In vitro delivery of calcium ions by nanogated mesoporous silica nanoparticles to induce cancer cellular apoptosis†
Derrick
Tarn‡
ab,
Chia-Jung
Yu‡
ab,
Jie
Lu‡
c,
Anna
Hartz
a,
Fuyuhiko
Tamanoi
cd and
Jeffrey I.
Zink
 *abd
*abd
aDepartment of Chemistry and Biochemistry, University of California, Los Angeles, California 90095, USA. E-mail: zink@chem.ucla.edu
bCalifornia NanoSystems Institute, University of California, Los Angeles, California 90095, USA
cDepartment of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, California 90095, USA
dJonsson Comprehensive Cancer Center, University of California, Los Angeles, California 90095, USA
First published on 7th August 2017
Abstract
A pH-operated nanogate on mesoporous silica nanoparticles (MSNs) capable of simultaneous dual-cargo delivery of both metal ions and organic cargo molecules was used to deliver both Hoechst 33342 nuclear stain and calcium ions to MiaPaCa-2 cancer cells. Delivery of calcium to the cell cytoplasm to induce cellular death is an attractive alternative to using traditional, more hazardous chemotherapy drugs. Treatment of cancer cells with Hoechst-loaded, calcium-latched nanogate-modified MSNs resulted in the autonomous release of both cargo types, as visualized by cell nuclei staining and calcium-induced apoptosis. Apoptosis was confirmed by observation of nuclear fragmentation, chromatin condensation, and western blotting for cleaved caspase-3.
Design, System, ApplicationA nanogated mesoporous silica nanoparticle system was designed to carry metal ions through aqueous and biological media at neutral pH and release the ions under acidic conditions. The system consists of nanoparticles with a total surface area of ∼1000 m2 g−1 that contain silanol groups available for grafting functional groups such as metal-binding ligands onto the surface. Synthetic pathways of ligand coverage were tested to optimize the number of bound metal ions, specifically calcium ions in this paper. The design considerations of the particles include small size (must be ∼100 nm in diameter or less), dispersibility in biological media, no intrinsic toxicity, and sensitivity to lysosomal pH (<6). This design enables many types of metal ions to be captured and released, and future applications may take advantage of this versatility for both therapeutic and imaging applications. The immediate application demonstrated in this paper is to kill cancer cells by releasing the calcium ions into their cytosol following endocytosis of the particles. A unique and important feature of this system is the possibility of cancer therapy without the use of harmful and costly anticancer drugs. |
Introduction
Mesoporous silica nanoparticles loaded with drugs and controlled by stimuli-responsive release mechanisms make it possible to deliver the drugs to cancer cells with no premature release.1–5 Ingenious methods of using external control6 (such as light,7,8 magnetic fields9–11 and ultrasound12) or autonomous control caused by cell-specific changes (such as pH,13–16 redox potential,17–19 enzymes20–22 and other biomolecules23,24) have been developed to reduce off-target release of drugs. Most cancer drugs have severe side effects.25–28 The goal of this research is to develop a method of delivering metabolically required innocuous metal ions into cells at high enough concentrations to disturb the function of specific enzymes or proteins or to destroy vital concentration gradients existing within cells and induce apoptosis. The metal ions should not be harmful to other cells even when released into the bloodstream and diluted after cell death. Specifically, delivering concentrated calcium ions into diseased cells would stimulate the apoptotic cycle, and cell death would occur without any adverse side effects on healthy cells.29 This treatment could be an alternative to using traditional, more hazardous and costly chemotherapy drugs.Calcium ions are essential to biological systems. Calcium storage is normally highly regulated inside cells and its concentration in the cytosol is kept at a low level.30 However, a large abundance of calcium in the cell cytoplasm induces a positive feedback loop that ultimately results in cell apoptosis.29,31 The challenge is how to carry Ca2+ into cells and release it there. It is too small to be trapped by most of the existing stimuli-responsive gating mechanisms. In prior work, we developed a gating mechanism we called a “swinging gate” that was composed of flexible chelating ligands attached to a pore surface.32 In the presence of a metal ion, two ligands bind the metal; when this chelation occurred over a pore opening, the binding resulted in “latching closed” the gate and prevented organic drug molecules from escaping. Acidification decreased the binding constant, unlatched the gate and resulted in drug release. The simultaneous release of the metal ion was considered a by-product of the desired release of the drug cargo.
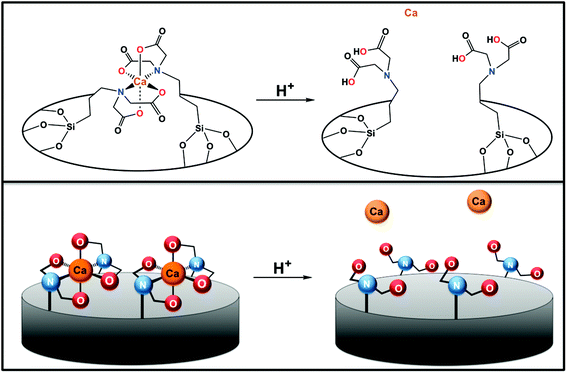
In this paper, we demonstrate that the decrease in pH from that of blood (pH 7.4) to that inside a lysosome after endocytosis of particles (pH <7) enables operation of the system33 and that particles designed to carry a large amount of Ca2+ efficiently induce apoptosis in cancer cells at very low particle concentrations. We make use of the calcium ion that latches the gate to induce apoptosis without using a toxic cargo (Fig. 1).
Experimental
Chemicals and characterization
Tetraethyl orthosilicate (TEOS; 99%, Aldrich), cetyltrimethylammonium bromide (CTAB; 98%, Aldrich), sodium hydroxide (Fisher), absolute ethanol (EtOH; Aldrich), methanol (Aldrich), hydrochloric acid (Fisher), dimethyl iminodiacetate (98%, Aldrich), 3-iodopropyltriethoxysilane (Gelest), N,N-diisopropylethylamine (Aldrich), CaCl2 (Aldrich), Tris buffer (Aldrich), Ca ICP standard (1000 ppm, Peak Performance) and Hoechst 33342 (2′-(4-ethoxyphenyl)-5-(4-methyl-1-piperazinyl)-2,5′-bi-1H-benzimidazole trihydrochloride; 98%, Aldrich) were used as-received. Anhydrous toluene was obtained by distillation from CaH2 under dry nitrogen. Moisture-sensitive reactions were carried out under an inert atmosphere of dry nitrogen.XRD measurements were made using a PANalytical X'Pert Pro powder diffractometer. Solid-state NMR was performed using a Bruker DSX300 spectrometer. TEM images were acquired using a Tecnai T12 microscope. IR measurements were made by using a JASCO Fourier transform infrared spectrophotometer. Hoechst release studies were conducted with an in-house time-resolved fluorescence spectroscopy setup. A Coherent 377 nm CUBE laser was used to excite the Hoechst dye, and the subsequent emission spectra were monitored using a Princeton Instruments Roper CCD detector cooled to liquid nitrogen temperature. ICP-OES measurements were made using an ICPE-9000 Shimadzu.
Synthesis of MCM-41 nanoparticles
NaOH solution (2 M, 870 μL, 1.7 mmol) and cetyltrimethylammonium bromide (CTAB, 0.25 g, 0.7 mmol) were mixed with deionized water (120 mL) in a 250 mL round bottom flask equipped with a magnetic stir bar. The temperature was kept constant at 80 °C for 1 hour and the stirring rate was 700 rpm. Tetraethyl orthosilicate (TEOS, 1.2 mL, 5.4 mmol) was added dropwise under continuous stirring for 2 hours at 80 °C. The solution color changed from clear to milky white. The solution was cooled down to room temperature and MSNs were collected by centrifugation (7830 rpm, 15 min). The MSNs were washed with methanol and distilled water until the washings showed a pH of 7. To extract the surfactant, the MSNs (120 mg) were suspended in methanol (75 mL) in a 250 mL round bottom flask and kept at room temperature under sonication and stirring. Concentrated HCl (12 M, 4.2 mL) was added to the suspension and the solution was refluxed for 6 hours. The MSNs were recollected by centrifugation (7830 rpm, 15 min) and washed with methanol and distilled water several times. Complete extraction of CTAB from the pores was confirmed through IR spectroscopy.Method I synthesis of (dimethyl iminodiacetate)-propyltriethoxysilane nanogated particles
Dimethyl iminodiacetate (1 mL, 6 mmol) and 3-iodopropyltriethoxysilane (1 mL, 5 mmol) were dissolved in anhydrous toluene (5 mL) in a 50 mL three-necked round bottom flask equipped with a reflux condenser, a N2 gas inlet, an outlet for gas, and a magnetic stir bar. Anhydrous toluene was prepared under N2 and refluxed with CaH2 for two to three hours. Catalytic amounts of N,N-diisopropylethylamine (30 μL) were added and the mixture was allowed to react under dry N2 overnight at reflux. In order to drive the reaction to completion, the mole ratio of dimethyl iminodiacetate and 3-iodopropyltriethoxysilane needed to be more than 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
In a 50 mL round bottom flask, 50 mg of MSNs that were washed with anhydrous toluene (2 × 10 mL) were suspended in anhydrous toluene dried over calcium hydride (10 mL). The as-synthesized (dimethyl iminodiacetate)-propyltriethoxysilane was added to the MSN suspension and heated overnight at reflux temperature. The functionalized particles were collected by centrifugation (7830 rpm, 10 min) and washed with toluene (2 × 10 mL) and methanol (2 × 10 mL).
To hydrolyze the ester to form the desired iminodiacetic acid-modified nanoparticles (IDA-MSNs), the dimethyl iminodiacetate-modified silica nanoparticles (50 mg) were suspended in doubly distilled water (50 mL) followed by slow addition of concentrated HCl (3 mL) to the reaction mixture. The mixture was allowed to reflux for 6 h. The particles were collected by centrifugation (7830 rpm, 10 min) and washed with methanol and distilled water until the pH of the washings was neutral.
Method II synthesis of (dimethyl iminodiacetate)-propyltriethoxysilane nanogated particles
For the attempt to increase the surface modification efficiency, the grafting of 3-iodopropyltrimethoxysilane (1 mL, 5 mmol) onto the MSNs was conducted first under dry N2 at 70 °C for 5.5 h. The idea was to minimize the polymerization of (dimethyl iminodiacetate)-propylsilane, which is synthesized during the nucleophilic substitution, and thus increase the grafting efficiency. The iodo group-modified MSNs were collected by centrifugation and washed with anhydrous toluene (5 × 10 mL). 3-Iodopropyltrimethoxysilane is sensitive to moisture in the air. If the reaction temperature exceeded 75 °C, the iodo groups degraded and a purple I2 precipitate was formed. To form the desired product, nucleophilic substitution was conducted. Dimethyl iminodiacetate (1 mL, 6 mmol) and catalytic amounts of N,N-diisopropylethylamine (30 μL) were added to a suspension of the modified MSNs in anhydrous toluene. The reaction was carried out at 70 °C under dry N2 for 8 h.Loading and containment of Hoechst 33342
IDA-MSNs (10 mg) were washed twice with methanol and doubly distilled water. The IDA-MSNs were centrifuged and suspended in a fresh solution of Hoechst 33342 in water (1 mM, 1 mL) and the mixture was stirred overnight to allow the dye to diffuse into the pores of the particles. The Hoechst 33342-loaded IDA-MSNs were collected by centrifugation.To close the nanogates on the IDA-MSNs with Ca2+, the Hoechst 33342-loaded IDA-MSNs were stirred in a buffered CaCl2 solution (300 μL, 160 mg mL−1, Tris buffer pH 8) overnight. The closed IDA-MSNs were collected by centrifugation (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min) and washed with doubly distilled water until the washing no longer exhibited fluorescence under UV light.
000 rpm, 10 min) and washed with doubly distilled water until the washing no longer exhibited fluorescence under UV light.
Measurement of dye release profiles activated by pH change
The release of the Hoechst 33342 dye from the particles was monitored by fluorescence spectroscopy. A solid bolus of particles was placed on one side of the bottom of a cuvette and a small stir bar was placed on the opposite side. The cuvette was slowly and carefully filled with doubly distilled water with minimal disturbance of the particles. The solution was stirred very slowly to distribute any escaped dye molecules throughout the cuvette. A 377 nm diode laser beam (14 mW) was directed through the solution above the sample to excite the dye that diffused out of the pores when the gate was opened. The emission spectrum and intensity of excited Hoechst dye fluorescence were monitored in real time using a monochromator with a CCD detector. A baseline was collected for over 2 hours to monitor any leakage of dye from the particles and any particles that became suspended due to stirring. After a stable baseline was established, a known quantity of dilute HCl was added to decrease the solution pH to 5. The fluorescence intensity as a function of time was plotted to measure the amount of dye released after the nanogate was opened. After complete release (i.e., when the intensity leveled off), the supernatant was collected by centrifugation (7830 rpm, 25 min) and analyzed through UV-vis to determine the weight percent release of the Hoechst dye.Loading and release of calcium ions from IDA-MSNs
To load the IDA-MSNs with Ca2+ ions, 10 mg IDA-MSNs were stirred in a buffered CaCl2 solution (300 μL, 160 mg mL−1, Tris buffer pH 8) overnight. To collect the calcium-loaded IDA-MSNs, the MSNs were centrifuged down and then washed with 1.5 mL Millipore water four times. The collected calcium-loaded IDA-MSNs were re-suspended in 1 mL Millipore water. To obtain the uptake capacity, the calcium ion concentration of the loading solution before and after centrifugation was converted into mass and was divided by the mass of MSNs.The triggered calcium ion release was measured by suspending the calcium-loaded IDA-MSNs in 1.5 mL pH 5 HCl solution overnight. Particles were centrifuged down and the amount of calcium ions in the resulting supernatant was determined by ICP-OES. 1000 ppm Ca standard solution was used to make standard solutions of 0.1 to 50 ppm for generating a calibration curve.
1 mL of each centrifuged supernatant including those obtained after the loading and washing steps was diluted 10 times with 2% HNO3 to make 10 mL solutions for ICP-OES analysis. The release capacity was then defined as the mass of calcium divided by that of MSNs.
Cell culture
Human cancer-cell line MiaPaca-2 was obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium (DMEM; GIBCO) or RPMI-1640 medium (Cellgro) supplemented with 10% fetal calf serum (Sigma), 2% L-glutamine, 1% penicillin, and 1% streptomycin stock solutions. The media were changed every three days, and the cells were passaged by trypsinization before confluence.Fluorescence and confocal microscopy
Cells were cultured overnight on a Lab-Tek chamber slide system (Nalge Nunc International). After the cells were incubated with nanoparticles for 3 hours, they were washed with PBS and examined via fluorescence microscopy (Carl Zeiss).Cell death assay
The cytotoxicity assay was performed by using a cell counting kit from Dojindo Molecular Technologies, Inc. Cells were seeded in 96-well plates (5000 cells per well) and incubated in fresh culture medium at 37 °C in a 5% CO2/95% air atmosphere for 24 h. The cells were then washed with PBS and the medium was changed to a fresh medium containing Hoechst-loaded, calcium-latched IDA-MSNs, a control (IDA-modified MSNs), or a second control (IDA-modified MSNs loaded with Hoechst but unlatched) at the indicated concentrations. After 24 h, the cells were washed with PBS to remove nanoparticles that were not taken up by the cells, and the cells were then incubated in fresh medium for an additional 48 h. The cells were washed with PBS and incubated in DMEM with 10% WST-8 solution for another 2 h. The absorbance of each well was measured at 450 nm with a plate reader. Because the absorbance is proportional to the number of viable cells in the medium, the viable cell number was determined by using a previously prepared calibration curve (Dojindo Co.).Apoptosis assay
Cell death was also examined by using the propidium iodide and Hoechst 33342 double-staining method. The cells were stained with propidium iodide/Hoechst 33342 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 5 min and then examined using fluorescence microscopy. For the western blot assay, proteins collected from cell lysates were separated by gel electrophoresis on a polyacrylamide gel containing SDS and then transferred to nitrocellulose membranes. The membranes were blocked with Tris-buffered saline (TBS) containing 5% (w/v) skimmed milk. After being washed with TBS containing 0.1% Tween 20 (Sigma), the membranes were incubated overnight at room temperature with caspase-3 (BD Biosciences) and cleaved caspase-3 antibodies (Cell Signaling) diluted with TBS. After being washed, the membranes were incubated for 2 h at room temperature with the second antibody (Santa Cruz Biotechnology, CA). Bands were detected with an ECL system (Amersham Pharmacia Biotech K.K., UK).
1) for 5 min and then examined using fluorescence microscopy. For the western blot assay, proteins collected from cell lysates were separated by gel electrophoresis on a polyacrylamide gel containing SDS and then transferred to nitrocellulose membranes. The membranes were blocked with Tris-buffered saline (TBS) containing 5% (w/v) skimmed milk. After being washed with TBS containing 0.1% Tween 20 (Sigma), the membranes were incubated overnight at room temperature with caspase-3 (BD Biosciences) and cleaved caspase-3 antibodies (Cell Signaling) diluted with TBS. After being washed, the membranes were incubated for 2 h at room temperature with the second antibody (Santa Cruz Biotechnology, CA). Bands were detected with an ECL system (Amersham Pharmacia Biotech K.K., UK).
Results and discussion
1. Synthesis of nanoparticles with gates latched by calcium ions
Iminodiacetic acid (IDA) has chelating properties similar to those of one-half of an ethylenediaminetetraacetic acid (EDTA) molecule (Fig. 1).34 When IDA is attached to silica with a flexible linker, two neighboring IDA molecules can bind strongly to a metal ion. The calculated size of fully stretched calcium cross-linked IDA is about 1.8 nm.32 It is probable that multiple IDA molecules are attached around the pore entrances; there are at least six available silanols around a perfectly shaped pore and likely more around a roughened pore entrance. One IDA cannot perfectly cover the exact center of a pore, but one or more located over the pore entrance can easily block cargos. The binding constant of IDA with various metal centers exhibits a ∼103-fold decrease upon changing the pH from neutral to 5 as a result of protonation of the carboxylate chelating groups.35 This property is the basis for the mechanism of operation of pH-responsive IDA nanovalves. A wide variety of metal ions can be used; in this paper, Ca2+ is chosen because it is a nutritional requirement at normal physiological concentrations but is toxic to cells at high intracellular concentrations. It has a binding constant to IDA on the order of 102 and can be released when the ligand is protonated at ∼pH 4–6.The MCM-41 MSNs necessary for the calcium delivery platform were synthesized according to published methods.32 In a typical synthesis of 100 nm MCM-41 MSNs, the base-catalyzed sol–gel synthesis was conducted at pH 12 and 80 °C. As shown in Fig. S1a and b,† the average size of the MSNs as measured from the TEM images was between 90–110 nm and thus in the desired size range for biomedical applications. The pore size was estimated to be about 2 nm. In Fig. S1c,† the hexagonal arrangement of pores in the MSNs, which can be seen in the TEM images, was also confirmed by small-angle X-ray scattering. The data show the typical 2D hexagonal (p6mm) pattern of MCM-41. The reflections can be indexed to (100), (110), (200) and (210) planes. The d-spacing is 4.2 nm.
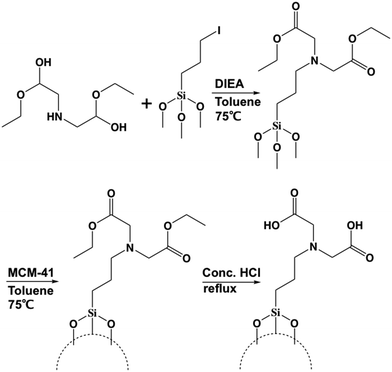
In order to modify the MSN surface with IDA, the first step is to synthesize (diethyl iminodiacetate)-trimethylsilane via nucleophilic substitution. The second step is to modify the MSN surface with the (diethyl iminodiacetate)-trimethylsilane from the first step. The third step is to convert the ester groups on the (diethyl iminodiacetate)-trimethylsilane to carboxylic acid groups. The final iminodiacetic acid-modified MSNs (IDA-MSNs) were formed. The scheme is shown in Fig. 2.
The successful surface modification of MSNs with IDA groups was confirmed by FT-IR as shown in Fig. S2.† Noticeable in the spectrum of the MSNs with the (diethyl iminodiacetate)propyl moiety (blue line) are the aliphatic C–H stretching vibrations of the ethyl ester groups at 2978 and 2937 cm−1 and the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration at 1732 cm−1. The disappearance of the aliphatic C–H stretching vibrations in the spectrum of the IDA-functionalized particles (pink line) and the remaining C
O stretching vibration at 1732 cm−1. The disappearance of the aliphatic C–H stretching vibrations in the spectrum of the IDA-functionalized particles (pink line) and the remaining C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration indicate the successful conversion of the ester groups to free carboxylic acids. The successful functionalization was further supported by solid-state 13C–{1H}-CP-MAS-NMR. The spectrum is shown in Fig. S3.† The carbonyl peak appears at 168 ppm, the corresponding α-carbon peak at 46 ppm and the aliphatic carbon peaks between −3 and 50 ppm.
O stretching vibration indicate the successful conversion of the ester groups to free carboxylic acids. The successful functionalization was further supported by solid-state 13C–{1H}-CP-MAS-NMR. The spectrum is shown in Fig. S3.† The carbonyl peak appears at 168 ppm, the corresponding α-carbon peak at 46 ppm and the aliphatic carbon peaks between −3 and 50 ppm.
To optimize the IDA surface coating of the particles and therefore the amount of calcium ions that can be loaded onto the particles, two functionalization syntheses were explored. The first procedure utilized the functionalization pathway discussed above. The second functionalization procedure involved first the modification of the surface of the MSNs with the linker, 3-iodopropyltrimethoxysilane, and then attachment of dimethyl iminodiacetate by nucleophilic substitution as shown in Fig. S4.† The idea was to minimize the polymerization of (diethyl iminodiacetate)-propylsilane and thus increase the grafting efficiency. As will be discussed in the following section, the first procedure produced particles that bound the largest amount of calcium ions, and these particles were used in all of the following studies.
2. Binding and release of calcium ions and Hoechst dye molecules
In order to test the capability of IDA-MSNs to carry Ca2+, the IDA-MSNs were suspended in a CaCl2 solution overnight. After the calcium ions were chelated, the loaded IDA-MSNs were collected by centrifugation and washed with water several times to make sure that the adsorbed Ca2+ was washed off as shown in Fig. S5.† The particles were suspended in pH 5 HCl solution overnight to release all of the Ca2+ and the amount of the released ions was measured by ICP-OES. The nanoparticles synthesized via the first functionalization pathway had an average release capacity of 0.42 μmol per mg of particles with a maximum in one of the batches of 0.54 μmol per mg of particles (Table S1†). Nanoparticles that were synthesized via the second pathway had a much smaller average release capacity of 0.17 μmol per mg of particles. The former particles were chosen for all of the following studies.3. Calcium-latched Hoechst-loaded IDA-MSNs
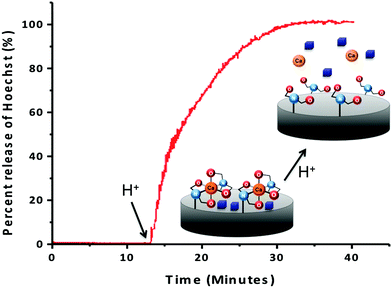
To confirm that the IDA groups on the MSNs' surfaces coordinated to calcium can trap and release cargo molecules from the pores, the IDA-MSNs were loaded with Hoechst 33342, a dye commonly used as a nuclear stain in cells that was chosen as the probe due to its ease of detection by fluorescence spectroscopy. After suspending the IDA-MSNs in a concentrated solution of the dye overnight to allow the dye to diffuse into the pores, the Hoechst-loaded IDA-MSNs were latched shut by adding the calcium ions. Introducing the Ca2+ ion latches the nanogates together by bringing two IDA units together (Fig. 3). After the pores of the MSNs were shut, the excess dye molecules adsorbed on the MSNs' surfaces and excess calcium ions were washed away with fresh doubly distilled water until the supernatant no longer exhibited any fluorescence.In order to monitor the function of the IDA nanogates, a real-time release profile of Hoechst dye from Ca2+ ion-latched IDA-MSNs was generated. The fully assembled and latched nanoparticles were placed in a corner of a glass cuvette and an excitation beam (377 nm) was used to excite any dye released into the supernatant. The corresponding emission spectrum was collected to generate a time-resolved release profile. As shown in Fig. 3, before adding the acidic solution into the cuvette, no dye was released into the supernatant. However, after acidification to trigger the release, the intensity went up. To verify whether the calcium latch is necessary to hold back the Hoechst cargo, a control experiment was performed with IDA-MSNs loaded with Hoechst but unlatched with calcium ions. However, after being subjected to the same washing steps as those for the calcium-latched IDA-MSNs, the Hoechst molecules were washed out. When being acidified to pH 5, only a barely noticeable absorbance was observed by UV-vis.
4. In vitro autonomous pH-activated release of cargo
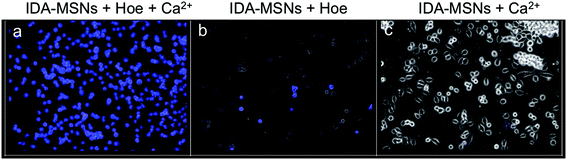
To test the autonomous release of the cargo triggered by the pH of the IDA-MSNs, MiaPaCa-2 cells were treated for three hours with a sample of IDA-modified MSNs loaded with the Hoechst 33342 nuclear stain latched with calcium. As shown in Fig. 4, the lysosomal pH causes cargo release as illustrated by the blue fluorescence nuclear staining. To verify whether the calcium latch is necessary to hold back the Hoechst cargo, a control experiment was performed where the cells were treated with IDA-MSNs loaded with Hoechst but unlatched. In this sample, no Hoechst nuclear staining was observed because all of the Hoechst cargo had been washed out of the nanoparticle pores during the washing steps (Fig. 4b). This indicates that the calcium latch is responsible for successful cargo trapping inside the MSN pores. Cells treated with this sample show no fluorescence staining, confirming that the observed fluorescence in Fig. 4a is due to released Hoechst. It is noteworthy to mention that signs of cellular apoptosis were present in the cells treated with calcium-latched IDA-MSNs. In this Hoechst-stained sample, bright, condensed fluorescence was observed in a number of treated cells (Fig. 4a), signifying collapsed nuclei and indicating the progression of cell death through an apoptotic mechanism.5. Inhibition of lysosomal acidification and suppression of cargo release
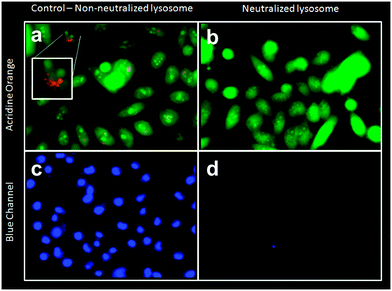
The pH-activated mechanism of release was further corroborated by testing for cargo release after inhibiting acidification of the cells' lysosomes. MiaPaCa-2 cells were stained with acridine orange, a pH-indicating dye, before treatment with bafilomycin A1 to neutralize the lysosomes. In Fig. 5, the neutral cellular components of cells stained with acridine orange appear with green fluorescence (Fig. 5b), while the acidic lysosomal compartments appear orange/red under confocal microscopy (Fig. 5a). Once treated with bafilomycin A1, the cells appear uniform with green fluorescence (Fig. 5b), indicating that the acidification of lysosomes has been inhibited. Dosing the pH-neutralized cells with IDA-MSNs loaded with Hoechst and latched with calcium resulted in very little nuclear staining (Fig. 5d). Dosing a control group of cells not treated with bafilomycin with the same group of IDA-MSNs resulted in Hoechst nuclear staining (Fig. 5c). These experiments indicated that the inhibition of acidification of the cell lysosomes blocked the release of Hoechst and that the cell lysosomal compartments must be acidic in order to autonomously release the cargo from loaded IDA-MSNs, in agreement with our solution study results.6. Apoptosis induced by intracellular calcium ion release
The large abundance of calcium delivered by IDA-MSNs into the cell cytoplasm induces a positive feedback loop that ultimately results in cell apoptosis (Fig. 6).29 The release of calcium into the cytoplasm causes mitochondrial release of cytochrome C. Upon finding its way to the endoplasmic reticulum (ER) where the majority of intracellular calcium is stored, binding of cytochrome C to receptors on the ER causes endoplasmic release of calcium, resulting in additional cytochrome C release from the mitochondria. This positive feedback loop results in toxic concentrations of cytoplasmic calcium, ultimately causing the formation of an apoptosome and cell death.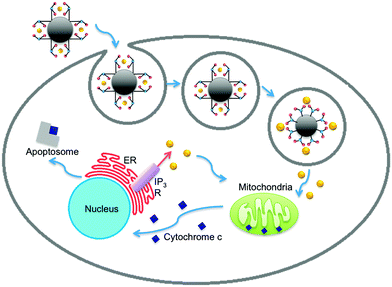 | ||
| Fig. 6 The role of calcium in orchestrating cellular apoptosis.29–31 Release of the death receptor Bid or influx of calcium into the cell cytoplasm causes the mitochondria to release cytochrome C. Some of the released cytochrome C ultimately binds to receptors on the endoplasmic reticulum (ER), causing an outflux of calcium to the cytoplasm. An increased presence of calcium in the cytoplasm causes the mitochondria to release additional cytochrome C, generating a positive feedback loop. When calcium concentrations reach cytotoxic levels, an apoptosome is assembled which leads to cell apoptosis. | ||
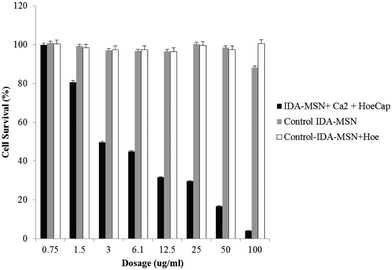
The cell viability assay was conducted using a cell counting kit. Cells were seeded in 96-well plates and incubated in fresh culture medium. The cells were then washed with PBS and the medium was changed to a fresh medium containing Hoechst-loaded, calcium-latched IDA-MSNs or a control (IDA-modified MSNs) or a second control (IDA-MSNs loaded with Hoechst but unlatched) at the indicated concentrations. After twenty-four hours, the cells were washed with PBS to remove nanoparticles that were not taken up by the cells, and the cells were incubated in fresh medium for an additional forty-eight hours. The cells were then washed with PBS and incubated in DMEM for another two hours. The absorbance of each well was measured at 450 nm with a plate reader.
While control samples of unlatched Hoechst 33342-loaded IDA-MSNs and plain IDA-MSNs showed no toxicity within the tested dosage range, a sample of Hoechst-loaded, calcium-latched IDA-MSNs demonstrated a high amount of dose-dependent cell killing (Fig. 7). Exclusion of calcium from the control samples resulted in no cell toxicity, suggesting that the observed cell killing is due to controlled delivery and release of calcium into the cytosol. It is important to note that the chelating properties of unloaded IDA-MSNs do not cause cell toxicity even at high dosages (100 μg mL−1), making this design a viable option for pharmaceutical applications.
7. Verification of apoptosis
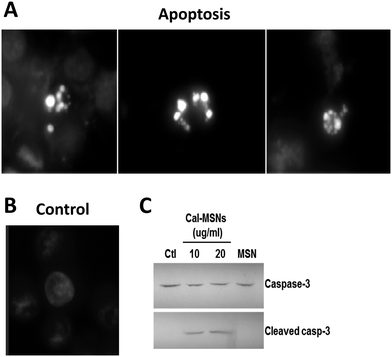
The cells treated with calcium-latched IDA-MSNs above were examined for apoptosis by two methods: nuclear double staining with propidium iodide and Hoechst 33342 solution and western blotting for cleaved caspase-3 expression. For the first method, the cells were stained with propidium iodide/Hoechst 33342 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 5 min and then examined by fluorescence microscopy. As shown in Fig. 8A, nuclear fragmentation and chromatin condensation were observed in the cells treated with calcium-latched IDA-MSNs. For the western blot assay, proteins collected from cell lysates were separated by gel electrophoresis on a polyacrylamide gel containing SDS and then transferred to nitrocellulose membranes. The membranes were probed with caspase-3 and cleaved caspase-3 antibodies. As can be seen in Fig. 8B, the appearance of a cleaved caspase-3 band was detected after the treatment with calcium-latched IDA-MSNs.
1) for 5 min and then examined by fluorescence microscopy. As shown in Fig. 8A, nuclear fragmentation and chromatin condensation were observed in the cells treated with calcium-latched IDA-MSNs. For the western blot assay, proteins collected from cell lysates were separated by gel electrophoresis on a polyacrylamide gel containing SDS and then transferred to nitrocellulose membranes. The membranes were probed with caspase-3 and cleaved caspase-3 antibodies. As can be seen in Fig. 8B, the appearance of a cleaved caspase-3 band was detected after the treatment with calcium-latched IDA-MSNs.
Summary
IDA-modified MSNs were engineered and used to deliver both calcium and Hoechst 33342 nuclear stain to MiaPaCa-2 cancer cells. Treatment of the cells with IDA-MSNs resulted in lysosomal localization of the nanoparticles. The intrinsic low lysosomal pH allows for the autonomous release of both cargo types, as visualized by Hoechst nuclear staining and calcium-induced apoptosis. The delivery of calcium to cancer cells resulted in cancer cell killing at low particle concentrations while control particles that lacked calcium were found to be non-toxic. This design opens the possibility of a safer alternative to using traditional toxic and costly chemotherapy drugs.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grant NIH R01Ca133677 (DT, FT, JL and JIZ) and the UCLA Zink Student and Research Support fund (C-JY).References
- Z. Li, J. C. Barnes, A. Bosoy, J. F. Stoddart and J. I. Zink, Chem. Soc. Rev., 2012, 41, 2590 RSC.
- C. Argyo, V. Weiss, C. Bräuchle and T. Bein, Chem. Mater., 2014, 26, 435–451 CrossRef CAS.
- M. Vallet-Regí, F. Balas and D. Arcos, Angew. Chem., Int. Ed., 2007, 46, 7548–7558 CrossRef PubMed.
- F. Tang, L. Li and D. Chen, Adv. Mater., 2012, 24, 1504–1534 CrossRef CAS PubMed.
- Y. Chen, H. Chen and J. Shi, Adv. Mater., 2013, 25, 3144–3176 CrossRef CAS PubMed.
- B. Ruehle, P. Saint-Cricq and J. I. Zink, ChemPhysChem, 2016, 17, 1769–1779 CrossRef CAS PubMed.
- T. M. Guardado-Alvarez, L. S. Devi, J.-M. Vabre, T. A. Pecorelli, B. J. Schwartz, J.-O. Durand, O. Mongin, M. Blanchard-Desce and J. I. Zink, Nanoscale, 2014, 6, 4652 RSC.
- J. Liu, W. Bu, L. Pan and J. Shi, Angew. Chem., Int. Ed., 2013, 52, 4375–4379 CrossRef CAS PubMed.
- P. Saint-Cricq, S. Deshayes, J. I. Zink and A. M. Kasko, Nanoscale, 2015, 7, 13168–13172 RSC.
- A. H. Lu, E. L. Salabas and F. Schüth, Angew. Chem., Int. Ed., 2007, 46, 1222–1244 CrossRef CAS PubMed.
- B. Rühle, S. Datz, C. Argyo, T. Bein and J. I. Zink, Chem. Commun., 2016, 52, 1843–1846 RSC.
- J. L. Paris, M. V. Cabanas, M. Manzano and M. Vallet-Regí, ACS Nano, 2015, 9, 11023–11033 CrossRef CAS PubMed.
- M. Xue, X. Zhong, Z. Shaposhnik, Y. Qu, F. Tamanoi, X. Duan and J. I. Zink, J. Am. Chem. Soc., 2011, 133, 8798–8801 CrossRef CAS PubMed.
- H.-Y. Chiu, W. Deng, H. Engelke, J. Helma, H. Leonhardt and T. Bein, Sci. Rep., 2016, 6, 25019 CrossRef CAS PubMed.
- F. Yu, X. Tang and M. Pei, Microporous Mesoporous Mater., 2013, 173, 64–69 CrossRef CAS.
- J. Sun, C. Hong and C. Pan, J. Phys. Chem. C, 2010, 114, 12481–12486 CAS.
- B. Y. Lee, Z. Li, D. L. Clemens, B. J. Dillon, A. A. Hwang, J. I. Zink and M. A. Horwitz, Small, 2016, 12, 3690–3702 CrossRef CAS PubMed.
- T. M. Guardado-alvarez, L. S. Devi, M. M. Russell, B. J. Schwartz and I. Zink, J. Am. Chem. Soc., 2013, 135, 14000–14003 CrossRef CAS PubMed.
- M. W. Ambrogio, T. A. Pecorelli, K. Patel, N. M. Khashab, A. Trabolsi, H. A. Khatib, Y. Y. Botros, J. I. Zink and J. F. Stoddart, Org. Lett., 2010, 12, 3363–3366 CrossRef PubMed.
- A. Popat, S. B. Hartono, F. Stahr, J. Liu, S. Z. Qiao and G. Qing Lu, Nanoscale, 2011, 3, 2801 RSC.
- C. Hu, P. Huang, Z. Zheng, Z. Yang and X. Wang, Langmuir, 2017, 33, 5511–5518 CrossRef CAS PubMed.
- J. Liu, B. Zhang, Z. Luo, X. Ding, J. Li, L. Dai, J. Zhou, X. Zhao, J. Ye and K. Cai, Nanoscale, 2015, 7, 3614–3626 RSC.
- B. Ruehle, D. L. Clemens, B.-Y. Lee, M. A. Horwitz and J. I. Zink, J. Am. Chem. Soc., 2017, 139, 6663–6668 CrossRef CAS PubMed.
- S. H. van Rijt, D. A. Bölükbas, C. Argyo, S. Datz, M. Lindner, O. Eickelberg, M. Königshoff, T. Bein and S. Meiners, ACS Nano, 2015, 9, 2377–2389 CrossRef CAS PubMed.
- N. Pabla and Z. Dong, Kidney Int., 2008, 73, 994–1007 CrossRef CAS PubMed.
- S. Miura, M. Maemondo, A. Iwashima, T. Harada, S. Sugawara, K. Kobayashi, A. Inoue, T. Nakagawa, Y. Takiguchi, H. Watanabe, T. Ishida, M. Terada, H. Kagamu, A. Gemma and H. Yoshizawa, Invest. New Drugs, 2017, 35, 227–234 CrossRef CAS PubMed.
- D. Schmid, G. E. Jarvis, F. Fay, D. M. Small, M. K. Greene, J. Majkut, S. Spence, K. M. McLaughlin, K. D. McCloskey, P. G. Johnston, A. Kissenpfennig, D. B. Longley and C. J. Scott, Cell Death Dis., 2014, 5, e1454 CrossRef CAS PubMed.
- K. Chatterjee, J. Zhang, N. Honbo and J. S. Karliner, Cardiology, 2010, 115, 155–162 CrossRef CAS PubMed.
- M. P. Mattson and S. L. Chan, Nat. Cell Biol., 2003, 5, 1041–1043 CrossRef CAS PubMed.
- S. Orrenius, B. Zhivotovsky and P. Nicotera, Nat. Rev. Mol. Cell Biol., 2003, 4, 552–565 CrossRef CAS PubMed.
- P. Pinton, Oncogene, 2010, 27, 6407–6418 CrossRef PubMed.
- D. Tarn, M. Xue and J. I. Zink, Inorg. Chem., 2013, 52, 2044–2049 CrossRef CAS PubMed.
- C. C. Scott and J. Gruenberg, BioEssays, 2011, 33, 103–110 CrossRef CAS PubMed.
- S. Chaberek Jr. and A. E. Martell, J. Am. Chem. Soc., 1952, 74, 5052–5056 CrossRef.
- H. Irving and D. H. Mellor, J. Chem. Soc., 1962, 5222–5237 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7me00046d |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2017 |