Detection of methylation on dsDNA using nanopores in a MoS2 membrane†
Abstract
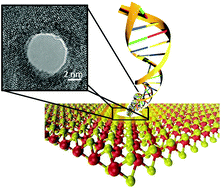
Methylation at the 5-carbon position of the cytosine nucleotide base in DNA has been shown to be a reliable diagnostic biomarker for carcinogenesis. Early detection of methylation and intervention could drastically increase the effectiveness of therapy and reduce the cancer mortality rate. Current methods for detecting methylation involve bisulfite genomic sequencing, which are cumbersome and demand a large sample size of bodily fluids to yield accurate results. Hence, more efficient and cost effective methods are desired. Based on our previous work, we present a novel nanopore-based assay using a nanopore in a MoS2 membrane, and the methyl-binding protein (MBP), MBD1x, to detect methylation on dsDNA. We show that the dsDNA translocation was effectively slowed down using an asymmetric concentration of buffer and explore the possibility of profiling the position of methylcytosines on the DNA strands as they translocate through the 2D membrane. Our findings advance us one step closer towards the possible use of nanopore sensing technology in medical applications such as cancer detection.


 Please wait while we load your content...
Please wait while we load your content...