Spectromicroscopic observation of a live single cell in a biocompatible liquid-enclosing graphene system†
Seong Uk
Yu‡
a,
Hwiwon
Lee‡
b,
Woo Jong
Cho
a,
Chulhyun
Kim
a,
Moon Cheol
Kang
c,
Hyun-Joon
Shin
d,
Namdong
Kim
*d,
Sei Kwang
Hahn
 *b and
Kwang S.
Kim
*b and
Kwang S.
Kim
 *a
*a
aDepartment of Chemistry, Ulsan National Institute of Science and Technology (UNIST), Ulsan 689-798, Korea. E-mail: kimks@unist.ac.kr
bDepartment of Materials Science and Engineering, POSTECH, Pohang 790-784, Korea. E-mail: skhanb@postech.ac.kr
cDepartment of Life Sciences, POSTECH, Pohang 790-784, Korea
dPohang Accelerator Laboratory, POSTECH, Pohang 790-784, Korea. E-mail: east@postech.ac.kr
First published on 17th October 2017
Abstract
On-the-spot visualization of biochemical responses of intact live cells is vital for a clear understanding of cell biology. The main obstacles for instant visualization of biochemical responses of living cells arise from the lack of a sophisticated detecting technique which can simultaneously provide chemical analysis tools and the biocompatible wet conditions. Here we introduce scanning transmission X-ray microscopy (STXM) combined with a liquid-enclosing graphene system (LGS), offering biocompatible conditions and improved X-ray absorption spectra to probe the chemical responses of live cells under wet conditions. This set-up enables us to probe a subtle change in absorption spectra depending on the oxidation state of a miniscule amount of oxygen in the functional groups present in each cell and its surroundings containing a minimal amount of liquid water. As an example of in situ biochemical responses of wet cells, chemical responses of a single Colo 205 cell are visualized and analyzed using X-ray absorption near the oxygen K-edge. This spectromicroscopic method using LGS can be applied to diverse biological samples under wet conditions for the analysis of their biochemical responses.
1 Introduction
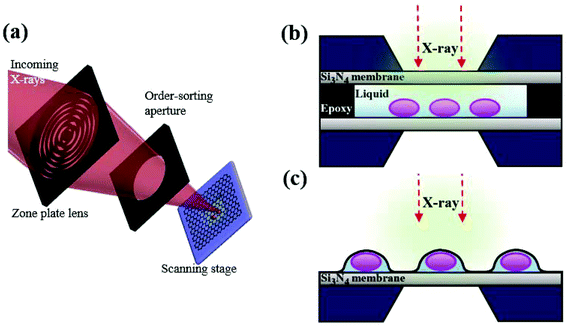
Recently, there have been several trials to image cells under wet conditions at high resolution using electron microscopy (EM) or transmission electron microscopy (TEM).1–3 These trials have overcome the traditional limitation of EM and TEM requiring samples to be fixed and dried, through covering biological samples in a liquid with graphene. The properties of graphene such as ultrathin thickness, flexibility and high thermal–electrical conductivity offer the optimal conditions for bio-samples to be observed through a microscope.4–6 Other two-dimensional (2D) materials, such as MoS2, MoSe2 and WS2 composed of 3 atomic layers, are not ultrathin like graphene except for a few cases such as h-BN sheets, while the h-BN sheet is unuseful as an insulator with respect to electron transfer and thermal energy transfer. Since graphene is composed of only carbon homogeneously, it does not have serious local stress, while other 2D materials composed of two or more elements experience serious scattering-induced local stress and local heating. Additionally, since graphene shows high electro- and thermal conductivity, it is stable without any rupture or burning upon X-ray heating. Therefore, graphene is indeed an optimal material for encapsulating live cells for X-ray imaging. Many trials with graphene have been improved to image live cells, but eventually ended up revealing only the morphological information on cells under wet conditions. To observe the biochemical responses of live cells, a few techniques such as fluorescence microscopy and confocal microscopy have been exploited. Despite successful observation in part, these techniques still have limitations, because antibody-labeled samples are required to be chemically fixed and often the cells need preprocessing with detergents to permeabilize cell membranes, which can cause artifacts.7In contrast, in situ scanning transmission X-ray microscopy (STXM) has a complementary potential to observe the biochemical specificity of live cells without the preprocessing of samples. X-ray absorption spectroscopy (XAS) has the advantage of higher resolution of chemical states with less damage to samples than energy dispersive spectroscopy (EDS) and electron energy loss spectroscopy (EELS). Furthermore, the temperature and vacuum levels of the sample environment for STXM can be easily controlled, offering moderate experimental conditions to biological samples.8 In this work, we combined a liquid-enclosing graphene system (LGS; Fig. 1) with STXM, which leads to successful observation of oxygen K-edge spectra from cancer cells without leakage and noise signals. In contrast to conventional hydrated biological samples prepared in a wet enclosure with silicon nitride (Si3N4) membranes (Fig. 1b), LGS offers highly thin stable liquid conditions without leakage during repetitive monitoring (Fig. S1†). This system provides a biocompatible environment over a few hours to live cells by perfectly confining the liquid without leakage, while the ultrathin sample thickness of LGS makes it possible to observe samples at high resolution, which is not workable for sample preparation methods using silicon nitride. Furthermore, the superior electrical and thermal conductivity of graphene layers may reduce the local charging and localized ray heating on a monitored cell surface,4 helping to preserve a cell's condition intact. However, applying a chemical stimulus to the cells is not easily manageable at present. Thus, the present techniques need to be developed further. Nevertheless, the strength of this technique is the detection of, e.g., reactive oxygen species (ROS) and iron oxide nanoparticles with chemical specificity in live cells without tagging. Given that the STXM technique works for samples under vacuum but suffers from a thick liquid layer, the technique with a graphene layer capsule is of importance for imaging bio-samples requiring a minimal wet condition.
Taking advantage of STXM combined with LGS, we observed the chemical variation of oxygen species from a single wet cell and analyzed absorption spectra near the oxygen K-edge (530–560 eV). Oxygen species, including ROS species such as peroxides, superoxide, singlet oxygen, and hydroxyl radicals, which are natural byproducts of metabolism, play important roles in homeostasis and cell signaling.9 We observed the oxygen K-edge spectra from a single wet cell enclosed by graphene, which can provide scientific evidence for biomolecular analysis. In addition, we also observed a single wet cell treated with iron oxide nanoparticles analyzed using the iron L-edge spectra (700–715 eV). Iron oxide nanoparticles have frequently been used in various experimental applications including immunoassay, cell separation, diagnostics, and drug delivery with suitable surface modifications.10 Observation of nanoparticles within a single wet cell offers opportunities for application in cellular biology, especially the change of the oxygen electronic state, responding to molecules loaded on the surface of nanoparticles. Because STXM analyzes a broad range of photon energies in high resolution, this system can be applied to diverse nanomaterials and biological samples under an intact condition for the analysis of their various biochemical responses.
2 Experimental
Synthesis of graphene using the chemical vapor deposition (CVD) process
Large-area graphene films were prepared by the CVD process using copper foil. Cu foil (99.999%, Alfa Aesar) was annealed at 1000 °C for 30 min under a hydrogen flow (20 standard cubic centimeters per minute (sccm)) to remove the metal oxide layer. While maintaining this temperature, methane gas was introduced at 50 sccm for 20 min at an operating pressure of 300 mTorr. After growth (methane flow), the sample was rapidly cooled to room temperature at a rate of 20 °C min−1 under the same hydrogen gas flow. A graphene bilayer has been grown mostly in our CVD setup (see the ESI† for Raman spectra).Transfer of graphene to a TEM copper grid
CVD-grown graphene films on Cu foil were covered with poly(methyl methacrylate) (PMMA; MW = 350 kg mol−1) and floated on a 0.1 M ammonium persulfate aqueous solution. After etching the Cu foil, graphene with the PMMA support was transferred to the holey carbon film on a 300 mesh copper grid, and then the PMMA support was removed with acetone.Calculation method
All calculations were carried out using the ORCA quantum chemistry program package. The geometries of all molecules were optimized using the B3LYP functional and the def2-TZVP basis set. The oxygen K-edge X-ray absorption spectra were calculated using TDDFT with constrained active space. A modified B3LYP functional, denoted BH0.58LYP, and the aug-cc-pVDZ basis set were employed, and the solvation effect in the aqueous environment was accounted for by a conductor-like screening model (COSMO, dielectric constant: ε = 80.4). Gaussian broadening of width 1.0 eV was applied to each of the absorption peaks obtained.STXM
All data were obtained using a STXM microscope at the 10A beamline at the Pohang Light Source. In STXM, X-rays were focused by a Fresnel zone plate (FZP) onto a sample. The outermost zone width of the FZP was 25 nm and the diffraction-limited size was 30 nm. A photon counting detector (composed of an X-ray-to-visible-light converting screen and a photo-multiplier tube) was placed behind the sample in order to measure the intensity of X-rays transmitted through the sample. The STXM image at a specific photon energy was obtained by scanning the sample perpendicular to the X-ray direction at the focal plane. To take the image, about ∼1 ms data acquisition time per pixel was required, and a few minutes or less than 1 minute was required for one STXM image. For XAS measurements, stacks of STXM images were obtained by changing the incident photon energy, while keeping the focal position at the sample plane, and then spectroscopic data from a specific point or area (in the lateral plane) were collected from the stack images through the aXis2000 software package. The energy resolution of the incident X-rays, defined by the beamline monochromator, was ∼0.1 eV at the photon energy of ∼530 eV. We obtained image stacks with the following scan parameters to minimize possible radiation damage. The stacks are composed of <100 images taken at different photon energies. Each image consists of <100 × 100 pixels in ∼1 ms dwell time per pixel, where the scanning step size is ∼200 nm.Cell culture and in situ LGS sample preparation
Colo 205 cells were cultured at 37 °C and 5% CO2 in RPMI 1640 medium containing 10% FBS and 1% antibiotics. For the sample preparation, the cell growth medium was replaced with a serum free medium. The cell-containing medium was dropped on a silicon nitride membrane. It was covered with a graphene-coated TEM grid. The surface tension and the capillary force induced by the evaporation of liquid between graphene and Si3N4 strengthen the tight encapsulation of cells.ROS release assay
Colo 205 cells were cultured at 37 °C and 5% CO2 in RPMI 1640 medium containing 10% FBS and 1% antibiotics. The cells were removed from the growth medium via centrifugation and resuspended in pre-warmed PBS containing the ROS indicator to provide a final working concentration of 10 μM. After incubation at 37 °C for 30 min, the cells were transferred to the RPMI 1640 medium and incubated in a 12-well plate (1 × 105 cells per well) in triplicate in a hypoxia chamber at 37 °C with or without oxygen supply. The hypoxic condition was created by extracting oxygen from the chamber using a vacuum pump. The level of ROS was detected using a fluorescence microplate reader at 485/538 nm.Confocal microscopy
Colo 205 cells were stained with the ROS indicator and DAPI for 30 min. Then, the cells were covered with graphene membranes. After incubation at 37 °C, Colo 205 cells were observed using a confocal microscope (Olympus FA1000) at 405 nm and 488 nm.Cell viability test
Colo 205 cells at a density of 2 × 104 cells per well were seeded on 96-well plates and cultured in 100 μL of serum free RPMI 1640 medium in septuplicate at 37 °C for 0, 1, 2, 4, and 12 h without oxygen supply. Then, 10 μL of ez-cytox reagent for measuring the cell viability was added for 30 min and the optical density was measured using a microplate reader at 450 nm.3 Results and discussion
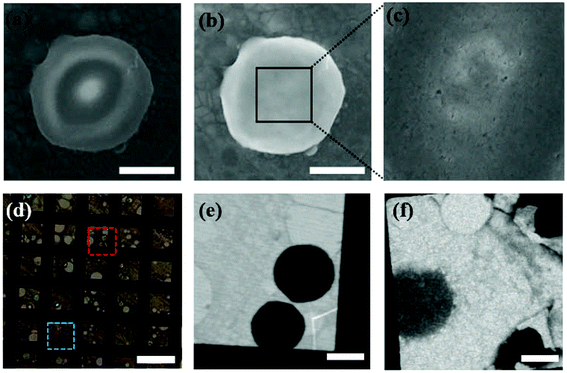
Fig. 1 shows a schematic illustration of our experimental set-up. We have covered graphene layers on Colo 205 cancer cells, which are suspension cell lines and have round cell shapes in solution. First, a graphene bilayer is synthesized by a standard CVD method5,6 and transferred onto a TEM copper grid. The flexibility and thickness of the covering material are critical for covering the biomaterials and reducing background effects. Although single layer CVD graphene would be the optimum for bio-samples due to its flexibility and ultrathin character, it intrinsically has some defects which can cause leakage of the liquid. Therefore, we have prepared a graphene bilayer which still shows flexibility and ultrathin character (∼0.7 nm). Then, Colo 205 cells pre-cultured in RPMI 1640 medium are simply dropped onto the silicon nitride substrate and covered with the graphene layer transferred to the copper grid. The evaporation of liquid between graphene and the silicon nitride substrate strengthens the tight encapsulation of cells. More details of the synthesis of graphene, cell culture, and LGS preparation are described in the Experimental section. To optimize the encapsulation of cells under an intact condition, we test three different types of copper grids: holey, quantifoil holey, and lacey carbon films on 300 mesh copper grids. When covered with holey and quantifoil holey carbon films, most of the cells are collapsed or crushed (see Fig. S2†). This indicates that the cell is very weak to the external stress. However, in the case of lacey carbon films, cells are intact without morphological changes. The higher graphene portion of the lacey carbon system offers higher flexibility, leading to the intact cell encapsulation. The atom-thick graphene layer offers higher flexibility suitable for the present experiment set-up requiring low-level background as well as stable wet conditions.We first observed the intact round-shaped cell morphology of Colo 205 cells encapsulated within the graphene layer through SEM (Hitachi, S4800) (Fig. 2a and b). When the voltage of SEM is elevated to 5–15 kV, graphene is damaged by an e-beam (5–10 μA, a pixel size of ∼20 nm), which induces leakage of the confined liquid (Fig. 2c). Note that the vacuum level of the SEM chamber is broken instantly when the cell gets damaged. The results clearly reveal that the cells under wet conditions are well encapsulated. Furthermore, optical microscopy observation of trypan blue treated cells in LGS also proves the wet conditions of the cells by different refractive indices arising from dried and dead cells stained with blue color (Fig. 2d). The STXM images in Fig. 2e and f corresponding to the wet and dried cells, respectively, show a distinctive contrast, as already verified by trypan blue treatment of cells in LGS. Meanwhile, a cell viability test in the hypoxic condition, which imitates the oxygen deficient condition in LGS, implies that cells in LGS can survive at least for a few hours before irradiation (Fig. S3†). Colo 205 cells at a density of 2 × 104 cells per well are seeded on 96-well plates and cultured in 100 μL of serum-free RPMI 1640 medium in septuplicate at 37 °C for 0, 1, 2, 4, and 12 h without oxygen supply. Then, 10 μL of ez-cytox reagent for measuring the cell viability is added for 30 min and the optical density is measured with a microplate reader at 450 nm.
To show that LGS enhances the absorption contrast by reducing the background effect, we have prepared a liquid system containing Colo 205 cells using a typical method for the microscope technique, which attaches two silicon nitride membranes with epoxy resin as in Fig. 1b. In the silicon nitride membrane system, the image at 520 eV for Colo 205 cells shows round-shaped cells (Fig. 3a) and that for crystalline graphene layers differentiates between monolayer and bilayer (Fig. 3c). However, their images at 535 eV (Fig. 3b and d) show that neither the cell nor the step edge of exfoliated graphene in the thick liquid medium is recognizable due to strong absorption of water at around 540 eV. The critical problem of these systems under wet conditions is that the region around the photon energies corresponding to strong absorption by water is not resolvable. Additionally, the cell in the silicon nitride system experiences severe morphological damage caused by irradiation. We find that the maintenance of the wet conditions of LGS is much more stable than that of silicon nitride since the high thermal conductivity of graphene layers hinders the evaporation of moisture and local burning/damage to cells.
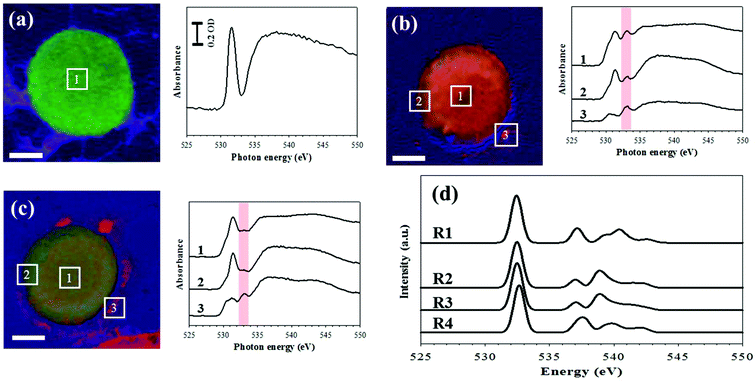
Thus, using the LGS system, we carried out STXM at the oxygen K-edge ranging from 520 to 560 eV. After incubation of Colo 205 cells in LGS at 37 °C for 0 h (cell A, Fig. 4a), 1 h (cell B, Fig. 4b) and 2 h (cell C, Fig. 4c), these three samples are mounted in a STXM chamber. First, the chamber is pumped down to ∼10−3 mbar carefully and then filled with 100 mbar helium. Incoming X-rays are focused on the sample surface by a zone plate lens. We count the transmitted photons through the sample with an energy resolution better than ∼0.1 eV. Thus, the chemical components can be resolved with such a spatial resolution inside the enclosure. Based on the results of XAS, the chemical composition maps are obtained by a linear combination of the three reference spectra (colored green for the pristine region of the cell at 531.5 eV, red for a new peak at 533 eV, and blue for the oxygen background at 538 eV), as shown in Fig. S4.† Least-squares fitting is performed by the linear combination of three reference X-ray absorption spectra using the aXis2000 software package. Without incubation (Fig. 4a), the cell is stained with green color and the XAS obtained from the square area in the image shows a sharp peak at 531.5 eV and a broad peak at around 540 eV. This feature of the O K-edge spectra stems mainly from the protein in the cell as shown in the previous report.11 The pre-edge peak at 531.5 eV is mainly due to the O 1s (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) → π* C
O) → π* C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O transitions of functional groups,12 while the broad peak whose intensity is saturated over the detection range (Fig. S5†) mostly arises from the O 1s (O–H) → σ* O–C and O 1s (C–O) → σ* O–C transitions.13,14 After 1 h incubation in Fig. 4b, the cell image visualizes the secretion of 533 eV in red color and the morphological change on the edge of the cell. We also found that the results of XAS, which correspond to the same numbered square area in the image, show a new peak at 533 eV corresponding to the O 1s-to-σ* (O–O) transition.15,16 However, after 2 h incubation in Fig. 4c, the intensity of red color decreases and the peak at 533 eV is smaller by 70% than that observed in the cell of Fig. 4b.
O transitions of functional groups,12 while the broad peak whose intensity is saturated over the detection range (Fig. S5†) mostly arises from the O 1s (O–H) → σ* O–C and O 1s (C–O) → σ* O–C transitions.13,14 After 1 h incubation in Fig. 4b, the cell image visualizes the secretion of 533 eV in red color and the morphological change on the edge of the cell. We also found that the results of XAS, which correspond to the same numbered square area in the image, show a new peak at 533 eV corresponding to the O 1s-to-σ* (O–O) transition.15,16 However, after 2 h incubation in Fig. 4c, the intensity of red color decreases and the peak at 533 eV is smaller by 70% than that observed in the cell of Fig. 4b.
For a better understanding of the experimental result, we have calculated XAS of the oxygen K-edge, hydrogen peroxide, and other possible oxygen containing molecules using time-dependent density functional theory (TDDFT).17–20 The lowest energy peak in Fig. 4d corresponds to the transition from oxygen 1s to the lowest unoccupied molecular orbital (LUMO). The peak at 533 eV for the 1s → σ* transition corresponds to hydrogen peroxide and peroxide-containing molecules (Fig. 4d). According to this calculation, the increase of peak intensity at 533 eV is closely related with ROS generation and the ROS effect on cells, in good agreement with the previous experimental report which identified the hydrogen peroxide peak at 533 eV.15,16 However, doubts could be cast on the O–O functional group concentration because it is too high to be produced in a cell. Stable chemical species are easily accumulated in the cell. The ROS released by Colo 205 cells may react with lipid or cell membranes, which produces O–O functional groups such as lipid peroxide or cyclic peroxide. It is well known that lipid peroxide is much more stable than typical ROS (H2O2). Thus, the concentration of molecules containing O–O functional groups can be presumably increased by the lipid or cyclic peroxide accumulation.
A LGS system perfectly isolates cells from the environment, which causes the hypoxic condition by blocking oxygen supply. For cellular survival, oxygen is an essential factor and the hypoxic condition causes extreme stress to the cells, which triggers cell death.21 The cell secretion is considered as the autophagosome or autolysosome produced during cell death. It is also well known that cells release ROS during a cell death process.22 The observed oxygen K-edge absorption images are likely to be related to ROS production or the structural change of cells such as lipid peroxidation of the cell membrane. Because the cells are completely blocked by the graphene layer without oxygen supply from the external environment, the ROS production of the cells decreases with increasing time. This result implies that most of the ROS is time-dependently decomposed. This observation would be explained by the result of the ROS release profile (Fig. S6†), in which the release of ROS is dramatically increased within 1 h and almost completely stops after another 1 h in the condition that oxygen is not supplied. The in situ STXM observation using LGS thus resolves the limitation of conventional methods to observe hydrated cells, and so it could be applied to other studies of biochemical responses.
In a biological environment, ROS which is normally generated as a natural byproduct of oxygen metabolism affects cell signaling and homeostasis. The generation of ROS, as a biochemical response of live cells, is dramatically increased by environmental stress such as a hypoxic condition or a radiation stimulus. Once the level of ROS is increased excessively, the detrimental oxidative stress causes senescence and death of cancer cells, damaging cell structures such as the membrane and DNA,23,24 which means chemical changes in oxygen containing functional groups. To experimentally confirm the chemical changes in oxygen containing functional groups, we have conducted a ROS release assay in a hypoxic condition and observed ROS release from cells in LGS using a confocal microscope (Fig. S6†). These results show highly increased ROS release from cells in LGS in a short time period. Otherwise, if the new peak at 533 eV near the oxygen pre-edge was not directly due to ROS itself, the change in the absorption spectra is regarded as the secondary effect of ROS release from the cell. ROS itself and the radiation-damaged macromolecules including cell organelles and lipid bilayer membrane are supposed to change the XAS of oxygen, as in Fig. 4.
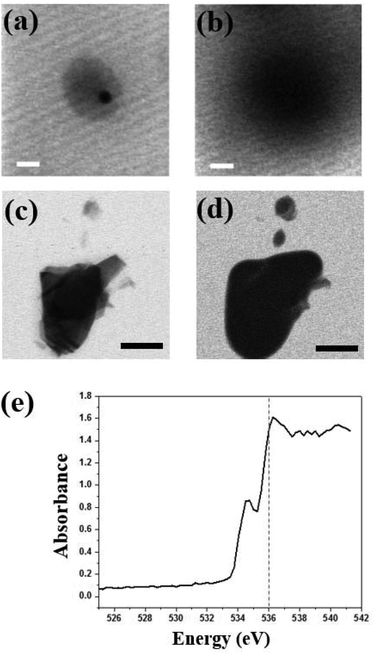
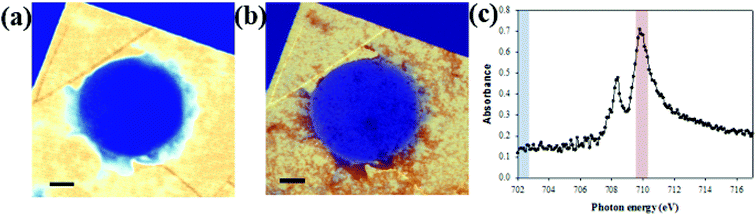
STXM can be used not only for oxygen species but also to observe iron elements within the cells. Colo 205 cells treated with ∼100 nm sized iron oxide nanoparticles (Sigma Aldrich) in LGS are observed at 702 eV and 710 eV (Fig. 5a and b). Fig. 5c shows the iron L-edge spectra (702–717 eV) of iron oxide with the strong absorption edge at 710 eV but negligible absorption at 702 eV. Iron oxide nanoparticles are seen selectively and clearly at 710 eV both inside and outside the cell in high resolution (Fig. 5b), but not at 702 eV (Fig. 5a). This result offers not only a high resolution image but also quantitative information on iron oxide nanoparticles penetrating a single wet cell. Because iron oxide nanoparticles have been widely used for various experimental applications including diagnostics and drug delivery with suitable surface modifications, it is important to observe the cellular uptake of iron oxide nanoparticles. In addition, the responses of a single wet cell to various biological or chemical samples, especially the changes of the oxygen electronic state, can be monitored using this system. Furthermore, since STXM analyzes a broad range of photon energies in high resolution, this system can be applied to diverse nanomaterials and biological samples in intact conditions for spatial analysis of changes in chemical composition arising from various biochemical responses.
4 Conclusions
STXM combined with LGS clearly visualizes important biochemical responses of a live single cell under biocompatible wet conditions. Without preprocessing of cells, the variation of the oxygen K-edge spectra from a single cancer cell in wet conditions under graphene layers was successfully monitored without signal interference by oxygen species in a small amount of water. By covering a graphene layer on the droplet of medium containing cells, we were able to confine the cells in LGS without liquid evaporation. In particular, owing to the minimal amount of water in LGS we were able to observe iron oxide nanoparticles within a single wet cell by tuning the photon energy resonantly, while the previous STXM using silicon nitride membranes showed some limitations due to the interference of strong oxygen signals from water. Because iron oxide nanoparticles can be modified with various biological agents including antibodies, this system can be applied to monitor various biochemical responses. In addition, this novel in situ X-ray microscopy imaging method capable of observing wet systems can be utilized to study real time changes in functional groups in diverse material systems such as catalytic reactions, polymeric reactions, and nanomaterial processing under wet conditions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank the Pohang Accelerator Laboratory. This research was supported by the National Research Foundation, Korea (National Honor Scientist Program, 2010-0020414; Bio & Medical Technology Development Program, 2012 M3A9C6049791; NRF-2014R1A1A2054592; NRF-2015R1A5A1009962) and KISTI (KSC-2016-C3-0074).References
- J. Park, H. Park, P. Ercius, A. F. Pegoraro, C. Xu, J. W. Kim, S. H. Han and D. A. Weitz, Nano Lett., 2015, 15, 4737–4744 CrossRef CAS PubMed.
- J. M. Yuk, J. Park, P. Ercius, K. Kim, D. J. Hellebusch, M. F. Crommie, J. Y. Lee, A. Zettl and A. P. Alivisatos, Science, 2012, 336, 61–64 CrossRef CAS PubMed.
- D. Shin, J. B. Park, Y.-J. Kim, S. J. Kim, J. H. Kang, B. Lee, S.-P. Cho, B. H. Hong and K. S. Novoselov, Nat. Commun., 2015, 6, 6068 CrossRef PubMed.
- A. A. Balandin, Nat. Mater., 2011, 10, 569–581 CrossRef CAS PubMed.
- S. Bae, H. Kim, Y. Lee, X. Xu, J.-S. Park, Y. Zheng, J. Balakrishnan, T. Lei, H. R. Kim, Y. I. Song, Y. J. Kim, K. S. Kim, B. Ozyilmaz, J. H. Ahn, B. H. Hong and S. Iijima, Nat. Nanotechnol., 2010, 5, 574–578 CrossRef CAS PubMed.
- K. S. Kim, Y. Zhao, H. Jang, S. Y. Lee, J. M. Kim, K. S. Kim, J.-H. Ahn, P. Kim, J.-Y. Choi and B. H. Hong, Nature, 2009, 457, 706–710 CrossRef CAS PubMed.
- S. Wilson and A. Bacic, Nat. Protoc., 2012, 7, 1716–1727 CrossRef CAS PubMed.
- N. Ohmer, B. Fenk, D. Samuelis, C.-C. Chen, J. Maier, M. Weigand, E. Goering and G. Schütz, Nat. Commun., 2015, 6, 6045 CrossRef CAS PubMed.
- B. D'Autreaux and M. B. Toledano, Nat. Rev. Mol. Cell Biol., 2007, 8, 813–824 CrossRef PubMed.
- D. Ling, N. Lee and T. Hyeon, Acc. Chem. Res., 2015, 48, 1276–1285 CrossRef CAS PubMed.
- S. Mitsunobu, M. Zhu, Y. Taeichi, T. Ohigashi, H. Suga, H. Makita, M. Sakata, K. Ono, K. Mase and Y. Takahashi, Chem. Lett., 2015, 44, 91–93 CrossRef.
- S. C. Ray, H. M. Tsai, J. W. Chiou, B. Bose, J. C. Jan, K. Krishna, W. F. Pong, D. Dasgupta and M. H. Tsai, J. Phys.: Condens. Matter, 2004, 16, 5713 CrossRef CAS.
- L. Å. Näslund, J. Lüning, Y. Ufuktepe, H. Ogasawara, P. Wernet, U. Bergmann, L. G. M. Pettersson and A. Nilsson, Phys. Chem. B, 2005, 109, 13835–13839 CrossRef PubMed.
- J. T. Francis and A. P. Hitchcock, J. Phys. Chem., 1992, 96, 6598–6610 CrossRef CAS.
- E. Rühl and A. P. Hitchcock, Chem. Phys., 1991, 154, 323–329 CrossRef.
- S. Thürmer, I. Unger, P. Slavíček and B. Winter, J. Phys. Chem. C, 2013, 117, 22268–22275 Search PubMed.
- F. Neese, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2012, 2, 73–78 CrossRef CAS.
- G. S. DeBeer, T. Petrenko and F. Neese, J. Phys. Chem. A, 2008, 112, 12936–12943 CrossRef PubMed.
- N. A. Besley, M. J. G. Peach and D. J. Tozer, Phys. Chem. Chem. Phys., 2009, 11, 10350–10358 RSC.
- S. Sinnecker, A. Rajendran, A. Klamt, M. Diedenhofen and F. Neese, Phys. Chem. A, 2006, 110, 2235–2245 CrossRef CAS PubMed.
- Y. Liu and B. Levine, Cell Death Differ., 2015, 22, 367–376 CrossRef CAS PubMed.
- D. D. Newmeyer and S. Ferguson-Miller, Cell, 2003, 112, 481–490 CrossRef CAS PubMed.
- R. A. Cairns, I. S. Harris and T. W. Mak, Nat. Rev. Cancer, 2011, 11, 85–95 CrossRef CAS PubMed.
- S. S. Sabharwal and P. T. Schumacker, Nat. Rev. Cancer, 2014, 14, 709–721 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7nr05223e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |