Open Access Article
Open Access ArticleViral nanoparticles decorated with novel EGFL7 ligands enable intravital imaging of tumor neovasculature†
Choi-Fong
Cho
ab,
Lihai
Yu
c,
Tienabe K.
Nsiama
c,
Alisha N.
Kadam
a,
Arun
Raturi
a,
Sourabh
Shukla
d,
Giulio A.
Amadei
c,
Nicole F.
Steinmetz
defg,
Leonard G.
Luyt
 dh and
John D.
Lewis
dh and
John D.
Lewis
 *a
*a
aTranslational Prostate Cancer Research Group, University of Alberta, 5-142C Katz Group Building, 114th St and 87th Ave, Edmonton, AB T6G 2E1, Canada. E-mail: jdlewis@ualberta.ca
bHarvey Cushing Neuro-Oncology Laboratories, Brigham and Women's Hospital, Harvard Medical School, Building of Transformative Medicine, 60 Fenwood Road, Boston, MA 02115, USA
cLondon Regional Cancer Program, 800 Commissioners Rd. E., London, ON, N6A 4L6 Canada
dDepartment of Biomedical Engineering, Case Western Reserve University, 10900 Euclid Ave., Cleveland, OH 44106, USA
eDepartment of Radiology, Case Western Reserve University, 10900 Euclid Ave., Cleveland, OH 44106, USA
fDepartment of Materials Science and Engineering, Case Western Reserve University, 10900 Euclid Ave., Cleveland, OH 44106, USA
gDepartment of Macromolecular Science and Engineering, Case Western Reserve University, 10900 Euclid Ave., Cleveland, OH 44106, USA
hDepartments of Chemistry, Oncology, Medical Imaging, The University of Western Ontario, London, ON, N6A 5C1 Canada
First published on 1st August 2017
Abstract
Angiogenesis is a dynamic process fundamental to the development of solid tumors. Epidermal growth factor-like domain 7 (EGFL7) is a protein whose expression is restricted to endothelial cells undergoing active remodeling that has emerged as a key mediator of this process. EGFL7 expression is associated with poor outcome in several cancers, making it a promising target for imaging or therapeutic strategies. Here, EGFL7 is explored as a molecular target for active neovascularization. Using a combinatorial peptide screening approach, we describe the discovery and characterization of a novel high affinity EGFL7-binding peptide, E7p72, that specifically targets human endothelial cells. Viral nanoparticles decorated with E7p72 peptides specifically target tumor-associated neovasculature with high specificity as assessed by intravital imaging. This work highlights the value of EGFL7 as a target for angiogenic vessels and opens the door for novel targeted therapeutic approaches.
1. Introduction
Deregulation of angiogenesis plays a major role in a number of human diseases, most notably cancer. Tumor neovasculature differs from normal mature vasculature due mainly to the rapid and uncontrolled growth typical of cancer development. Indeed, tumors with a high proportion of angiogenic vasculature have significantly poorer prognosis.1 Angiogenesis is now a well-established clinical therapeutic target, and anti-angiogenic therapies targeting the VEGF pathway have been approved for the treatment of solid tumors. For example, the anti-VEGF antibody bevacizumab is approved for use alone for glioblastoma or used in combination with other drugs to treat metastatic colorectal cancer, some non-small cell lung cancers, and metastatic renal cell cancer. Although angiogenesis inhibitors are amongst the most promising anticancer drug candidates, the vast majority of inhibitors in clinical development are focused on the VEGF pathway. Overall, the clinical efficacy of VEGF pathway inhibitors has not lived up to expectations,2–5 so it is imperative that alternative targets for angiogenesis be identified.The de novo generation of new blood vessels is a complex and tightly regulated process that is associated with a variety of specific angiogenic and growth factors. One of these, the 30 kDa epidermal growth factor-like domain 7 (EGFL7)6 is predominantly expressed in remodelling endothelium,7 and has been implicated in the regulation of key angiogenic pathways.8,9 EGFL7 is found principally in the endothelium during vascular development, and after vascular injury or during pregnancy,10–14 but is absent in quiescent endothelium in adults. EGFL7 is required for proper vascular structure formation,11,15 and over-expression of EGFL7 in mice leads to abnormal vasculature remodeling.14 Indeed, abnormal microvessel morphology is characteristic of tumor angiogenesis, and is closely linked with cancer-specific mortality.16 Elevated expression of EGFL7 has also been detected in a variety of solid tumors,17–19 and is associated with high tumor grade and poor prognosis in malignant glioma,17 hepatocellular carcinoma,20 breast cancer,20,21 ovarian cancer,22 pancreatic cancer23 and laryngeal squamous cell carcinoma.24These unique features make EGFL7 an attractive molecular target for imaging or targeted therapies.
EGFL7 has been investigated previously as a therapeutic target in cancer. Preclinical studies have shown that treatment using an antibody against EGFL7 in combination with anti-VEGF therapy has led to an improvement in efficacy and survival.25 While antibodies are widely utilized as cancer-targeting agents, peptides provide an attractive alternative because they offer many advantages including better pharmacokinetics and pharmacodynamics, improved tissue penetration and tumor biodistribution, as well as increased serum stability through structural modifications (i.e. incorporation of unnatural amino acids or cyclization).26 As targeting ligands for nanomedicines, peptides are particularly compelling due to their scalable synthesis, and straightforward incorporation into nanoparticle starting materials to achieve better uniformity and higher ligand density.27 We report here the discovery and characterization of a novel highly specific EGFL7-binding peptide, E7p72, which binds endothelial cells in an EGFL7 dependent manner. Decoration of near-infrared viral nanoparticles with E7p72 confers molecular specificity to blood vessels expressing EGFL7, allowing real-time intravital imaging of tumor neovasculature in vivo.
2. Methodology
2.1. OBOC library screening
1 g of OBOC peptide library beads were washed extensively before screening using 70% ethanol, distilled water and binding buffer (50 mM Na2HPO4, 150 mM NaCl, 2 mM CaCl2, 0.2 M L-arginine, pH 7.4). First, a library enrichment step to remove potential false positive beads was performed by mixing the OBOC library with magnetic beads coated with the anti-GST antibody and GST protein complex for 1 h at 37 °C with constant shaking at 200 rpm. A strong neodymium magnet (K&J Magnetics, PA, USA) was placed at the side of the vial for several minutes, and beads that did not respond to the magnet (enriched library) were transferred into another vial using a micropipette. The enriched library was washed five times with binding buffer. To screen for EGFL7-targeted peptides, 50 μg of the EGFL7-coated screening beads were mixed with the OBOC library under the conditions described above. Hits were collected by magnetic isolation as previously described, and washed extensively with ethanol and water. A secondary cell-based screen was performed using EGFL7-overexpressing HT1080-tdTomato human fibrosarcoma cells (red), and EGFL7-low MDA-MB-435-GFP human breast cancer cells (green) as previously described.30 Beads were visualized under an Olympus IX70 inverted fluorescent microscope, and the ones that associated with the red (high EGFL7 expression) HT1080-tdT cells, but not the green (low EGFL7 expression) MDA-MB-435-GFP cells, were manually isolated using a micropipette. Hits were collected, washed and the peptides were sequenced on-bead using MALDI TOF MS/MS as described.312.2. SPR characterization of EGFL7-binding peptides and nanoparticles
All procedures were done according to the SPRimager®II array instrument (GWC Technologies) guidelines. Briefly, amine-reactive SPR chip (GWC Technologies) was subjected to the Sulfo-SMCC crosslinker (Thermo Scientific) for functionalization with a maleimide group. Each peptide containing a Cys molecule was immobilized onto the gold-coated spots on the SPR chip. Different concentrations (1 nM, 20 nM, and 50 nM) of purified EGFL7 were introduced as a continuous flow over the chip for 10 min. Real-time images and percent change in reflectivity (%ΔR) were recorded and analyzed.For nanoparticles characterization, purified recombinant EGFL7 (Sino Biological Inc.) was diluted to a final concentration of 0.25 μM in 10 mM sodium acetate at pH 5, and coupled to a Biacore Sensor chip CM5 (GE Healthcare). Different concentrations of CPMV-PEG-E7p72 or CPMV-PEG nanoparticles (90, 45, 22.5, 11.3, 5.6, 2.8 and 1.4 μg mL−1) were injected into the Biacore 3000 instrument, and allowed to flow over the chip (flow rate: 30 μL min−1; injection time: 2 min). Dissociation curves were obtained by flowing binding buffer over the chip for 15 minutes. Chip regeneration was performed by exposing chip to a high salt solution (5 M NaCl) for 30 seconds in between samples. Response units were plotted over time to generate an association and dissociation curve.
2.3. Evaluation of FITC-E7p72 and FITC-E7-p74 uptake in fibrosarcoma cells by microscopy
Human HT1080 fibrosarcoma cells were detached using EDTA and resuspended in DMEM supplemented with 10% fetal bovine serum (FBS) into microcentrifuge tubes. Live cells were incubated with 3.3 μM of fluorescein-conjugated E7-p72, E7-p74 and control GAGDS peptide in suspension for 1 h at 37 °C. Blocking studies were conducted in the presence of 100× excess of non-fluoresceinated peptides. Cells were washed with PBS by centrifugation (1200 rpm for 1 minute). After three washes, PBS was replaced with DMEM growth media and cells were seeded onto coverslips. Once adhered, cells were washed with PBS and mounted for fluorescence imaging. Images were captured using the Zeiss AxioImager Z1 microscope. The average fluorescein intensity within each cell was quantified using Volocity, version 6.1.2 (PerkinElmer, Massachusetts, USA). All statistics were performed using a one-way ANOVA and Tukey post hoc test.The evaluation of E7-p72 uptake by HUVECs and SVECs using flow cytometry was performed using the same procedures.
2.4. Purification of CPMV and synthesis of targeted CPMV nanoparticles
CPMV was produced in black-eyed pea plants no. 5; two weeks after seeding, plants were infected through mechanical inoculation. Infected leaves were harvested 14–21 days after infection and stored at −80 °C. CPMV was extracted in yields of 1–2 mg per gram of infected leaf material using established methods.58 CPMV concentration was determined using UV-Vis absorbance (ε at 260 nm = 8.1 mg−1 mL cm−1), and virus particle structural integrity was confirmed by transmission electron microscopy (TEM) and size exclusion chromatography (SEC) using a Superose6 column on the ÄKTA Explorer chromatography system (GE Healthcare).A 3000 molar excess of NHS-Alexa Fluor (NHS-A647, Invitrogen) and a 3000 molar excess of NHS-(PEG)4-alkyne (Sigma) were added per CPMV particle in 0.1 M potassium phosphate buffer pH 7.0; the protein concentration was kept at 2 mg ml−1; the reaction was allowed to proceed at room temperature overnight. Excess NIR dyes and NHS-alkyne molecules were removed using 10 kDa cut-off centrifugal spin filters (Millipore); the filters were washed with buffer until there was no dye detectable in the flow-through. Next, CuAAC reaction was carried out: 3 molar equivalents of PEG-E7p72 peptide or PEG, were reacted to CPMV-alkyne (2 mg mL−1) in presence of CuSO4 (1.25 mM), tris(3-hydroxypropyltriazolylmethyl)amine (THPTA, 6.25 mM) (provided by courtesy of Prof. Finn, TSRI), amidoguanidine (12.5 mM), and ascorbic acid (12.5 mM). The reaction was stopped after two hours by adding 1% (w/v) saturated EDTA solution. Modified A647-CPMV-PEG-E7p72 (denoted CPMV-PEG-E7p72) and A647-CPMV-PEG (denoted CPMV-PEG), respectively were then purified using spin filters and re-suspended in buffer. Purified CPMV-PEG-E7p72 and CPMV-PEG were analyzed by UV/visible spectroscopy to determine the concentration of the nanoparticles, as well as to determine the degree of nanoparticle labeling (see below). The nanoparticles are stored in at 4 °C in the dark for further use.
2.5. Evaluation of CPMV uptake using flow cytometry
CPMV-PEG (control) and CPMV-PEG-E7p72 were stored in PBS at 4 °C before further use. EGFL7 knockdown in EA.hy926 endothelial cells and HT1080 fibrosarocma cells were achieved using siRNA using jetPRIME transfection reagent (Polyplus Transfection). After 48 h of transfection with either control or EGFL7 siRNA, cells were seeded onto a 6-well tissue culture plate to approximately 80% confluency. Cells were incubated with CPMV-PEG (control) or CPMV-PEG-E7p72 (1 μg mL−1) at 37 °C for 3 h. Cells were washed three times with PBS, and then detached using 2.5 mM EDTA. Cells were washed once, fixed with 4% paraformaldehyde, resuspended in flow buffer (PBS with 2 mM EDTA, 2% FBS and 0.05% sodium azide) and evaluated using the BD FACS calibur flow cytometer. Data analysis was performed using FCS express (version 3).2.6. Evaluation of CPMV uptake by endothelial cells using confocal microscopy
EA.hy926 endothelial cells were seeded onto 6-well tissue culture plate to approximately 80% confluency. Cells were incubated with CPMV-PEG or CPMV-PEG-E7p72 (1 μg mL−1) at 37 °C for 3 h. Cells were washed three times with PBS, fixed with 4% paraformaldehyde and imaged using confocal microscopy (Zeiss AxioImager Z1 confocal microscope). Data were analyzed and quantified using the Volocity software, version 6.1.2, and statistics were performed using a one-way ANOVA and Tukey's post hoc test.2.7. Intravital imaging of tumor neovasculature using targeted CPMV-PEG-E7p72 nanoparticles in avian embryos
HT1080 fibrosarcoma cell line that ubiquitously expressed the green fluorescent protein (GFP) was generated through transfection of cultured cells with plasmid encoding the GFP gene, and selected by neomycin without clonal propagation. Avian embryos were cultured as described in ref. 43. At day 9 of embryonic development, HT1080 tumor boli were established in the chorioallantoic membrane (CAM) of the embryos according to ref. 36. Embryos were placed back into the humidified incubator at 37 °C and the tumors were allowed to vascularize for approximately three days. Embryos were then injected with CPMV nanoparticles (20 μg) and rhodamine-lectin (0.17 mg) intravenously, and placed within a specialized avian embryo-imaging unit.43 GFP (488 nm solid state laser excitation, emission HQ535/50m), rhodamine (594 nm solid state laser excitation, emission HQ600/50m), and NIR (647 nm solid state laser excitation, emission HQ700/50m) channels were visualized using an intravital spinning-disk confocal microscopy using an upright Zeiss AxioImager Z1, LUDL filter wheels, a Yokogawa spinning disk head and a Hamamatsu 9100-12 ImageEM CCD camera. The Volocity software, version 6.1.2 (PerkinElmer) was used to control this set up. Z-Stack images were captured every two minutes. Images were flattened into a single image, and analyzed using the Volocity software, version 6.1.2 (PerkinElmer) or ImageJ (National Institutes of Health, USA). Two hours after injection, tumors were excised, washed in PBS and fixed in 10% sucrose and 4% formalin in PBS for 2 hours at 4 °C. Tumors were then washed and frozen in OCT (Tissue Tek) on dry ice. Frozen sections (5 μm) were prepared on positively charged histobond glass slides (VWR). Samples were mounted in ProLong Gold mounting media containing DAPI (Life Technologies) and imaged with a 60× objective using confocal microscopy.2.8. Evaluation of CPMV-PEG-E7p72 nanoparticle uptake in mice
All studies were performed according to protocols approved by the University of Alberta Animal Care and Use Committee. 8-weeks old mice were established with HT1080 fibrosarcoma tumors. HT1080 cells (1 × 106 cells) were injected subcutaneously into the flank. After two weeks (or when the tumor size reached 1000 mm3), animals were euthanized and tumors were excised, fixed in 4% formaldehyde and 10% sucrose overnight at 4 °C, and embedded in OCT medium. Frozen sections (10 μm) were prepared on a Leica cryomicrotome, and samples were stored at −80 °C for further analysis.For immunofluorescence staining, tumor sections were fixed in 4% formaldehyde at 25 °C. Samples were blocked using 10% goat serum and 1% BSA (in PBS + 0.02% Tween-20) for one hour at 25 °C. Tissues were incubated with a rat anti-CD31 antibody in blocking buffer (1 h at 25 °C) and washed four times (PBS + 0.02% Tween-20). Tissues were then incubated with the anti-rat AlexaFluor 488 antibody (Invitrogen) in blocking buffer (1 h at 25 °C in dark). Tissues were washed four times (PBS + 0.02% Tween-20), and mounted in ProLong Gold mounting media containing DAPI (Life Technologies). Tissues were imaged using the Zeiss AxioImager Z1 microscope upright confocal microscopy, and images were quantified using Volocity, version 6.1.2 (PerkinElmer, Massachusetts, USA).
For ex vivo staining, frozen tumor sections were fixed in 4% formaldehyde and incubated with 10 μg of CPMV-PEG-E7p72 or control CPMV-PEG nanoparticles. Samples were then blocked in 10% goat serum and 1% BSA (in PBS + 0.02% Tween-20) for one hour at 25 °C. The samples were then stained with CD31 antibody as described above.
2.9. PET imaging using [68Ga]-DOTA-E7p72
EGFL7-HT1080 cells and control HT1080 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% FBS in 6 well plates respectively. The number of cells in each well was counted using a microscope. [68Ga]-DOTA-E7p72 (100 μL, approx. 1 MBq mL−1) was added to each well with/without inhibitor (×100) and the cells were incubated at room temperature for 1 h. The medium in the well was decanted out. The cells on the bottom of well were washed twice by PBS (×1) and counted in a γ-counter. The radioactive counts based on the number of cells in each well was calculated.For in vivo studies, NOD/scid mice (male, 4–6 weeks age) were ordered from Charles River. All animal studies were performed following the Canadian Council on Animal Care guidelines and animal use protocol approved by Western University. NOD/scid mice were xenografted with EGFL7-HT1080 or MDA-MB-231 cells. The PET imaging study was conducted when the tumor grew to approx. 1 cm in diameter. [68Ga]-DOTA-E7p72 (10 MBq) was injected into the tail vein of the animal while under 1.5% isoflurane/O2 flow and the mouse was scanned for 60 min in dynamic mode using a small animal PET scanner (Explore Vista-DR, GE Healthcare). The PET image was reconstructed using Vista 4.7 software (GE Healthcare) and the SUV data in the tumor was calculated every 3 minutes based on the region-of-interest analysis on PET images.
2.10. Blocking study to test the specificity of FITC-E7p72 uptake in endothelial cells
EA.hy 926 were detached using EDTA and resuspended in DMEM supplemented with 10% FBS into microcentrifuge tubes. Live cells were incubated with 3.3 μM of fluorescein-conjugated E7-p72 and either 3 times molar excess of E7p72 peptide, 3× recombinant EGFL7 purified protein or 3× scramble peptide in suspension for 1 h at 37 °C. Cells were washed with PBS by gentle centrifugation (300g for 1 minute) three times. Cells were resuspended in 1 ml PBS supplemented with 2% FBS in polystyrene flow cytometry tubes and analysed using the Fortessa X-20 flow cytometer and analysed using FCS Express (version 4). For imaging, 100 μl of the cell suspension was supplemented with DMEM growth media and cells were seeded onto coverslips. Once adhered, cells were washed with PBS and cell nuclei were stained with Hoechst 33342 and mounted for fluorescence imaging. The Nikon A1+ confocal microscope was used to visualize the slides.3. Results
3.1. EGFL7 localizes to the lumen of tumor endothelium and is expressed on the surface of cancer cells
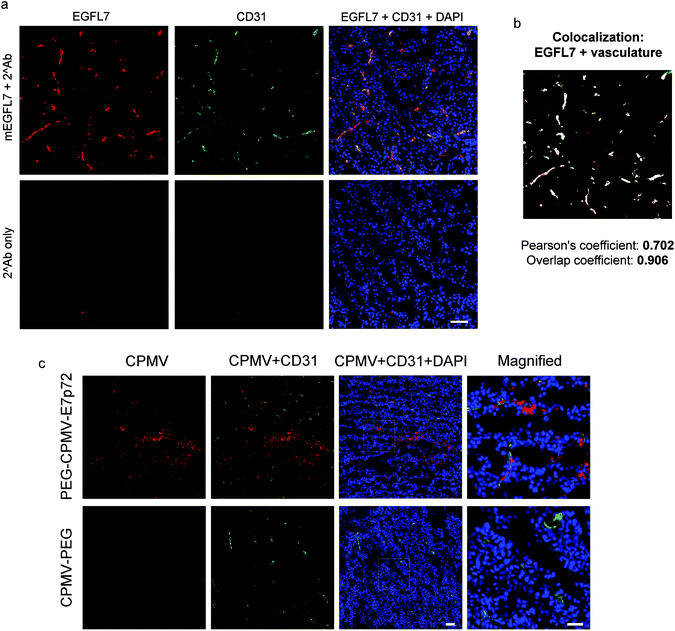
It has been reported previously that EGFL7 localizes to the extracellular matrix associated with the endothelial cells that express it.9,59 We sought to confirm for screening purposes that human HT1080 fibrosarcoma cells transfected with an EGFL7 expression plasmid would express EGFL7 on their surface. To assess this, we first performed a cell surface biotinylation experiment followed by immunoprecipitation of cell surface proteins using anti-biotin antibodies. EGFL7, but not tubulin, was present in the immunoprecipitate, confirming its localization on the cell surface (ESI Fig. 1a†). This was further confirmed by confocal imaging (ESI Fig. 1b†).We wanted to assess, for vascular targeting purposes, whether EGFL7 expressed by tumor endothelium is localized to the basal and/or lumenal surface. Sections of human breast cancer (MDA-MB-468) xenograft tumours in mice were co-stained with antibodies against CD31, an endothelial cell marker, and EGFL7. The image (ESI Fig. 1c†) and associated line scan (ESI Fig. 1d†) indicate that EGFL7 localizes to both the lumenal and basal surfaces of tumor endothelium.
3.2. EGFL7-targeting peptides identified through a high-throughput one bead one compound (OBOC) library screen
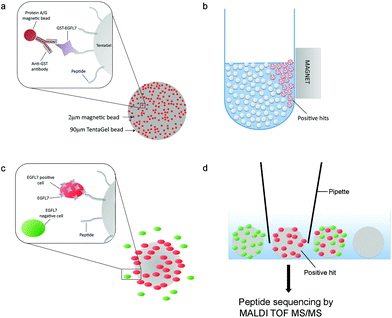
To identify novel peptide ligands of EGFL7, we performed a two-stage screen of an OBOC combinatorial peptide library using a magnetic/fluorescent bead-based approach and followed by a cell-based approach (Fig. 1). A linear octapeptide OBOC library was constructed on 90 μm Tentagel beads comprised of eighteen of the eukaryotic amino acids (minus cysteine and isoleucine) using a split-mix synthesis approach.28 This resulted in a library containing roughly 1017 distinct peptides, with each bead displaying 1013 copies of a unique peptide entity.Our peptide screening strategy was designed to maximize the likelihood of capturing high-affinity EGFL7 ligands with high stringency and low false positives. To achieve this, we employed two sequential approaches: (1) a magnetic/fluorescent ‘beads on a bead’ screening approach,29 and (2) a cell-based positive/negative selection screen30 (Fig. 1). The first approach exploited the tendency of small magnetic beads (2 μm diameter) decorated with EGFL7 to reversibly interact with the much larger peptide-coated TentaGel peptide library beads (90 μm diameter). EGFL7-GST was adsorbed onto magnetic screening beads via an anti-GST antibody (ESI Fig. 2†). We first performed a library enrichment step by removing all the library beads that associated with the magnetic beads displaying the anti-GST antibody and GST protein. Then, the EGFL7-displaying magnetic beads were mixed with the OBOC library and Tentagel beads displaying EGFL7-binding peptides were isolated by magnetic sorting. Roughly 500 hit beads were isolated in this first screen.
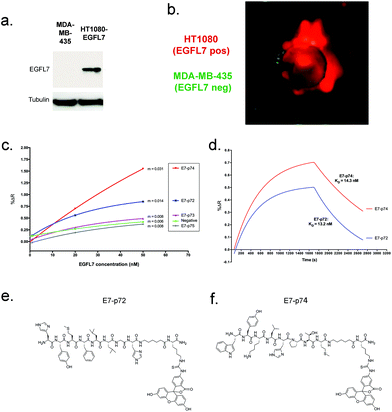
Using the positive hit library beads from the first screen, a second screen was employed to ensure that hit peptides recognized EGFL7 in its native cellular context and conformation. To do this, the Tentagel beads were mixed with live cells that either expressed EGFL7 (EGFL7+) or did not (EGFL7−) (Fig. 2a). To make the separation straightforward, the EGFL7+ cells were engineered to express tdTomato, while EGFL7− cells were transduced to express GFP (Fig. 2b). In this way, beads with a strong association for the red EGFL7+ cells, but not green EGFL7− cells, were readily identified using fluorescence microscopy and isolated manually (Fig. 2b). In total, 16 library beads were isolated and peptides were sequenced using a previously-developed on-bead mass spectrometry approach,31 leading to the identification of 8 unique peptide sequences. These peptides were prioritized based on our confidence in the sequence deconvolution, and four were selected for further characterization (Table 1).
| Peptide | Sequences |
|---|---|
| E7p72 | HMYFLLGH |
| E7p73 | SQSSMYPS |
| E7p74 | WYKLPTM |
| E7p75 | EWELHAEE |
3.3. Characterization of EGFL7-binding peptides
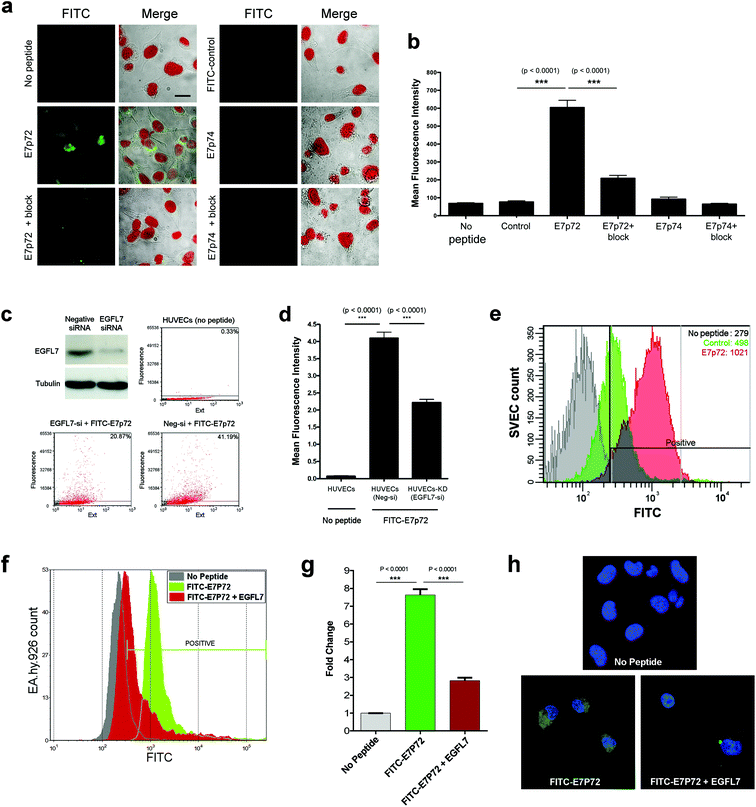
We re-synthesized the four lead peptides E7p72, E7p73, E7p74 and E7p75, and then characterized their specificity for EGFL7. The binding affinity for immobilized EGFL7 was first determined using surface plasmon resonance (SPR). Two peptides, E7p72 and E7p74, were observed to exhibit concentration-dependent affinity for EGFL7, while the association rate of E7p73 and E7p75 did not differ significantly from a control negative peptide (Fig. 2c). Kinetic binding analysis revealed the KD values for E7p72 and E7p74 to be 13.2 nM and 14.3 nM, respectively (Fig. 2d). Based on these findings, E7p72 and E7p74 were re-synthesized with a C-terminal fluorescein dye (Fig. 2e and f) to evaluate their ability to bind cells expressing EGFL7. We first confirmed that labeling of E7p72 with fluorescein did not hinder peptide binding to EGFL7 by SPR analysis (ESI Fig. 3†).Fluorescein-labeled E7p72 and E7p74 were first assessed using a panel of HT1080 cancer cell lines engineered to express no, low or high EGFL7 (ESI Fig. 4a and b†). The binding and uptake of the peptides to these cells was assessed by fluorescence microscopy and flow cytometry. While binding of FITC-E7p72 to HT1080 cells was highly dependent on EGFL7 expression (Fig. 4a and b and ESI Fig. 4c–e†), little to no specific binding was observed with FITC-E7p74 (Fig. 4a and b). The specificity of E7p72 for EGFL7 was further confirmed by blocking with 100 molar excess of unlabeled E7p72 peptide, which resulted in a significant reduction in the binding of FITC-E7p72 to HT1080 EGFL7high (Fig. 4a and b). The binding of E7p72 to EGFL7 was then assessed using human umbilical vein endothelial cells (HUVEC), which normally express high levels of EGFL7 that can be reduced by siRNA knockdown (Fig. 4c). Flow cytometry analysis showed that HUVECs incubated with FITC-E7p72 demonstrate significant binding and uptake (Fig. 3c and d). When EGFL7 expression was knocked down, uptake was reduced by more than 50% (Fig. 3d). We then also evaluated the affinity of E7p72 for murine EGFL7, which shares 73% identity with human EGFL7. Flow cytometry analysis revealed that FITC-E7p72 showed 2× selectivity for mouse endothelial cells (SVECs) compared to control peptide (Fig. 3e). Finally, we quantified human endothelial cell (EA.hy926) uptake of FITC-E7p72 in the presence of 3× molar excess of purified EGFL7 protein. Flow cytometry and confocal microscopy analysis showed nearly 70% reduction in FITC-E7p72 uptake by endothelial cells in the presence of EGFL7 (Fig. 3f–h). Taken together, these data indicate that E7p72 specifically targets EGFL7 expressed in human and mouse endothelial cells.10
To further assess the specificity of E7p72 for EGFL7, a radiotracer derived from E7p72 was synthesized, and its ability to target EGFL7 in vivo was evaluated by positron emission tomography (PET) imaging. First, a metal chelator DOTA was placed at the C-terminus of the peptide through a Lys side-chain, with a short linker to separate the lysine-metal complex from the targeting peptide. A non-radioactive (69/71Ga) surrogate was used as a chromatographic standard during radiolabelling. Radiolabeled [68Ga]-DOTA-E7p72 (ESI Fig. 5a†) was prepared using the short-lived positron emitter 68Ga (t1/2 = 68 min) in a 53% yield (decay-corrected), >95% purity and 9.4 GBq μmol−1 specific activity (ESI Fig. 5c†). In vitro evaluation showed a significantly higher uptake of [68Ga]-DOTA-E7p72 in HT1080-EGFL7 overexpressing cells compared to regular HT1080 cells. Additional evidence for EGFL7 specific uptake was proven by blocking protein-mediated uptake through competition with an excess of unlabeled E7p72 peptide (ESI Fig. 5b†). To investigate EGFL7 targeting in vivo, we established two xenograft models using NOD/scid mice. The first was a breast cancer xenograft consisting of MDA-MB-231 cells, which express low levels of human EGFL7 (ESI Fig. 5d†) yet grow rapidly and recruit significant EGFL7-expressing neovasculature. The second xenograft model consisted of HT1080 tumor cells with high expression of EGFL7, which grows just as rapidly and has significant expression of EGFL7 (ESI Fig. 5d†). [68Ga]-DOTA-E7p72 was injected i.v. and dynamic PET scans were carried out over a 60 minutes time period. The xenograft tumors were readily visualized in both models; however, the HT1080-EGFL7 tumor displayed a much higher uptake of the radiotracer (ESI Fig. 5e and f†). Standard uptake values (SUV) were calculated throughout the time course of the dynamic scan (ESI Fig. 5g†). The HT1080-EGFL7 (EGFL7high) tumor was found to have a SUV reaching 0.6 shortly after injection, with sustained tumor uptake over 60 min dropping only to 0.5. The MDA-MB-231 (EGFL7low) tumor (ESI Fig. 5g†) also had robust initial uptake (up to a SUV of nearly 0.4), but this was followed by a more rapid washout. This suggests that while EGFL7 targeting is occurring initially in both tumors, E7p72 was retained at higher levels in the EGFL7-overexpressing tumor over time.
3.4. Development of EGFL7-targeted near infrared viral nanoparticles
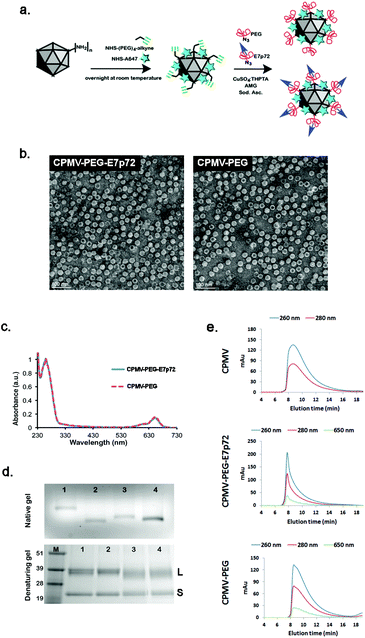
We next sought to validate the potential of E7p72 peptides to mediate the targeting and visualization of neovasculature by nanoparticles. Viral nanoparticles (VNPs) based on the cowpea mosaic virus (CPMV) offer a compelling biocompatible platform for intravital imaging of tumors and their associated vasculature.32–36 CPMV nanoparticles label blood vessels in animal models34 through a specific interaction with endothelial cell surface vimentin.37 We have previously shown that this interaction can be blocked by PEGylating CPMV,38,39 and that VNPs can then be engineered to be selective by decorating the exterior surface with targeting peptides.32,33 We expected, therefore, that the incorporation of E7p72 peptide onto the surface PEGylated CPMV VNPs would render them selective for endothelial cells expressing EGFL7.The CPMV viral capsid displays 300 lysine side chains on its surface, which can be covalently modified with diverse functional groups.40 To create a molecular targeted formulation, we engineered multivalent VNPs to display polyethylene glycol (PEG) polymers to shield them from the mononuclear phagocyte system (MPS), Alexa Fluor 647 near infrared fluorescent dyes for intravital imaging, and E7p72 peptides to confer specificity for EGFL7. The bioconjugation of VNPs was carried out using a two-step approach. First, surface lysines were modified using NHS ester chemistry to introduce NIR dyes and alkyne handles. Then, the alkyne handles were conjugated with either azide-containing PEG2000 (CPMV-PEG) or PEG2000-E7p72 peptide (CPMV-PEG-E7p72) so that the targeting peptide was displayed on the outer surface (Fig. 4a).
The engineered VNPs were characterized first using transmission electron microscopy (TEM), which confirmed that the VNPs remained intact after chemical modification (Fig. 4b). Using UV/visible spectroscopy, we determined that each formulation was labeled with ∼25 NIR dyes per virus, resulting in exceptionally bright particles with no detectable quenching (Fig. 4c). The control and targeted formulations were then analyzed using native and denaturing gel electrophoresis. Each yielded a single band indicating particles and subunits with expected sizes with no aggregation or unexpected cross-linking. The increased mobility observed in the CPMV-647-alkyne intermediate compared with native CPMV is consistent with the lysine modifications, where conjugation with either NIR dyes or alkynes substantially reduced the positive charges on the viral surface, and therefore accelerated its migration towards the anode41 (Fig. 4d). Conjugation with PEG and PEG-E7p72 increased the size of the particles, resulting in reduced electrophoretic mobility compared with CPMV-alkyne (Fig. 4d). The VNP formulations were analyzed by size exclusion chromatography (SEC) using fast protein liquid chromatography (FPLC) and a Superose6 column. A typical elution profile was observed for CPMV with an elution time of approximately 17 minutes and an A260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) A280 ratio of 1.7 ± 0.1 (Fig. 4e), indicating native intact CPMV particles. CPMV-PEG-E7p72 eluted in a sharper, left-shifted peak with an elution time of 15.5 minutes (Fig. 4e), consistent with the expected increase in overall size. The elution profile of CPMV-PEG nanoparticles followed a similar trend as CPMV-PEG-E7p72, where sharpening of the peak was observed (Fig. 4e).
A280 ratio of 1.7 ± 0.1 (Fig. 4e), indicating native intact CPMV particles. CPMV-PEG-E7p72 eluted in a sharper, left-shifted peak with an elution time of 15.5 minutes (Fig. 4e), consistent with the expected increase in overall size. The elution profile of CPMV-PEG nanoparticles followed a similar trend as CPMV-PEG-E7p72, where sharpening of the peak was observed (Fig. 4e).
3.5. VNPs displaying E7p72 peptides specifically label human endothelial cells
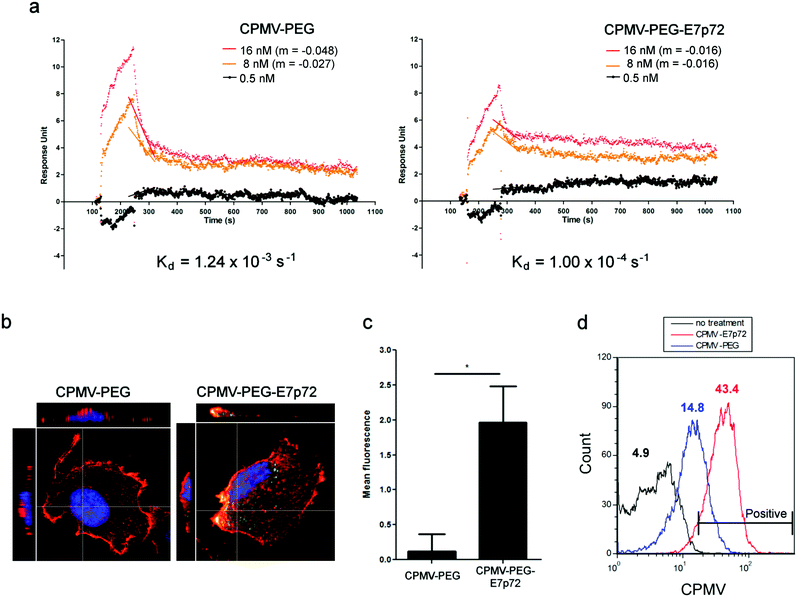
The binding of CPMV-E7p72 and CPMV-PEG VNP formulations to recombinant EGFL7 protein was characterized using SPR analysis. Our analysis indicated that while both nanoparticles associated with EGFL7 at relatively similar rates, the dissociation of CPMV-PEG-E7p72 (Kd = 1.00 × 10−4 s−1) was significantly slower compared to CPMV-PEG (Kd = 1.24 × 10−3 s−1) (Fig. 5a). This suggests that CPMV-PEG-E7p72 associated more strongly with immobilized EGFL7 compared to the non-targeted CPMV-PEG nanoparticles. We then evaluated the specificity of the VNP formulations to human endothelial cells. High-resolution confocal imaging revealed that CPMV-PEG-E7p72 VNPs are endocytosed and internalized by endothelial cells at a rate 6 times higher than that observed for control CPMV-PEG VNPs (Fig. 5b and c).3.6. EGFL7-targeted nanoparticles bind tumor neovasculature
CPMV-PEG-E7p72 was internalized by endothelial cells, while little or no uptake was observed for CPMV-PEG nanoparticles. Flow cytometry analysis also confirmed that endothelial cell uptake of CPMV-PEG-E7p72 was more than 3 times higher than control CPMV-PEG nanoparticle (Fig. 5d). Taken together, these data show that CPMV-PEG-E7p72 VNPs specifically target endothelial cells in an EGFL7-dependent manner.A xenograft mouse model of human fibrosarcoma (HT1080) was used to test the specificity of CPMV-PEG-E7p72 for EGFL7. Tumors were implanted subcutaneously and allowed to grow to 1000 mm3. Immunofluorescence staining of tumor cryo-sections showed that the tumor endothelium expressed a high level of EGFL7 (Fig. 6a). Further analyses revealed the Pearson's correlation coefficient and overlap coefficient to be 0.702 and 0.906, respectively, indicating strong co-localization between EGFL7 expression and the tumor-associated neovasculature (Fig. 6b). Ex vivo staining of the tumor cryo-sections with the VNPs revealed that CPMV-PEG-E7p72, but not control CPMV-PEG nanoparticles, bound to the tumor-associated blood vessels (Fig. 6c).
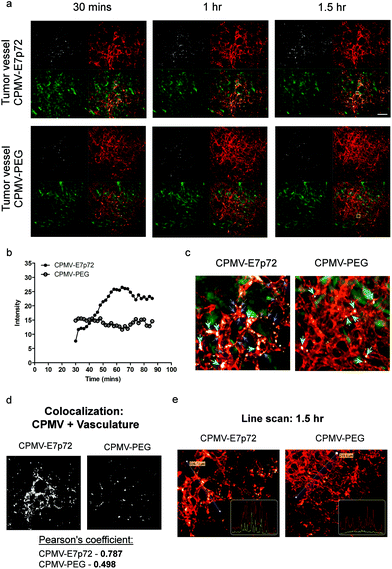
We have shown previously that VNPs based on CPMV are useful tools for intravital imaging in animal models. Intravital imaging in the ex ovo avian embryo model in particular allows for high resolution temporal analysis of uptake of nanoparticles in tumors.32,34–36,42,43 To test if CPMV-PEG-E7p72 can target the tumor neovasculature in vivo, we established human HT1080-GFP fibrosarcoma tumors in the CAM of ex ovo embryos, which form large, highly vascularized tumors. Vasculature is visualized in vivo using a fluorescent lectin.42–44 Following intravenous administration of either CPMV-PEG-E7p72 or control CPMV-PEG nanoparticles, the embryo was placed in a specialized imaging unit for immobilization of the CAM to allow for high-resolution real-time confocal imaging of the tumor and its associated vasculature.43–45 Intravital imaging over several hours revealed that CPMV-PEG-E7p72 VNPs accumulate significantly in tumor endothelium over 90 minutes (Fig. 7a and b; ESI Movie 1†). In contrast, CPMV-PEG VNPs circulated within the vasculature with no significant uptake by the endothelium (Fig. 7a and b; ESI Movie 2†). In quiescent vessels distal from the tumor, our intravital imaging experiments revealed little to no uptake of either the targeted or control VNPs (ESI Fig. 6†). We did observe some uptake of both targeted and non-targeted VNPs by macrophages in the tumor and normal vasculature (ESI Fig. 6; ESI Movie 1 and 2†), consistent with previous studies.46 CPMV-PEG and CPMV-PEG-E7p72 appear to have a similar biodistribution profile in most organs, with the exception of a significant elevation in spleen uptake observed in case of CPMV-PEG-E7p72 (ESI Fig. 7†). Higher fluorescence signal was also observed in the PC3 flank tumor in mice injected with CPMV-PEG-E7p72 compared with control nanoparticles.
We observed that CPMV-PEG-E7p72 VNPs were internalized by the tumor endothelium in what appears to be the endosomal compartment (Fig. 7c), which can also be observed in the confocal images of the fixed tissue (ESI Fig. 8a†). CPMV-PEG-E7p72 colocalized strongly with the tumor endothelium (Pearson's correlation coefficient: 0.787) 90 minutes after administration (Fig. 7d). In contrast, no colocalization was observed for CPMV-PEG (Pearson's correlation coefficient: 0.498). A line profile through the tumor endothelium highlights the close co-localization of CPMV-PEG-E7p72 with the endothelial lectin labeling (Fig. 7e). In contrast, this association was not observed with the control CPMV-PEG VNPs. Quantitative analysis of the images of fixed tissue sections revealed that CPMV-PEG-E7p72 was taken up in tumor endothelium 40 times greater than the control CPMV-PEG nanoparticle (ESI Fig. 8b†).
4. Discussion
Given the importance of angiogenesis in the pathogenesis of diseases such as cancer, it would be greatly beneficial to be able to specifically target angiogenesis for the purpose of molecular imaging or therapy. In this work, we focused our efforts on the novel angiogenic factor EGFL7, a protein that is highly upregulated in angiogenic endothelium, but has not been directly implicated in VEGF-mediated signaling. In our search for novel ligands targeting EGFL7, we have undertaken a stringent combinatorial peptide library screen that integrates several complementary approaches that we developed previously.29,30 This has resulted in the discovery of a novel peptide ligand, E7p72, with high affinity and specificity for human EGFL7 that labels endothelial cells in an EGFL7-dependent manner. To demonstrate the utility of this EGFL7-directed ligand for molecular targeting, we have engineered fluorescent viral nanoparticles decorated with E7p72 peptides and evaluated their capacity to specifically label tumor neovasculature in two animal models. Real-time intravital confocal imaging of human tumors in ex ovo avian embryos revealed specific accumulation of nanoparticles in the tumor-associated endothelium within an hour of injection, allowing for rapid identification of the tumor neovasculature. These results establish E7p72 as a potent targeting ligand for angiogenic vasculature, and suggest its potential for applications in non-invasive imaging and drug delivery strategies.EGFL7 is an emerging therapeutic target in cancer. Given the advent of precision medicine approaches, imaging agents based on EGFL7 targeting ligands may be useful to predict the efficacy of EGFL7-targeted therapies, or to monitor treatment with these agents. A recent study in metastatic colorectal cancer (mCRC) revealed that low expression of endothelial-specific EGFL7 in tumor samples is significantly correlated to better response rate to first-line chemotherapy and bevacizumab, indicating that EGFL7 expression has predictive value.51 The ability of E7p72 to detect EGFL7 in the tumor vasculature indicates that this peptide can potentially serve as a companion diagnostic with bevacizumab. This suggests that for patients where biopsy samples are difficult or impossible to obtain (i.e. presence of micrometastases), whole-body imaging of EGFL7 expression with E7p72 may provide valuable information for deciding the most appropriate therapeutic option. A correlation between EGFL7 expression and survival has also been observed in patients with mCRC,51 ovarian cancer22 and pancreatic cancer,23 highlighting the potential prognostic value of E7p72.
We have chosen to use the viral nanoparticle platform for our targeting studies, as these are naturally occurring nanomaterials, making them biocompatible and biodegradable compared with other synthetic nanoparticles, which potentially render them safer for use in humans.47 Viral-based nanotechnology has received significant attention over the last decade and making major strides towards the clinic. They are extremely robust, stable, and compatible with a broad range of conjugation chemistries. While we have focused on the use of VNPs for optical imaging in this study, other viral nanoparticles bearing iron oxide or gadolinium for magnetic resonance imaging (MRI),48,49 and radioisotopes for positron emission tomography (PET) have also been described.50 Furthermore, the viral capsid can be easily modified (either chemically or by genetic engineering) to allow for the incorporation of a diverse range of payload (i.e. toxins or drugs), thereby opening doors to the generation of new therapeutics.
Over the past decade, peptides have gained extensive interest because of their standard and cost-effective synthesis protocols, good efficacy and safety profiles, as well as high specificity and potency.52 Currently, more than 60 peptide drugs have been approved by the FDA, and around 400 peptides are being evaluated in clinical trials.53 Several of these peptides are or being developed for applications in oncology.54 Some of the most popular anti-angiogenesis peptides include the RGD peptide, which functions through binding the integrin αvβ3 and αvβ5,55 as well as the NGR peptide, which targets the aminopeptidase N.56 Although integrins and aminopeptidase N are overexpressed in tumor cells and their associated vasculature, they are also found in several types of normal tissues (i.e. breast, lung, kidney and skin).55,56 EGFL7 serves as a unique marker because it is almost exclusively expressed by remodeling endothelial cells in the vasculature system, and is virtually absent from mature blood vessels in normal adult tissues.7 Targeting the vasculature can be advantageous because tumor cells are genetically unstable and heterogeneous, and they are more difficult to target from a drug delivery perspective. Meanwhile, the tumor-associated endothelium is genetically stable and highly accessible for targeting.57 EGFL7-targeted nanoparticles should provide a robust platform for future imaging and drug delivery efforts.
5. Conclusions
In summary, angiogenesis is a dynamic process that is fundamental to cancer, and a high proportion of angiogenic vessels is closely linked with poor prognosis. EGFL7 is a key mediator of angiogenesis, and its expression is highly upregulated in remodeling endothelium, such as in cancers. EGFL7 expression is associated with poor outcome in several cancers, making it a promising target for imaging or therapeutic strategies. We have described the discovery and characterization of a novel high-affinity EGFL7-specific peptide, called E7p72. We have shown that E7p72 is taken up by endothelial and tumor cells in an EGFL7-dependent manner. PET imaging using a 68Ga-labeled E7p72 radiotracer has also demonstrated in vivo targeting of EGFL7. As assessed by intravital imaging, viral nanoparticles decorated with E7p72 specifically accumulate in the tumor neovasculature in vivo. E7-p72 is, to our knowledge, the first peptide that binds EGFL7 and targets the remodelling endothelium. This work highlights the potential of EGFL7 as a target for cancer imaging and opens the door for novel molecular targeted therapeutic approaches.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This study was supported by Prostate Cancer Canada Grant 2011-742 to JDL, the Ontario Institute for Cancer Research (OICR) and the London Regional Cancer Program to LGL, and the National Science Foundation Grant CMMI NM 333651 to NFS. This work was funded in part by NIH R21-EB020946 (to NFS and JDL). CFC was supported by CIHR Fellowship 1122374. JDL holds the Frank and Carla Sojonky Chair in Prostate Cancer Research funded by the Alberta Cancer Foundation. We thank Dr Marek Michalak and Dr Jody Groenendyk (Dept. of Biochemistry, University of Alberta) for providing us with access to the Biacore 3000 instrument and Dr Xuejun Sun and George Huang (Dept. of Oncology, University of Alberta) for access to the flow cytometry facility. We also thank Dr Michael Kovacs for access to the cyclotron facility, Dr Ting-Yim Lee for access to small animal PET camera and Jennifer Hadway for assistance with animal models (Lawson Health Research Institute). We thank Amy M. Wen (Case Western Reserve University) for performing the TEM imaging. We acknowledge the generosity of Dr Sean Lawler (Brigham and Women's Hospital, Harvard Medical School) for editing the manuscript.References
- S. B. Fox and A. L. Harris, Invest. New Drugs, 1997, 15, 15–28 CrossRef CAS PubMed
.
- O. L. Chinot, W. Wick, W. Mason, R. Henriksson, F. Saran, R. Nishikawa, A. F. Carpentier, K. Hoang-Xuan, P. Kavan, D. Cernea, A. A. Brandes, M. Hilton, L. Abrey and T. Cloughesy, N. Engl. J. Med., 2014, 370, 709–722 CrossRef CAS PubMed
.
- M. R. Gilbert, J. J. Dignam, T. S. Armstrong, J. S. Wefel, D. T. Blumenthal, M. A. Vogelbaum, H. Colman, A. Chakravarti, S. Pugh, M. Won, R. Jeraj, P. D. Brown, K. A. Jaeckle, D. Schiff, V. W. Stieber, D. G. Brachman, M. Werner-Wasik, I. W. Tremont-Lukats, E. P. Sulman, K. D. Aldape, W. J. Curran Jr. and M. P. Mehta, N. Engl. J. Med., 2014, 370, 699–708 CrossRef CAS PubMed
.
- I. L. Ray-Coquard, J. Domont, E. Tresch-Bruneel, E. Bompas, P. A. Cassier, O. Mir, S. Piperno-Neumann, A. Italiano, C. Chevreau, D. Cupissol, F. Bertucci, J. O. Bay, O. Collard, E. Saada-Bouzid, N. Isambert, C. Delcambre, S. Clisant, A. Le Cesne, J. Y. Blay and N. Penel, J. Clin. Oncol., 2015, 33, 2797–2802 CrossRef CAS PubMed
.
- M. Pinter, G. Ulbrich, W. Sieghart, C. Kölblinger, T. Reiberger, S. Li, A. Ferlitsch, C. Müller, J. Lammer and M. Peck-Radosavljevic, Radiology, 2015, 277, 903–912 CrossRef PubMed
.
- K. Bambino, L. A. Lacko, K. A. Hajjar and H. Stuhlmann, Genesis, 2014, 52, 657–670 CrossRef CAS PubMed
.
- D. Nichol and H. Stuhlmann, EGFL7: a unique angiogenic signaling factor in vascular development and disease, Blood, 2012, 119, 1345–1352 CrossRef CAS PubMed
.
- M. Massimiani, L. Vecchione, D. Piccirilli, P. Spitalieri, F. Amati, S. Salvi, S. Ferrazzani, H. Stuhlmann and L. Campagnolo, Mol. Hum. Reprod., 2015, 21, 435–451 CrossRef CAS PubMed
.
- I. Nikolic, N. D. Stankovic, F. Bicker, J. Meister, H. Braun, K. Awwad, J. Baumgart, K. Simon, S. C. Thal, C. Patra, P. N. Harter, K. H. Plate, F. B. Engel, S. Dimmeler, J. A. Eble, M. Mittelbronn, M. K. Schäfer, B. Jungblut, E. Chavakis, I. Fleming and M. H. Schmidt, Blood, 2013, 121, 3041–3050 CrossRef CAS PubMed
.
- M. J. Fitch, L. Campagnolo, F. Kuhnert and H. Stuhlmann, Dev. Dyn., 2004, 230, 316–324 CrossRef CAS PubMed
.
- L. H. Parker, M. Schmidt, S. W. Jin, A. M. Gray, D. Beis, T. Pham, G. Frantz, S. Palmieri, K. Hillan, D. Y. Stainier, F. J. De Sauvage and W. Ye, Nature, 2004, 428, 754–758 CrossRef CAS PubMed
.
- F. Soncin, V. Mattot, F. Lionneton, N. Spruyt, F. Lepretre, A. Begue and D. Stehelin, EMBO J., 2003, 22, 5700–5711 CrossRef CAS PubMed
.
- L. Campagnolo, A. Leahy, S. Chitnis, S. Koschnick, M. J. Fitch, J. T. Fallon, D. Loskutoff, M. B. Taubman and H. Stuhlmann, Am. J. Pathol., 2005, 167, 275–284 CrossRef CAS PubMed
.
- D. Nichol, C. Shawber, M. J. Fitch, K. Bambino, A. Sharma, J. Kitajewski and H. Stuhlmann, Blood, 2010, 116, 6133–6143 CrossRef CAS PubMed
.
- A. Durrans and H. Stuhlmann, J. Angiog. Res., 2010, 2, 4 CrossRef PubMed
.
- L. A. Mucci, A. Powolny, E. Giovannucci, Z. Liao, S. A. Kenfield, R. Shen, M. J. Stampfer and S. K. Clinton, J. Clin. Oncol., 2009, 27, 5627–5633 CrossRef PubMed
.
- C. H. Huang, X. J. Li, Y. Z. Zhou, Y. Luo, C. Li and X. R. Yuan, J. Cancer Res. Clin. Oncol., 2010, 136, 1737–1743 CrossRef CAS PubMed
.
- F. Wu, L. Y. Yang, Y. F. Li, D. P. Ou, D. P. Chen and C. Fan, Hepatology, 2009, 50, 1839–1850 CrossRef CAS PubMed
.
- S. Delfortrie, S. Pinte, V. Mattot, C. Samson, G. Villain, B. Caetano, G. Lauridant-Philippin, M. C. Baranzelli, J. Bonneterre, F. Trottein, C. Faveeuw and F. Soncin, Cancer Res., 2011, 71, 7176–7186 CrossRef CAS PubMed
.
- C. Fan, L. Y. Yang, F. Wu, Y. M. Tao, L. S. Liu, J. F. Zhang, Y. N. He, L. L. Tang, G. D. Chen and L. Guo, Int. J. Biol. Markers, 2013, 28, 71–83 CrossRef CAS PubMed
.
- G. Philippin-Lauridant, M. C. Baranzelli, C. Samson, C. Fournier, S. Pinte, V. Mattot, J. Bonneterre and F. Soncin, Int. J. Oncol., 2013, 42, 1367–1375 CrossRef CAS PubMed
.
- J. Oh, S. H. Park, T. S. Lee, H. K. Oh, J. H. Choi and Y. S. Choi, J. Gynecol. Oncol., 2014, 25, 334–341 CrossRef PubMed
.
- L. Zhou, J. Li, Y. P. Zhao, J. C. Guo, Q. C. Cui, W. X. Zhou, T. P. Zhang, W. M. Wu, L. You and H. Shu, Hepatobiliary Pancreatic Dis. Int., 2014, 13, 523–528 CrossRef CAS
.
- J. J. Li, X. M. Yang, S. H. Wang and Q. L. Tang, J. Laryngol. Otol., 2011, 125, 1152–1157 CrossRef PubMed
.
- L. Johnson, M. Huseni, T. Smyczek, A. Lima, S. Yeung, J. H. Cheng, R. Molina, D. Kan, A. De Mazière, J. Klumperman, I. Kasman, Y. Zhang, M. S. Dennis, J. Eastham-Anderson, A. M. Jubb, O. Hwang, R. Desai, M. Schmidt, M. A. Nannini, K. H. Barck, R. A. Carano, W. F. Forrest, Q. Song, D. S. Chen, L. Naumovski, M. Singh, W. Ye and P. S. Hegde, J. Clin. Invest., 2013, 123, 3997–4009 CAS
.
- A. K. Sato, M. Viswanathan, R. B. Kent and C. R. Wood, Curr. Opin. Biotechnol, 2006, 17, 638–642 CrossRef CAS PubMed
.
- Z. Cheng, A. Al Zaki, J. Z. Hui, V. R. Muzykantov and A. Tsourkas, Science, 2012, 338, 903–910 CrossRef CAS PubMed
.
- K. S. Lam, S. E. Salmon, E. M. Hersh, V. J. Hruby, W. M. Kazmierski and R. J. Knapp, Nature, 1991, 354, 82–84 CrossRef CAS PubMed
.
- C. F. Cho, G. A. Amadei, D. Breadner, L. G. Luyt and J. D. Lewis, Nano Lett., 2012, 12, 5957–5965 CrossRef CAS PubMed
.
- C. F. Cho, B. Behnam Azad, L. G. Luyt and J. Lewis, ACS Comb. Sci., 2013, 15, 393–400 CrossRef CAS PubMed
.
- G. A. Amadei, C. F. Cho, J. D. Lewis and L. G. Luyt, J. Mass Spectrom., 2010, 45, 241–251 CrossRef CAS PubMed
.
- N. F. Steinmetz, A. L. Ablack, J. L. Hickey, J. Ablack, B. Manocha, J. S. Mymryk, L. G. Luyt and J. D. Lewis, Small, 2011, 7, 1664–1672 CrossRef CAS PubMed
.
- F. M. Brunel, J. D. Lewis, G. Destito, N. F. Steinmetz, M. Manchester, H. Stuhlmann and P. E. Dawson, Nano Lett., 2010, 10, 1093–1097 CrossRef CAS PubMed
.
- J. D. Lewis, G. Destito, A. Zijlstra, M. J. Gonzalez, J. P. Quigley, M. Manchester and H. Stuhlmann, Nat. Med., 2006, 12, 354–360 CrossRef CAS PubMed
.
- N. F. Steinmetz, C. F. Cho, A. Ablack, J. D. Lewis and M. Manchester, Nanomedicine, 2011, 6, 351–364 CrossRef CAS PubMed
.
- C. F. Cho, A. Ablack, H. S. Leong, A. Zijlstra and J. D. Lewis, J. Visualized Exp., 2011, 52, 2808 Search PubMed
.
- K. J. Koudelka, G. Destito, E. M. Plummer, S. A. Trauger, G. Siuzdak and M. Manchester, PLoS Pathog., 2009, 5, e1000417 Search PubMed
.
- G. Destito, R. Yeh, C. S. Rae, M. G. Finn and M. Manchester, Chem. Biol., 2007, 14, 1152–1162 CrossRef CAS PubMed
.
- N. F. Steinmetz and M. Manchester, Biomacromolecules, 2009, 10, 784–792 CrossRef CAS PubMed
.
- V. Hong, S. I. Presolski, C. Ma and M. G. Finn, Angew. Chem., Int. Ed., 2009, 48, 9879–9883 CrossRef CAS PubMed
.
- N. F. Steinmetz, D. J. Evans and G. P. Lomonossoff, ChemBioChem, 2007, 8, 1131–1136 CrossRef CAS PubMed
.
- H. S. Leong, N. F. Steinmetz, A. Ablack, G. Destito, A. Zijlstra, H. Stuhlmann, M. Manchester and J. D. Lewis, Nat. Protoc., 2010, 5, 1406–1417 CrossRef CAS PubMed
.
- C. F. Cho, S. Shukla, E. J. Simpson, N. F. Steinmetz, L. G. Luyt and J. D. Lewis, Methods Mol. Biol., 2014, 1108, 211–230 CAS
.
- H. S. Leong, A. E. Robertson, K. Stoletov, S. J. Leith, C. A. Chin, A. E. Chien, M. N. Hague, A. Ablack, K. Carmine-Simmen, V. A. McPherson, C. O. Postenka, E. A. Turley, S. A. Courtneidge, A. F. Chambers and J. D. Lewis, Cell Rep., 2014, 8, 1558–1570 CrossRef CAS PubMed
.
- E. Arpaia, H. Blaser, M. Quintela-Fandino, G. Duncan, H. S. Leong, A. Ablack, S. C. Nambiar, E. F. Lind, J. Silvester, C. K. Fleming, A. Rufini, M. W. Tusche, A. Brüstle, P. S. Ohashi, J. D. Lewis and T. W. Mak, Oncogene, 2012, 31, 884–896 CrossRef CAS PubMed
.
- A. Agrawal and M. Manchester, Biomacromolecules, 2012, 13, 3320–3326 CrossRef CAS PubMed
.
- N. F. Steinmetz, Viral nanoparticles as platforms for next-generation therapeutics and imaging devices, Nanomedicine, 2010, 6, 634–641 CrossRef CAS PubMed
.
- E. A. Anderson, S. Isaacman, D. S. Peabody, E. Y. Wang, J. W. Canary and K. Kirshenbaum, Nano Lett., 2006, 6, 1160–1164 CrossRef CAS PubMed
.
- S. Qazi, L. O. Liepold, M. J. Abedin, B. Johnson, P. Prevelige, J. A. Frank and T. Douglas, Mol. Pharm., 2013, 10, 11–17 CrossRef CAS PubMed
.
- S. Shukla and N. F. Steinmetz, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7, 708–721 CrossRef CAS PubMed
.
- T. F. Hansen, B. S. Nielsen, F. B. Sorensen, A. Johnsson and A. Jakobsen, Mol. Cancer Ther., 2014, 13, 2238–2245 CrossRef CAS PubMed
.
- K. Fosgerau and T. Hoffmann, Drug Discovery Today, 2015, 20, 122–128 CrossRef CAS PubMed
.
- A. A. Kaspar and J. M. Reichert, Drug Discovery Today, 2013, 18, 807–817 CrossRef CAS PubMed
.
- J. Thundimadathil, J. Amino Acids, 2012, 2012, 967347 Search PubMed
.
- E. V. Rosca, J. E. Koskimaki, C. G. Rivera, N. B. Pandey, A. P. Tamiz and A. S. Popel, Curr. Pharm. Biotechnol., 2011, 12, 1101–1116 CAS
.
- L. Guzman-Rojas, R. Rangel, A. Salameh, J. K. Edwards, E. Dondossola, Y. G. Kim, A. Saghatelian, R. J. Giordano, M. G. Kolonin, F. I. Staquicini, E. Koivunen, R. L. Sidman, W. Arap and R. Pasqualini, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 1637–1642 CrossRef CAS PubMed
.
- T. Boehm, J. Folkman, T. Browder and M. S. O'Reilly, Nature, 1997, 390, 404–407 CrossRef CAS PubMed
.
- J. Wellink, Methods Mol. Biol., 1998, 81, 205–209 CAS
.
- E. Lelièvre, A. Hinek, F. Lupu, C. Buquet, F. Soncin and V. Mattot, EMBO J., 2008, 27(12), 1658–1670 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Real-time intravital confocal microscopy showing E7p72-nanoparticle accumulation in vivo, purification of recombinant EGFL7, surface plasmon resonance analysis of FITC-labeled E7p72 binding to EGFL7, peptide uptake analysis in HT1080 cells using confocal microscopy, PET imaging using radiolabeled E7p72, real-time analysis of E7p72-nanoparticle distribution in quiescent vessels, and supplementary methodologies. See DOI: 10.1039/c7nr02558k |
| This journal is © The Royal Society of Chemistry 2017 |