, X. Yang, Y. Yang, L. Ying, H. Wang and K. Wang, Analyst, 2015, 140, 1004–1007 RSC.
- A. Gole and C. J. Murphy, Langmuir, 2005, 21, 10756–10762 CrossRef CAS PubMed.
- C. Wang, Y. Chen, T. Wang, Z. Ma and Z. Su, Chem. Mater., 2007, 19, 5809–5811 CrossRef CAS.
- V. Pavlov, Y. Xiao, B. Shlyahovsky and I. Willner, J. Am. Chem. Soc., 2004, 126, 11768–11769 CrossRef CAS PubMed.
- J. Liu and Y. Lu, Adv. Mater., 2006, 18, 1667–1671 CrossRef CAS.
- J. Liu, D. Mazumdar and Y. Lu, Angew. Chem., Int. Ed., 2006, 118, 8123–8127 CrossRef.
- J. Liu and Y. Lu, Org. Biomol. Chem., 2006, 4, 3435–3441 CAS.
- C. Guarise, L. Pasquato, V. De Filippis and P. Scrimin, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 3978–3982 CrossRef CAS PubMed.
- Y. L. J. Liu, Anal. Chem., 2004, 76, 627–632 CrossRef PubMed.
- R. Liu, R. Liew, J. Zhou and B. Xing, Angew. Chem., Int. Ed., 2007, 46, 8799–8803 CrossRef CAS PubMed.
- Y. Choi, N. Ho and C. Tung, Angew. Chem., Int. Ed., 2007, 46, 707–709 CrossRef CAS PubMed.
- Z. Wang, R. Levy, D. G. Fernig and M. Brust, J. Am. Chem. Soc., 2006, 128, 2214–2215 CrossRef CAS PubMed.
- X. Xu, M. S. Han and C. a. Mirkin, Angew. Chem., Int. Ed., 2007, 46, 3468–3470 CrossRef CAS PubMed.
- Y. Chen, Y. Xianyu, J. Wu, B. Yin and X. Jiang, Theranostics, 2016, 6, 969 CrossRef CAS PubMed.
- K. Sato, M. Onoguchi, Y. Sato, K. Hosokawa and M. Maeda, Anal. Biochem., 2006, 350, 162–164 CrossRef CAS PubMed.
- D. Liu, W. Chen, J. Wei, X. Li, Z. Wang and X. Jiang, Anal. Chem., 2012, 84, 4185–4191 CrossRef CAS PubMed.
- X. Lou, Y. Xiao, Y. Wang, H. Mao and J. Zhao, ChemBioChem, 2009, 10, 1973–1977 CrossRef CAS PubMed.
- R. Cao, B. Li, Y. Zhang and Z. Zhang, ChemComm, 2011, 47, 12301–12303 RSC.
- W. Zhao, W. Chiuman, J. C. F. Lam, M. a. Brook and Y. Li, Chem. Commun., 2007, 3729–3731 RSC.
- J. Oishi, Y. Asami, T. Mori, J. H. Kang, T. Niidome and Y. Katayama, Biomacromolecules, 2008, 9, 2301–2308 CrossRef CAS PubMed.
- J. Oishi, Y. Asami, T. Mori, J. H. Kang, M. Tanabe, T. Niidome and Y. Katayama, ChemBioChem, 2007, 8, 875–879 CrossRef CAS PubMed.
- G. Chen, Y. Xie, H. Zhang, P. Wang, H. Y. Cheung, M. Yang and H. Sun, RSC Adv., 2014, 4, 6560 RSC.
- H. S. Aldewachi, N. Woodroofe, S. Turega and P. H. E. Gardiner, Talanta, 2017, 169, 13–19 CrossRef CAS PubMed.
- B. Wu, F. Zou, X. Wang, K. Koh, K. Wang and H. Chen, Anal. Methods, 2017, 9, 2153–2158 RSC.
- M. Schollbach, F. Zhang, F. Roosen-Runge, M. W. a. Skoda, R. M. J. Jacobs and F. Schreiber, J. Colloid Interface Sci., 2014, 426, 31–38 CrossRef CAS PubMed.
- W. Zhao, W. Chiuman, M. A. Brook and Y. Li, ChemBioChem, 2007, 8, 727–731 CrossRef CAS PubMed.
- W. Zhao, W. Chiuman, J. C. Lam, S. A. McManus, W. Chen, Y. Cui, R. Pelton, M. A. Brook and Y. Li, J. Am. Chem. Soc., 2008, 130, 3610–3618 CrossRef CAS PubMed.
- Y. L. W. Zhao, W. Chiuman, J. Lam, S. A. McManus, W. Chen, Y. Cui, R. Pelton and M. A. Brook, J. Am. Chem. Soc., 2008, 130, 3610–3618 CrossRef PubMed.
- Z. Wu, H. Zhao, Y. Xue, Q. Cao, J. Yang, Y. He, X. Li and Z. Yuan, Biosens. Bioelectron., 2011, 26, 2574–2578 CrossRef CAS PubMed.
- R. Baron, M. Zayats and I. Willner, Anal. Chem., 2005, 77, 1566–1571 CrossRef CAS PubMed.
- Z. Gao, K. Deng, X. Wang, M. Miró and D. Tang, ACS Appl. Mater. Interfaces, 2014, 6, 18243–18250 CAS.
- L. Rodriguez-Lorenzo, R. de la Rica, R. a. Alvarez-Puebla, L. M. Liz-Marzan and M. M. Stevens, Nat. Mater., 2012, 11, 604–607 CrossRef CAS PubMed.
- D. Liu, J. Yang, H. F. Wang, Z. Wang, X. Huang, Z. Wang, G. Niu, A. R. Hight Walker and X. Chen, Anal. Chem., 2014, 86, 5800–5806 CrossRef CAS PubMed.
- R. de la Rica and M. M. Stevens, Nat. Nanotechnol., 2012, 7, 821–824 CrossRef CAS PubMed.
- R. De Larica, R. M. Fratila, A. Szarpak, J. Huskens and A. H. Velders, Angew. Chem., Int. Ed., 2011, 50, 5704–5707 CrossRef CAS PubMed.
- D. Cecchin, R. de la Rica, R. E. S. Bain, M. W. Finnis, M. M. Stevens and G. Battaglia, Nanoscale, 2014, 6, 9559–9562 RSC.
- H. Liu, P. Rong, H. Jia, J. Yang, B. Dong, Q. Dong, C. Yang, P. Hu, W. Wang, H. Liu and D. Liu, Theranostics, 2016, 6, 54–64 CrossRef CAS PubMed.
- J. N. Anker, W. P. Hall, O. Lyandres, N. C. Shah, J. Zhao and R. P. Van Duyne, Nat. Mater., 2008, 7, 442–453 CrossRef CAS PubMed.
- T. A. Taton, C. A. Mirkin and R. L. Letsinger, Science, 2000, 289, 1757–1760 CrossRef CAS PubMed.
- L. Guo, Y. Xu, A. R. Ferhan, G. Chen and D. H. Kim, J. Am. Chem. Soc., 2013, 135, 12338–12345 CrossRef CAS PubMed.
- J. I. L. Chen, Y. Chen and D. S. Ginger, J. Am. Chem. Soc., 2010, 132, 9600–9601 CrossRef CAS PubMed.
- G. B. Kim, J. O. Lee and Y. Kim, Sens. Actuators, B, 2017, 246, 271–277 CrossRef CAS.
- P. Valentini, R. Fiammengo, S. Sabella, R. Cingolani and P. P. Pompa, ACS Nano, 2013, 7, 5530–5538 CrossRef CAS PubMed.
- L. Yuan, X. Wang, Y. Fang, C. Liu, D. Jiang, X. Wo, W. Wang and H. Chen, Anal. Chem., 2016, 88, 2321–2326 CrossRef CAS PubMed.
- J. H. Kang, Y. Asami, M. Murata, H. Kitazaki, N. Sadanaga, E. Tokunaga, S. Shiotani, S. Okada, Y. Maehara, T. Niidome, M. Hashizume, T. Mori and Y. Katayama, Biosens. Bioelectron., 2010, 25, 1869–1874 CrossRef CAS PubMed.
- R. Hong, G. Han, J. M. Fernandez, B.-j. Kim, N. S. Forbes and V. M. Rotello, J. Am. Chem. Soc., 2006, 128, 1078–1079 CrossRef CAS PubMed.
- L. Duchesne, G. Wells, D. G. Fernig, S. A. Harris and R. Levy, ChemBioChem, 2008, 9, 2127–2134 CrossRef CAS PubMed.
- W. Wang, X. Fan, S. Xu, J. J. Davis and X. Luo, Biosens. Bioelectron., 2015, 71, 51–56 CrossRef CAS PubMed.
- P. Free, C. P. Shaw and R. Lévy, Chem. Commun., 2009, 5009–5011 RSC.
- S. Paterson and R. de la Rica, Analyst, 2015, 140, 3308–3317 RSC.
- S. Cheng, J. Tian, B. Zheng, H. Wei, Y. Ding and J. Zhao, Anal. Methods, 2017, 9, 5407–5413 RSC.
- H. Xu, X. Mao, Q. Zeng, S. Wang, A. Kawde and G. Liu, Anal. Chem., 2009, 81, 669–675 CrossRef CAS PubMed.
- H. Huang, Q. Zhang, J. Luo and Y. Zhao, Anal. Methods, 2012, 4, 3874 RSC.
- X. Wang, Y. Xu, X. Xu, K. Hu, M. Xiang, L. Li, F. Liu and N. Li, Talanta, 2010, 82, 693–697 CrossRef CAS PubMed.
- X. Wang, Y. Xu, Y. Chen, L. Li, F. Liu and N. Li, Anal. Bioanal. Chem., 2011, 400, 2085–2091 CrossRef CAS PubMed.
- C. K. Chen, C. C. Huang and H. T. Chang, Biosens. Bioelectron., 2010, 25, 1922–1927 CrossRef CAS PubMed.
- Z. Chen, Y. Tan, C. Zhang, L. Yin, H. Ma, N. Ye, H. Qiang and Y. Lin, Biosens. Bioelectron., 2014, 56, 46–50 CrossRef CAS PubMed.
- A. J. Welser K, R. Adsley, B. M. Moore and W. C. Chan, Analyst, 2011, 136, 29–41 RSC.
- M. M. Raja, A. Raja, M. M. Imran, A. M. Santha and K. Devasena, Biotechnology, 2011, 10, 51–59 CrossRef CAS.
- M. Pompili, G. Addolorato, G. Pignataro, C. Rossi, C. Zuppi, M. Covino, A. Grieco, G. Gasbarrini and G. L. Rapaccini, J. Gastroenterol. Hepatol., 2003, 18, 288–295 CrossRef CAS PubMed.
- D. J. Gunnersen and B. E. Haley, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 11949–11953 CrossRef CAS.
- T. S. Hauck, S. Giri, Y. Gao and W. C. Chan, Adv. Drug Delivery Rev., 2010, 62, 438–448 CrossRef CAS PubMed.
- Y. Zhang and J. Y. Ying, Anal. Chem., 2015, 87, 10193–10198 CrossRef CAS PubMed.
- S. Y. Liu and F. Lin, Lab Chip, 2016, 16, 943–958 RSC.
- C. Guarise, L. Pasquato and P. Scrimin, Langmuir, 2005, 21, 5537–5541 CrossRef CAS PubMed.
- R. Chandrawati and M. M. Stevens, Chem. Commun., 2014, 50, 5431–5434 RSC.
- Z. Zhen, L. Tang, H. Long and J. Jiang, Anal. Chem., 2012, 84, 3614–3620 CrossRef CAS PubMed.
- J. H. Choi, H. S. Kim, J. W. Choi, J. W. Hong, Y. K. Kim and B. K. Oh, Biosens. Bioelectron., 2013, 49, 415–419 CrossRef CAS PubMed.
- D. Aili, M. Mager, D. Roche and M. M. Stevens, Nano Lett., 2011, 11, 1401–1405 CrossRef CAS PubMed.
- F. Xia, X. Zuo, R. Yang, Y. Xiao, D. Kang, A. Vallee-Belisle, X. Gong, J. D. Yuen, B. B. Y. Hsu, A. J. Heeger and K. W. Plaxco, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 10837–10841 CrossRef CAS PubMed.
- E. Sharon, E. Golub, A. Niazov-Elkan, D. Balogh and I. Willner, Anal. Chem., 2014, 86, 3153–3158 CrossRef CAS PubMed.
- R. Selegard, K. Enander and D. Aili, Nanoscale, 2014, 6, 14204–14212 RSC.
- N. Xia, X. Wang, X. Wang and B. Zhou, Materials, 2016, 9, 857 CrossRef PubMed.
- J. Deka, A. Mojumdar, P. Parisse, S. Onesti and L. Casalis, Sci. Rep., 2017, 7, 44358 CrossRef CAS PubMed.
- M. Nilam, A. Hennig, W. M. Nau and K. Assaf, Anal. Methods, 2017, 9, 2784–2787 RSC.
- A. G. Kanaras, Z. Wang, M. Brust, R. Cosstick and A. D. Bates, Small, 2007, 3, 590–594 CrossRef CAS PubMed.
- G. Wang, X. He, G. Xu, L. Chen, Y. Zhu, X. Zhang and L. Wang, Biosens. Bioelectron., 2013, 43, 125–130 CrossRef CAS PubMed.
- D. Liu, W. Chen, Y. Tian, S. He, W. Zheng, J. Sun, Z. Wang and X. Jiang, Adv. Healthcare Mater., 2012, 1, 90–95 CrossRef CAS PubMed.
- Z. Wu, Z. K. Wu, H. Tang, L. J. Tang and J. H. Jiang, Anal. Chem., 2013, 85, 4376–4383 CrossRef CAS PubMed.
- N. Uehara, M. Fujita and T. Shimizu, Anal. Sci., 2009, 25, 267–273 CrossRef CAS PubMed.
- P. Chen, R. Selegard, D. Aili and B. Liedberg, Nanoscale, 2013, 5, 8973–8976 RSC.
- J. Oishi, X. Han, J. H. Kang, Y. Asami, T. Mori, T. Niidome and Y. Katayama, ChemBioChem, 2007, 8, 875–879 CrossRef CAS PubMed.
- J. Oishi, X. Han, J. H. Kang, Y. Asami, T. Mori, T. Niidome and Y. Katayama, Anal. Biochem., 2008, 373, 161–163 CrossRef CAS PubMed.
- W. Zhang, Y. Tang, J. Liu, L. Jiang, W. Huang, F. W. Huo and D. Tian, J. Agric. Food Chem., 2015, 63, 39–42 CrossRef CAS PubMed.
- Y. C. Chuang, J. C. Li, S. H. Chen, T. Y. Liu, C. H. Kuo, W. T. Huang and C.-S. Lin, Biomaterials, 2010, 31, 6087–6095 CrossRef CAS PubMed.
- Y. C. Chuang, W. T. Huang, P. H. Chiang, M. C. Tang and C. S. Lin, Biosens. Bioelectron., 2012, 32, 24–31 CrossRef CAS PubMed.
- T. Serizawa, Y. Hirai and M. Aizawa, Mol. Biosyst., 2010, 6, 1565–1568 RSC.
- C. M. Li, S. J. Zhen, J. Wang, Y. F. Li and C. Z. Huang, Biosens. Bioelectron., 2013, 43, 366–371 CrossRef CAS PubMed.
- C. J. Kim, D. I. Lee, C. Kim, K. Lee, C. H. Lee and I. S. Ahn, Anal. Chem., 2014, 86, 3825–3833 CrossRef CAS PubMed.
- J. Deng, Q. Jiang, Y. Wang, L. Yang, P. Yu and L. Mao, Anal. Chem., 2013, 85, 9409–9415 CrossRef CAS PubMed.
- A. L. Garner, J. L. Fullagar, J. A. Day, S. M. Cohen and K. D. Janda, J. Am. Chem. Soc., 2013, 135, 10014–10017 CrossRef CAS PubMed.
- L. Lu and Y. Xia, Anal. Chem., 2015, 87, 8584–8591 CrossRef CAS PubMed.
- Z. Zeng, S. Mizukami and K. Kikuchi, Anal. Chem., 2012, 84, 9089–9095 CrossRef CAS PubMed.
- N. R. Tiwari, Adv. Biosci. Biotechnol., 2010, 1, 322–329 CrossRef CAS.
- J. Wang, L. Wu, J. Ren and X. Qu, Small, 2012, 8, 259–264 CrossRef CAS PubMed.
- J. Lin, C. Chang, Z. Wu and W. Tseng, Anal. Chem., 2010, 82, 8775–8779 CrossRef CAS PubMed.
- Y. Pan, M. Guo, Z. Nie, Y. Huang, Y. Peng, A. Liu, M. Qing and S. Yao, Chem. Commun., 2012, 48, 997 RSC.
- J. Wang, L. Wu, J. Ren and X. Qu, Nanoscale, 2014, 6, 1661–1666 RSC.
- L. Zhang, J. Zhao, J. Jiang and R. Yu, Chem. Commun., 2012, 48, 10996–10998 RSC.
- T. Liu, J. Zhao, D. Zhang and G. Li, Anal. Chem., 2010, 82, 229–233 CrossRef CAS PubMed.
- F. Cheng, Y. He, X.-J. Xing, D.-D. Tan, Y. Lin, D.-W. Pang and H.-W. Tang, Analyst, 2015, 140, 1572–1577 RSC.
- Z. Ban, C. J. Bosques and R. Sasisekharan, Org. Biomol. Chem., 2008, 6, 4290–4292 CAS.
- Y. He, F. Cheng, D. Pang and H. Tang, Microchim. Acta, 2016, 184, 101–106 CrossRef.
- L. Zhang, S. Zhang, W. Pan, Q. Liang and X. Song, Biosens. Bioelectron., 2016, 77, 144–148 CrossRef CAS PubMed.
- L. Saa, R. Grinyte, A. Sanchez-iglesias, L. M. Liz-marzan and V. Pavlov, Appl. Mater. Interfaces, 2016, 8, 11139–11146 CrossRef CAS PubMed.
- A. I. Nossier, O. S. Mohammed, R. R. Fakhr El-deen, A. S. Zaghloul and S. Eissa, Talanta, 2016, 161, 511–519 CrossRef CAS PubMed.
- S. Chen, M. Svedendahl, R. P. Van Duyne and M. Käll, Nano Lett., 2011, 11, 1826–1830 CrossRef CAS PubMed.
- W. Cheng, Y. Chen, F. Yan, L. Ding, S. Ding, H. Ju and Y. Yin, Chem. Commun., 2011, 47, 2877–2879 RSC.
- J. M. Bergen, H. A. von Recum, T. T. Goodman, A. P. Massey and S. H. Pun, Macromol. Biosci., 2006, 6, 506–516 CrossRef CAS PubMed.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2018 |