Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploring the links between peptoid antibacterial activity and toxicity†‡
H. L.
Bolt
a,
G. A.
Eggimann
a,
C. A. B.
Jahoda
b,
R. N.
Zuckermann
c,
G. J.
Sharples
*b and
S. L.
Cobb
 *a
*a
aDepartment of Chemistry, Durham University, South Road, Durham, DH1 3LE, UK. E-mail: s.l.cobb@durham.ac.uk
bSchool of Biological and Biomedical Sciences, Durham University, Durham DH1 3LE, UK. E-mail: gary.sharples@durham.ac.uk
cMolecular Foundry, Lawrence Berkeley National Laboratory, Berkeley, California, USA
First published on 1st February 2017
Abstract
Peptoids are a promising class of antimicrobial agents with reported activities against a range of both Gram-positive and Gram-negative bacteria, fungi and most recently parasites. However, at present the available toxicity data is somewhat limited and as such rationally designing effective antimicrobial peptoids can be challenging. Herein, we present the toxicity profiling of a series of linear peptoids against mammalian cell lines (HaCaT and HepG2). The cytotoxicity of the peptoid library has then been correlated with their antibacterial properties against Gram-positive and Gram-negative bacteria and also to the hydrophobicity of the peptoid sequences. The work presented provides valuable data to aid in the future rational design of antimicrobial peptoids.
The growing prevalence of antibiotic resistance has intensified demand for novel antimicrobials to replace or complement existing treatments for infectious diseases. Recent incentives such as the 10 × '20 Initiative and the Global Antimicrobial Resistance Research Innovation Fund encourage investment and a commitment to the development of new antibacterial drugs.1 Given the effectiveness of the innate immune system in providing the first line of defence against infection, considerable research effort has focused on investigating the activities of antimicrobial peptides (AMPs), with a view to their deployment as templates for innovative therapeutic design.2–5 AMPs typically contain fewer than 50 amino acids, are cationic and play a fundamental role in host defence, functioning as both antimicrobial agents and modulators of the inflammatory response.6–11 AMPs display potent antimicrobial activity against a range of clinically important pathogens, including bacterial, fungal and parasitic species.12–16 However, despite promising biological properties, AMPs are highly susceptible to degradation by host proteinases, which has hindered their exploitation as novel therapeutics including limited success in clinical trials.2–8,15–19
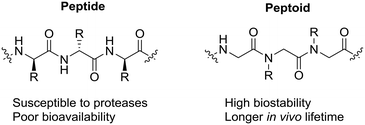
In the search for peptidomimetics that retain potent antimicrobial activity yet also exhibit enhanced proteolytic stability, α-peptoids (N-substituted glycines) have emerged as highly promising candidates. In α-peptoids, the side chain of each residue (monomer) is located on the nitrogen in the amide backbone (Fig. 1). This allows peptoids to keep many of the advantageous properties of AMPs (e.g. amphilicity) but, with the inclusion of a tertiary amide backbone, significantly improve their resistance to enzymatic degradation.20
Peptoids have been shown to have potency against a wide variety of Gram-positive and negative bacteria,21–30 parasites31–33 and fungi.25,34–36 These studies highlight the clear potential that peptoids offer as a new class of antimicrobials but in order to progress their clinical development a more detailed study of their toxicity towards mammalian cells needs to be undertaken.
Peptoid toxicity is often evaluated by haemolytic activity, however, these assays cannot necessarily be used to predict toxicity more generally against mammalian cells. For example, many peptoids in the literature are based upon peptoid (NLysNspeNspe)4 (compound 13, Table 1) which exhibits excellent antimicrobial properties, with a reported selectivity ratio of >6 and acceptable 50% haemolytic dose (HD50 of 100 μM).23 However, more recently, another publication reports in vitro mammalian assays and observed toxicity of peptoid 13 at 5.1 μM against NIH 3T3 murine fibroblasts.37 The Olsen group have also demonstrated that α-peptide β-peptoid hybrids show negligible haemolytic activity, but cause severe membrane alterations to human erythrocytes at low concentrations via microscopy.31 Therefore, there is a clear need to scrutinise the relationship between peptoid antimicrobial activity and their toxicity towards mammalian cells in greater detail.
| Peptoid sequence | Ref. | ED50 (μM) | MIC (μM) | Average selectivity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HaCaT | HepG2 | E. coli | P. aeruginosa | S. aureus | S. epidermidis | E. coli | P. aeruginosa | S. aureus | S. epidermidis | |||
| (NahNpheNphe)4 | 1 | 33 | >100 | >100 | 13 | >100 | 2 | 6 | 8 | 1 | 50 | 17 |
| (NahNpheNphe)3 | 2 | 33 | >100 | >100 | 50 | >100 | 6 | 3 | 2 | 1 | 17 | 33 |
| (NahNpheNphe)2 | 3 | 33 | >100 | >100 | >100 | >100 | >100 | >100 | 1 | 1 | 1 | 1 |
| (NLysNpheNphe)4 | 4 | 33, 37 | 36 | >100 | 13 | 50 | 3 | 2 | 6 | 2 | 23 | 34 |
| (NLysNpheNphe)3 | 5 | 33 | >100 | >100 | 50 | >100 | 25 | 6 | 2 | 1 | 4 | 17 |
| (NLysNpheNphe)2 | 6 | 33 | >100 | >100 | >100 | >100 | >100 | >100 | 1 | 1 | 1 | 1 |
| (NaeNpheNphe)4 | 7 | 33 | >100 | >100 | 13 | 50 | 2 | 6 | 8 | 2 | 50 | 17 |
| (NaeNpheNphe)3 | 8 | 33 | >100 | >100 | >100 | >100 | 13 | >100 | 1 | 1 | 8 | 1 |
| (NaeNpheNphe)2 | 9 | 33 | >100 | >100 | >100 | >100 | >100 | >100 | 1 | 1 | 1 | 1 |
| (NahNspeNspe)4 | 10 | 33 | 23 | 41 | 25 | 50 | 2 | 2 | 2 | 1 | 16 | 17 |
| (NahNspeNspe)3 | 11 | 33 | >100 | >100 | 25 | >100 | 3 | 2 | 4 | 1 | 33 | 50 |
| (NahNspeNspe)2 | 12 | 33 | >100 | >100 | >100 | >100 | >100 | 25 | 1 | 1 | 1 | 4 |
| (NLysNspeNspe)4 | 13 | 21, 23, 33 | 20 | 29 | 25 | 50 | 2 | 1 | 1 | 1 | 13 | 25 |
| (NLysNspeNspe)3 | 14 | 23, 33 | >100 | >100 | 13 | >100 | 2 | 2 | 8 | 1 | 50 | 50 |
| (NLysNspeNspe)2 | 15 | 23, 33 | >100 | >100 | >100 | >100 | >100 | 25 | 1 | 1 | 1 | 4 |
| (NaeNspeNspe)4 | 16 | 33 | 26 | 41 | >100 | 50 | 2 | 2 | 0 | 1 | 17 | 17 |
| (NaeNspeNspe)3 | 17 | 33 | >100 | >100 | 25 | >100 | 2 | 13 | 4 | 1 | 50 | 8 |
| (NaeNspeNspe)2 | 18 | 33 | >100 | >100 | >100 | >100 | >100 | >100 | 1 | 1 | 1 | 1 |
| (NLysNpmbNpmb)4 | 19 | 41 | >100 | >100 | >100 | 3 | 2 | 1 | 1 | 24 | 36 | |
| (NLysNpcbNpcb)4 | 20 | 18 | 22 | >100 | >100 | 25 | 6 | 0 | 0 | 1 | 4 | |
| (NLysNpcbNpcb)3 | 21 | 22 | 23 | 50 | 25 | 3 | 2 | 0 | 1 | 8 | 12 | |
| (NLysNpfbNpfb)4 | 22 | 46 | 30 | 13 | 25 | 2 | 1 | 3 | 2 | 19 | 38 | |
| (NLysNpfbNpfb)3 | 23 | >100 | 45 | 13 | 25 | 3 | 3 | 6 | 3 | 24 | 24 | |
| (NLysNmfbNmfb)4 | 24 | 25 | 17 | 25 | 25 | 6 | 3 | 1 | 1 | 4 | 7 | |
| (NLysNmfbNmfb)3 | 25 | 64 | 43 | 13 | 25 | 6 | 2 | 4 | 3 | 9 | 27 | |
| (NLysNpfbNspe)4 | 26 | 20 | 26 | 13 | 13 | 2 | 2 | 2 | 2 | 12 | 12 | |
| (NLysNpfbNspe)3 | 27 | 52 | 36 | 25 | 25 | 3 | 2 | 2 | 2 | 15 | 22 | |
| [(NLysNpfbNpfb)(NLysNspeNspe)]2 | 28 | >100 | 55 | 6 | 50 | 2 | 1 | 13 | 2 | 39 | 78 | |
| (NLysNspeNspe)(NLysNpfbNpfb)(NLysNspeNspe) | 29 | 65 | 43 | 13 | 25 | 3 | 2 | 4 | 3 | 21 | 28 | |
| (NamyNspeNspe)[(NLysNspeNspe)]3 | 30 | 12 | 15 | 50 | 50 | 2 | 2 | 0 | 0 | 7 | 7 | |
| (NamyNspeNspe)2(NLysNspeNspe)2 | 31 | 20 | 18 | >100 | >100 | 2 | 1 | 0 | 0 | 10 | 19 | |
| (NLysNspeNspe)2(NamyNspeNspe)(NLysNspeNspe) | 32 | 20 | 22 | >100 | >100 | 6 | 2 | 0 | 0 | 4 | 11 | |
| (NhArgNpheNphe)4 | 33 | >100 | ND | 6 | 25 | ND | 2 | 17 | 4 | — | 50 | |
| (NhArgNspeNspe)4 | 34 | 20 | 12 | 6 | 13 | 1 | 1 | 3 | 2 | 16 | 16 | |
| (NhArgNspeNspe)3 | 35 | ND | ND | 6 | 50 | 2 | 2 | — | — | — | — | |
| (NhArgNmfbNmfb)4 | 36 | 28 | 21 | 13 | 13 | 2 | 2 | 2 | 2 | 12 | 12 | |
| (NhArgNmfbNmfb)3 | 37 | ND | ND | 6 | 25 | 2 | 1 | — | — | — | — | |
| (NhArgNhLeuNspe)4 | 38 | ND | ND | 13 | 25 | 1 | 2 | — | — | — | — | |
| (NhArgNhLeuNspe)3 | 39 | ND | ND | ND | >100 | 3 | 2 | — | — | — | — | |
| [(NamyNspeNspe)(NhArgNspeNspe)]2 | 40 | 31 | 24 | >100 | >100 | 3 | 6 | 0 | 0 | 9 | 5 | |
| (NLysNspeNspe)2(NhArgNspeNspe)2 | 41 | >100 | ND | 17 | 34 | 17 | ND | 6 | 3 | 6 | — | |
| (NhArgNspeNspe)2(NLysNspeNspe)2 | 42 | 15 | ND | 17 | 17 | 17 | ND | 1 | 1 | 1 | — | |
| (NLysNspeNspe)(NhArgNspeNspe)(NLysNspeNspe)2 | 43 | 33 | ND | 17 | 17 | 17 | ND | 2 | 2 | 2 | — | |
| [(NhArgNspeNspe)(NLysNspeNspe)]2 | 44 | 33 | ND | 17 | 67 | 17 | ND | 2 | 0 | 2 | — | |
In this study we report the synthesis of one of the largest single library of antimicrobial peptoids published to date, in order to undertake a structure–activity relationship (SAR) of the wide variety of chemical functionalities present. Some of the sequence motifs and monomers represented here have not yet been reported in any antibacterial peptoids. This study focuses on the antibacterial potency of these peptoids against representative Gram-positive and Gram-negative species and significantly, we also examine the toxicity of this library using therapeutic indices against representative mammalian cell lines to evaluate the potential of peptoids as novel anti-infective compounds.
Results and discussion
Library design
We previously described the antiparasitic activities of a small library of linear peptoids.33 In this work, 18 peptoids from this first library have been retested to investigate their antibacterial activity and toxicity profiles (peptoids 1–18, Table 1), which to date have not been reported. An additional 26 novel peptoids were also synthesised to conduct a broader SAR study (peptoids 19–44, Table 1). This library represents the largest library of anti-infective peptoids published to our knowledge.Since the presumed mode of action of AMPs is by membrane disruption, the peptoid sequences selected contain a variety of side chain functionalisation to help elucidate the features necessary for activity. A defined secondary structure is thought to promote the antimicrobial action of both peptoids and peptides, therefore all peptoids tested here were designed around a repeating trimer motif where the third monomer was typically a cationic monomer (a peptoid helix turn is reported to occur every three residues in a polyproline type I helix) and often included the α-chiral Nspe or N(S-phenylethyl) glycine monomer in order to help induce an amphipathic secondary structure.23,38
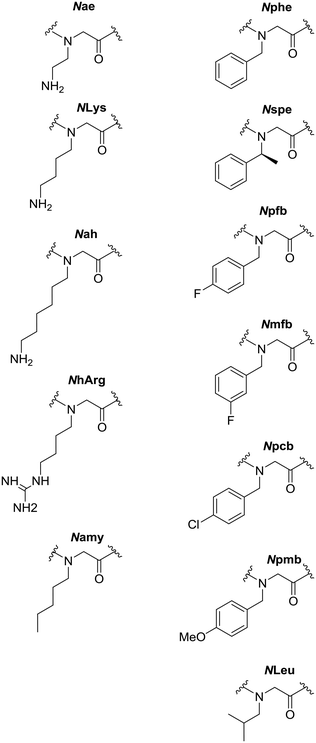
All peptoids were either synthesised manually on-resin or using an automated synthesiser using the submonomer method of synthesis at room temperature. Compounds between six and twelve residues in length were made, corresponding to peptoids with an overall positive charge of +2, +3 or +4.39,40 This extended library includes a wide range of monomers, such as those containing alkyl and substituted aromatic residues, including chlorinated and fluorinated peptoids. In addition, we have included peptoids that have different cationic functionalities in the sequence to further probe the relationship between charge and activity (Fig. 2).
Previously we described the activity of peptoids that contain lysine monomers (amino-functionalised cationic residues).32,33 It has been suggested that arginine-containing peptoids may have increased membrane permeability41 and so a small number of arginine analogues of our lysine library were generated. In addition, we recently described the synthesis of peptoids with mixed lysine and arginine functionalities, which are some of the first examples of this class of compound to be reported in the literature.42
Antimicrobial activity
The activity of our peptoid library was assessed against both bacterial and mammalian cells to gain a broader understanding of the factors that lead to effective and selective antimicrobial peptoids. We screened for broad-spectrum antibacterial activity against Gram-negative (Escherichia coli and Pseudomonas aeruginosa) and Gram-positive (Staphylococcus aureus and S. epidermidis) bacteria. The toxicity of the library was also assessed against two mammalian cell lines; HaCaT spontaneously transformed aneuploid keratinocytes were employed to model toxicity against human skin cells and HepG2, a cell line derived from a human liver carcinoma, were used as a study on polarised human hepatocytes. Detailed methods for these assays can be found in the ESI.‡In order to compare the activity of our compounds between the minimum inhibitory concentration (MIC) values obtained for bacteria and the effective dose (ED50) measurements against eukaryotic cells, all results have been presented in μM units. As seen from the data summarised in Table 1, many peptoids within the library have low MIC values against both Gram-negative and Gram-positive bacteria, ranging from the most active at less than 1 μM to inactive peptoids with no activity even at 100 μM. Some of the MIC values obtained are within the same range of selected natural antimicrobial peptides described in the literature. For example, the AMP cecropin A was shown to kill 90% of E. coli at 2 μM (ref. 43) and peptoids 33, 34, 35 and 37 all have an MIC of 6 μM against E. coli. Magainin 2, an amphibian derived AMP, was reported to have MICs against E. coli, S. epidermidis and S. aureus of 5 μg mL−1 (2 μM), 10 μg mL−1 (4 μM) and 50 μg mL−1 (20 μM) respectively and many of the peptoids in Table 1 exhibit even better antibacterial activities.43,44
Unsurprisingly, the activity of most of the peptoids is significantly greater against the Gram-positive species (S. aureus and S. epidermidis) than the Gram-negative E. coli and P. aeruginosa. This differential activity is probably due to the presence of the lipopolysaccharide-rich outer membrane of Gram-negatives, which presents a significant permeability barrier to many hydrophobic molecules.45,46 Certain compounds within the library displayed selectivity for particular bacterial species, For example, compound 17 had an MIC of 25 μM against E. coli, but >100 μM against the other Gram-negative bacterium, P. aeruginosa or 20 which has an MIC of 6 μM against S. epidermidis but only moderate activity of 25 μM against S. aureus. However, no Gram-negative specific antibacterial peptoids were identified. Any sequences that can selectively target Gram-negative bacteria are highly sought after due to rising concerns over antibiotic resistance.47–49
Structure–activity relationships: simple library
Factors necessary for robust activity against the protozoan intracellular parasite Leishmania mexicana were previously determined and include sequence length and inclusion of chiral monomers.31 Side chain length of cationic residues was also identified as an important feature for efficacy, with Nae or NLys displaying improved antiparasitic activity over the longer Nah residue.33 The same features are replicated here in the activity of the peptoid library against differing bacterial species. The longest 12 residue peptoids (1, 4, 7, 10, 13 and 16) were always more active than their 9 residue analogues (2, 5, 8, 11, 14 and 17), which were themselves more active than the 6 residue sequences (i.e.3, 6, 9, 12, 15 and 18), conclusions that agree with those from the Barron group.23 Hexapeptoids 3, 6, 9, 12, 15 and 18 showed generally limited antibacterial activity against the bacteria tested with MICs of 100 μM, although it is interesting to note that S. epidermidis did show some increased sensitivity to peptoids 12 and 15 (Table 1).Interestingly, the effect of monomer chirality was less important than with L. mexicana in achieving antibacterial activity; in many cases, sequences comprised exclusively from achiral monomers had comparable or better activity than analogues containing the chiral Nspe building block. For example, comparing peptoid 4 to 13 which are achiral and chiral analogues of the same sequence, we observe similar MIC values against P. aeruginosa (50 μM for both) and S. aureus (MIC 3 μM and 2 μM). However, against E. coli, the achiral peptoid 4 has an MIC of 12 μM compared to 25 μM for the chiral equivalent 13. A similar pattern is apparent with peptoids 7 and 16 (sequences with Nae and either Nphe or Nspe respectively), with the peptoids having similar activity against both P. aeruginosa and S. aureus but the achiral sequence 7 shows better activity against E. coli (MIC 13 μM and >100 μM respectively).
When comparing sequences containing different amino-functionalised cationic monomers, Nae, NLys and Nah, it is not possible to draw a simple conclusion about the optimum length of side chain for best antibacterial activity since promising activity is evident with all three cationic residues. Peptoids based upon the NxNpheNphe motif 1 (Nah), 4 (NLys) and 7 (Nae) all have an MIC of 13 μM against E. coli and good activity against both Staphylococcus species. However, when comparing the chiral analogues of the same motif (Nspe replaced Nphe), the sequences with Nah and NLys (10 and 13) have an MIC of 25 μM against E. coli but the peptoid with the shortest Nae residue (16) has negligible activity. For peptoids 10, 13 and 16, the activity against P. aeruginosa, S. aureus and S. epidermidis is similar regardless of the choice of cationic monomer (Table 1).
Effects of aromatic building block substitution
A selection of substituted aromatic monomers were included in a similar repeating motif of two aromatic monomers followed by the charged NLys residue to determine their impact on biological activity. Monomer substitutions included a methoxy group (Npmb), chlorine (Npcb) or fluorine in both para and meta positions (Npfb, Nmfb). The effect of these substitutions on anti-Leishmanial activity were reported recently.32 It was shown that halogenated monomers (in particular fluorinated ones) improve the efficacy of peptoid sequences against amastigotes. Based upon the success of this approach, we concentrated our efforts on the antibacterial screening of halogenated sequences.Methoxy substituted peptoids were only tested at the longest 12 residue length (peptoid 19) and show a negligible effect against the two Gram-negative species and no improvement in action against the Gram-positive bacteria compared to the unsubstituted analogue (peptoid 4). Addition of chlorine in the para position improves activity against L. mexicana axenic amastigotes, however decreases antibacterial activity (i.e. compounds 20cf.4 with an MIC of >100 μM and 13 μM with E. coli) and also significantly increases the toxicity of the sequences to mammalian cells (ED50 HepG2 22 μM and >100 μM respectively).
Fluorinated peptoid sequences were more successful at targeting the various bacteria. Peptoids with exclusively achiral monomers (Npfb or Nmfb) and those with a mixture of chiral and achiral building blocks (Npfb and Nspe) were tested. Those sequences containing the achiral monomers (peptoids 22–25) have marginally improved antibacterial activities compared to analogues of the same length with Nphe rather than fluorinated monomers (peptoids 4–6). There seems to be no significant difference between monomers substituted in the para or meta position. Interestingly, the 9-residue peptoids 23 and 25 have a similar level of broad spectrum antibacterial activity as those with 12 residues (22 and 24), but the former show reduced toxicity to mammalian cells. The 6-residue sequences display reduced activity against bacteria. In this case, it appears that the shorter 9-residue sequences 23 and 25 may prove to be better antibacterial candidates with a larger therapeutic window between activity and toxicity (Table 1)
The simple chiral sequences (i.e.13) are more potent, but also more toxic than the achiral Nphe peptoids (4–6). Given that the fluorinated, achiral monomers exhibit an increase in antibacterial activity compared to the unsubstituted analogues, the Npfb monomer was also placed into the following motif: NLysNpfbNspe to examine whether the activity of chiral sequences could be modulated (peptoids 26 and 27). Further iterations were also synthesised where the Npfb and Nspe monomers were placed in a coblock manner (i.e. sequences 28 and 29). In both templates, the longest sequences showed the best broad-spectrum antibacterial activity. In particular, the 12-residue block peptoid (28) is promising, with reduced toxicity compared to sequences made of Nspe or Npfb exclusively and improved activity against E. coli (MIC 6 μM), S. aureus and S. epidermidis (MICs 2 μM and <1 μM).
Peptoids containing alkyl chains
To probe the relationship between net charge and hydrophobicity of a peptoid and its biological activity, analogues of peptoid 13 were synthesised where the cationic NLys monomer was substituted by the alkyl monomer Namy. By replacing the charged amino group with a methyl group, the overall compound charge is reduced, however the number of atoms in the side chain and overall molecular weight remains unaltered. In these analogues (peptoids 30–32), the charge is replaced at just the N terminal end of the sequence or at two positions within the sequence.When the antibacterial properties are considered, there is little difference in efficacy against the Gram-positive bacteria tested, however, the substitutions lead to a reduction in overall activity against Gram-negatives. The parent sequence 13 shows moderate activity against E. coli and P. aeruginosa (MICs of 25 μM and 50 μM respectively) and potent micromolar activity against the Gram-positives. However, peptoids 30–32 have MICs of 50 μM or >100 μM against E. coli and similar reductions in activity are seen for P. aeuriginosa.
Substitution of arginine-instead of lysine-residues
As described in the literature,31,41,50 arginine-containing peptoids are known to increase membrane permeability and antibacterial activity. Hence, sequences containing arginine peptoid monomers were included in the library. This allowed the comparison of differently functionalised cationic residues on the peptoid sequences (i.e. the amino NLys peptoids or the guanidine groups of NhArg). It has also been suggested that arginine in peptide sequences can improve antibacterial potency, although this is also linked with enhanced toxicity.26,31 In an attempt to modulate the biological properties of the peptoid library, for the first time, peptoids with both lysine and arginine in the same sequence were evaluated against bacterial targets using the new methodology developed in our group.42This sublibrary of peptoids can be split between sequences exclusively containing arginine residues (peptoids 33–40) and those that contain a mixture of both lysine and arginine-type side chains (peptoids 41–44). In these compounds NhArg was introduced, which is the equivalent side chain to the NLys residue, with 4 carbons in the backbone and the terminal guanidine moiety.
In contrast to peptoids that contain amino-functionalised NLys residues the NhArg sequences tend to have an increased toxicity to the mammalian cell lines tested but do also display improved activity against the bacteria tested. For example, when comparing the fluorinated peptoids 36 and 37 to their lysine-equivalents (24 and 25 respectively) we see an approximate 2-fold increase in antibacterial activity for all species tested; for the longest 12-residue sequence (NxNmfbNmfb)4 activity against E. coli is 13 μM in 36 with NhArg, compared to 25 μM in 24 (NLys). Against S. aureus the arginine-type peptoid has an MIC of 2 μM contrasting with 6 μM for the lysine equivalent. However, in sequences that were inactive with lysine residues, replacement by NhArg does not make the sequence active (see peptoid 40, where the sequence is not active against Gram-negative bacteria at any concentration), but in these cases the inclusion of the guanidine group does increase the toxicity.
As predicted, sequences with a combination of lysine and arginine-type residues show a balance between toxicity to mammalian cells and antibacterial activity compared to sequences containing NLys or NhArg residues exclusively. For example, in related peptoids containing all arginine residues (34), all lysine residues (13) and both lysine/arginine monomers within the same sequence (peptoids 42–44), we see toxicity to the HaCaT keratinocytes at 11 μM, 20 μM and 15–33 μM respectively. The general antibacterial activity follows a similar trend, for example, against E. coli the lysine-only peptoid 13 has the lowest activity at 25 μM, the arginine-only peptoid 34 has the most potent activity with an MIC of 6 μM and the mixed sequences 42–44 have intermediate activity at 17 μM.
The observation that guanidine-only peptoids display the most potent biological activities is in agreement with previous studies into arginine-rich peptides, which are able to bind membrane-bound lipids more readily than their amino-functionalised lysine equivalents.31,51 In this case it was proposed that the arginine-type side chains can form bidentate hydrogen bonds with the phospholipid head groups. This conclusion was also reached in studies with antimicrobial peptide–peptoid hybrids containing both lysine and arginine.31,51
Toxicity
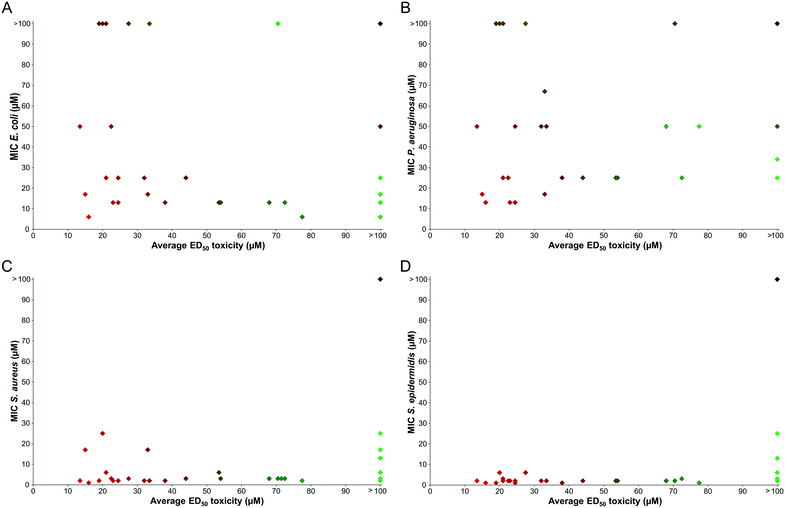
From the antibacterial MIC determination in Table 1, multiple promising peptoids were identified that showed little or no toxicity to either of the mammalian cell lines tested. For example, compounds 4, 7, 23 and 28 display negligible toxicity to HaCaT or HepG2 at the highest concentrations used and are also broad spectrum antibacterial agents. However, many of the sequences generated did display significant toxicity to mammalian cells, and in general these compounds were similarly toxic to both HaCaT and HepG2. On the whole, as the antimicrobial action of a compound increases, the associated toxicity is also increased. This is a problem found in other recent studies that focused on the biological applications of peptoids, however, attention is frequently not directed towards the issue of toxicity.26,29,37The selectivity of sequences is highlighted as a particular challenge for the design of antimicrobial peptoids. To explore the relationship between activity and toxicity a comparison was made between the MIC values against each bacterial species versus the average toxicity of each peptoid to HaCaT and HepG2 (see Fig. 3).
A handful of peptoids were identified that show respectable antibacterial activity and also display low toxicity to mammalian cells. These compounds have the potential to be future selective antibiotic compounds. However, a large proportion of the peptoids within this library do display significant toxicity. There are also many sequences that are toxic to mammalian cells, but show little activity against Gram-negative bacteria. It is likely that the external lipopolysaccharide layer on the outer membrane of both E. coli and P. aeruginosa (absent in Gram positive bacteria) prevents these peptoids from reaching the cell membrane. To investigate a possible explanation for this observation, the hydrophobicity of the compounds was considered.
Hydrophobicity
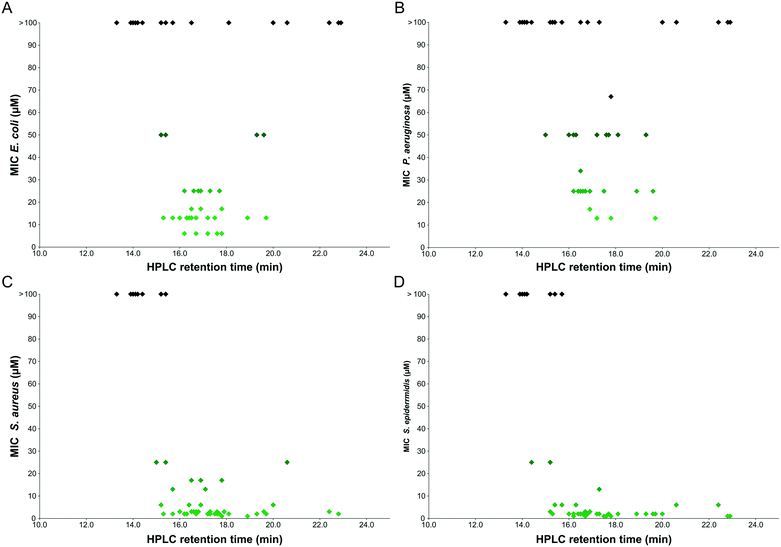
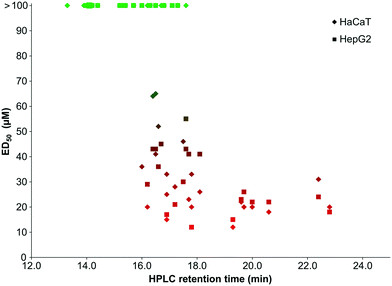
The hydrophobicity of our library was assessed using reverse-phase HPLC retention times as in Fig. 4. Although this provides only a relatively crude measure of hydrophobicity and may not translate directly into predictions about how a compound will interact with the cell membranes of biological systems, certain interesting trends were evident from the analysis.Since the activity of peptoids is much greater against Gram-positive bacteria than Gram-negative bacteria, the graphs are predominantly populated by peptoids for E. coli and P. aeruginosa due to their higher MIC values. There appears to be a linear relationship between activity and retention time for the Gram-positive bacteria, where the peptoids at longer retention times (therefore more hydrophobic) have the lowest MIC values against S. aureus or S. epidermidis.
However, for the Gram-negative bacteria, there is no clear correlation between hydrophobicity and antibacterial activity of the peptoid library. Some compounds with the longest retention times (presumably the most hydrophobic) are inactive against these bacteria, whereas others with shorter retention times display good activity. It has previously been suggested that highly hydrophobic sequences may have lower activities due to self-association, preventing sufficient contact with the cell membrane.29 However, this may be an oversimplification since the same results are not observed with Gram-positive bacteria, where the same hydrophobic peptoid sequences result in low MIC values.
The discovery that peptoids with high hydrophobicity are not always the most potent against E. coli and P. aeruginosa, but against S. aureus a linear relationship is seen between hydrophobicity and activity is corroborated by a recent report.29 However, the lack of a consensus between hydrophobicity and activity in published libraries, highlights the need for the research community to develop additional tools to help predict peptoid properties and likely efficacy.
There also seems to be a correlation between the toxicity of our peptoid library to mammalian cells and compound hydrophobicity, with a similar profile of toxicity for both HaCaT and HepG2 (see Fig. 5). The least hydrophobic compounds are generally the least toxic, whereas those with the highest retention times show the lowest ED50 values.
In the literature, HPLC retention times are used to rationalise antimicrobial activity21,23,29 and also for in silico predictions of activity.37 From the data presented here, it is suggested that HPLC retention time alone cannot estimate or rationalise activity. For Gram-negative bacteria, HPLC retention time is not predictive of activity and although peptoids with longer retention times may have an increased efficacy against Gram-positive bacteria, these compounds may also have a concomitant and undesirable increased toxicity.
Other parameters to evaluate the activity of antimicrobial peptoids and guide the design of bioactive peptoids need to be considered. We recently proposed hydrophobicity measurements determined via partitioning experiments (i.e. log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D) as an improved approach to rationalise the biological activity of peptoids.52
D) as an improved approach to rationalise the biological activity of peptoids.52
Conclusions
This study presents a large and varied library of peptoids that were specifically designed to mimic natural antimicrobial peptides. Many of the peptoids generated show potential as antimicrobial compounds, with broad-spectrum activity against a wide variety of bacterial and parasitic targets and may be promising leads in the future for antibiotics that can combat the increasing problem of resistance.Most of the peptoids were considerably more active towards Gram positive species than Gram-negatives in keeping with their differing envelope structures. Hence the peptoid library has much better therapeutic indices against S. aureus and S. epidermidis. The best activity against E. coli reported was 6 μM for peptoids 28, 33, 34, 35 and 37 and 13 μM against P. aeruginosa for peptoids 26, 34 and 36. Many peptoids also showed micromolar activities against S. aureus and S. epidermidis with both species showing a similar pattern of sensitivity; S. aureus did show a tendency to be more resistant than S. epidermidis possibly due to minor differences in cell surface charge and hydrophobicity.53,54 In many cases, P. aeruginosa also proved more peptoid-resistant than E. coli. This difference may reflect variation in lipid composition or the capacity of P. aeruginosa to form biofilms and has been observed previously with AMPs, such as LL-37.55
Factors contributing to enhanced antimicrobial activity include the overall length of sequence, with longer, 12-residue peptoids typically displaying the best activity, although in many cases the 9-residue peptoids also display broad spectrum activity against bacteria. Substitution of fluorine in peptoid monomers also enhances activity and in many cases, achiral sequences displayed potency, especially against the Gram-positive species. Interestingly, from our library most compounds that are active against the bacteria but exhibit no/low toxicity to mammalian cells contain achiral monomers, or at least a reduced frequency of chiral monomers within the sequence compared to the standard NxNspeNspe motif. Whether through the presumed increase in hydrophobicity, or through other effects, the addition of chiral Nspe residues in a sequence often has a detrimental effect on overall toxicity. Since many of the achiral sequences show potent antibacterial activity and these peptoids are not expected to form fully folded peptoid helices, secondary structure is perhaps not as important as predicted.
It is generally stated that the mode of action for AMPs/peptoids is cell membrane disruption.47,56,57 For certain compounds, particularly the more hydrophobic compounds that are both active and toxic, this may be the case. However, the differences exhibited by some compounds, particularly those with negligible toxicity to mammalian cells yet good activity against bacteria, may indicate that cell membrane disruption is not the only mechanism at work.46 Current work within our group is now focussing on investigating the molecular mechanism of a selection of these compounds to elucidate the factors necessary for antimicrobial selectivity and potency.
The peptoid community clearly has the requisite tools to design and synthesise effective antimicrobial peptoid sequences with potential for clinical application, however, the challenge now is to focus on increasing pathogen selectivity while minimising host cell toxicity.
Data availability
In addition to the material provided in the ESI underlying research data for this paper is also available in accordance with the EPSRC open data policy from doi: 10.15128/r1zg64tk92g.Experimental
Materials and reagents
Abbreviations for reagents are as follows: tert-butoxycarbonyl (Boc); 9-fluorenylmethoxylcarbonyl (Fmoc); trifluoroacetic acid (TFA); triisopropylsilyl (TIPS); N,N-dimethylformamide (DMF); N,N-diisopropylcarbodiimide (DIC); dimethylsulphoxide (DMSO). Solvents and reagents were purchased from commercial sources and used without further purification unless otherwise stated. Rink amide resin (typical loading level 0.6–0.8 mmol g−1) was purchased from Merck4Biosciences. DMF was purchased from AGTC Bioproducts (National Diagnostics). Piperidine, bromoacetic acid and TFA were purchased from Sigma Aldrich. The amine building blocks were sourced from Sigma Aldrich or TCI Europe.Peptoid synthesis procedures
Peptoids in this library were synthesised both manually and on an automated synthesiser. Protocols for each synthesis method follow.Manual linear peptoid synthesis
Fmoc-protected Rink Amide resin (normally 100 mg, 0.1 mmol, typical loading between 0.6–0.8 mmol g−1) was swollen in DMF (at least 1 h at room temperature, overnight preferred) in a 20 mL polypropylene Bond Elut SPPS cartridge fitted with two polyethylene frits (Crawford Scientific). The resin was deprotected with piperidine (20% in DMF v/v, 2 × 20 min) and washed with DMF (3 × 2 mL). The resin was treated with bromoacetic acid (1 mL, 0.6 M in DMF) and DIC (0.2 mL, 50% v/v in DMF) for 20 min at room temperature on a shaker platform at 400 rpm (Radleys Technology). The resin was washed with DMF (3 × 2 mL), before the desired amine sub-monomer was added (1 mL, 1.5 M in DMF) and allowed to react for 60 min on the shaker. The resin was again washed with DMF (3 × 2 mL) and the bromoacetylation and amine displacement steps were repeated until the final sub-monomer had been added and the desired peptoid sequence obtained. Resin was washed with DCM and the final cleavage from resin was achieved using a TFA cleavage cocktail (4 ml; TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIPS
TIPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 95
H2O, 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5) on the shaker at 400 rpm for 60 min. The resin was removed by filtration and the cleavage cocktail removed in vacuo. The crude product was precipitated in diethyl ether (30 mL) and the precipitate retrieved by centrifuge for 15 min at 5000 rpm. The ether phase was decanted and the crude product dissolved in a mixture of acidified H2O and MeCN and lyophilised to a powder before purification.
2.5) on the shaker at 400 rpm for 60 min. The resin was removed by filtration and the cleavage cocktail removed in vacuo. The crude product was precipitated in diethyl ether (30 mL) and the precipitate retrieved by centrifuge for 15 min at 5000 rpm. The ether phase was decanted and the crude product dissolved in a mixture of acidified H2O and MeCN and lyophilised to a powder before purification.
Automated linear peptoid synthesis
Automated peptoid synthesis using an Aapptec Apex 396 synthesiser. Fmoc-protected Rink Amide resin (0.1 mmol, loading 0.54 mmol g−1) was swollen in DMF (2 mL, 2 min, 475 rpm at RT) and deprotected with 4-methylpiperidine (20% in DMF v/v, 1 mL for 1 min, 475 rpm at RT; then 2 mL for 12 min, 475 rpm at RT). The resin was treated with haloacetic acid solution (either bromo- or chloroacetic acid, 1 mL, 0.6 M in DMF) and DIC (0.18 mL, 50% v/v in DMF) for 20 min at 475 rpm, RT. The resin was washed with DMF (2 mL DMF for 1 min at 475 rpm, ×5) before the desired amine sub-monomer was added (1 mL, 1.5 M in DMF) and shaken for 60 min at 475 rpm. The resin was washed again with DMF (2 mL DMF for 1 min at 475 rpm, ×5) and the acetylation and amine displacement steps were repeated until the desired sequence was achieved. The resin was shrunk in diethyl ether and peptoids cleaved off the resin using a TFA cleavage cocktail (4 ml; TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIPS
TIPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 95
H2O, 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5) for 30–60 min on an orbital shaker at 250 rpm, RT. The cocktail was filtered from the resin and evaporated in vacuuo and the resulting residue precipitated in diethyl ether (∼20 ml). The crude peptoid was obtained via centrifugation (15 min, 4000 rpm, 5 °C) and the ether layer decanted to yield the crude product as a powder. Peptoids were lyophilised before purification by semi-preparative RP-HPLC.
2.5) for 30–60 min on an orbital shaker at 250 rpm, RT. The cocktail was filtered from the resin and evaporated in vacuuo and the resulting residue precipitated in diethyl ether (∼20 ml). The crude peptoid was obtained via centrifugation (15 min, 4000 rpm, 5 °C) and the ether layer decanted to yield the crude product as a powder. Peptoids were lyophilised before purification by semi-preparative RP-HPLC.
Addition of NhArg and NnArg residues to sequence
To introduce arginine-type residues during the submonomer procedure, the appropriate unprotected diamine was added under normal submonomer coupling conditions (1.5 M amine in DMF, 60 min, room temperature) in place of the mono N-Boc diamine and the resin washed with DMF (3 × 2 mL). Dde-OH (10 eq. wrt resin in the minimum volume of DMF) was added to the resin and placed on the shaker at RT for 60 min and the resin washed well with DMF (3 × 2 mL). Subsequent peptoid couplings were made as normal until the desired sequence was achieved, including any extra Dde-protected residues.After synthesis of the linear peptoid sequence, on resin deprotection of the Dde group was undertaken using 2% hydrazine in DMF (4 × 4 ml × 3 min) and the resin washed with DMF (3 × 2 mL). Guanidinylation of the free amines was achieved using pyrazole-1-carboxamide (6 eq. per free amine, in the minimum amount of DMF) and DIPEA (6 eq. per free amine) on the shaker at 400 rpm, RT for 60 min. The resin was washed with DCM (3 × 2 mL) and shrunk in ether prior to cleavage from the resin, as above.
Purification by preparative RP-HPLC
Preparative RP-HPLC was performed with a semi-preparative Perkin Elmer Series 200 lc pump fitted with a 785A UV/vis detector using a SB-Analytical ODH-S optimal column (250 × 10 mm, 5 μm); flow rate 2 ml min−1; λ = 250 nm, where a linear gradient from solvent A to B applied (A = 0.1% TFA in 95% H2O and 5% MeCN, B = 0.1% TFA in 5% H2O and 95% MeCN).Characterisation
Peptoids were characterised by accurate LC-MS (QToF mass spectrometer and an Acquity UPLC from Waters Ltd) using an Acquity UPLC BEH C8 1.7 μm (2.1 mm × 50 mm) column with a flow rate of 0.6 ml min−1 and a linear gradient of 5–95% of solvent B over 3.8 min (A = 0.1% formic acid in H2O, B = 0.1% formic acid in MeCN). Peptide identities were also confirmed by MALDI-TOF mass spectra analysis (Autoflex II ToF/ToF mass spectrometer Bruker Daltonik GmBH) operating in positive ion mode using an α-cyano-4-hydroxycinnamic acid (CHCA) matrix. Data processing was done with MestReNova version 8.1.Analytical RP-HPLC was carried out using a Perkin Elmer Series 200 lc pump fitted with a series 200 UV/vis detector and autosampler using a SB-Analytical ODH-S optimal column (100 × 1.6 mm, 3.5 μm); flow rate 1 ml min−1; λ = 220 nm, linear gradient elution 0–100% of solvent B over 30 min (A = 0.05% TFA, 95% H2O, 5% MeCN, B = 0.03% TFA, 5% H2O, 95% MeCN).
Biological assays
Antibacterial MIC determination
Please note – ED50, the median effective dose, was defined as the dose that kills 50% of cells.Escherichia coli K-12 wild-type strain (W3110/ATCC27325, F−, λ−, rpoS(Am), rph-1, Inv(rrnD-rrnE)), Pseudomonas aeruginosa PA01 (ATCC 15692) Staphylococcus aureus (3R7089 strain Oxford/ATCC9144) and Staphylococcus epidermidis (laboratory strain from clinical isolate) were selected for bacteriological studies as representative Gram-negative (E. coli and P. aeruginosa) and Gram-positive (S. aureus and S. epidermidis) species. Bacterial cultures were prepared by streaking bacterial strains onto LB agar plates with an inoculation loop and incubated overnight at 37 °C. A single colony was selected and placed in 5 mL of Iso-sensitest broth (Oxoid, ThermoScientific) and incubated with shaking for 16–18 h at 37 °C to provide liquid cultures for testing.
MIC values were obtained according to the protocol described by J. M. Andrews et al.58 and were conducted in 96-well plates (Sarstedt). Bacteria were grown from overnight cultures in Iso-sensitest broth to an A650nm of 0.07 equivalent to a 0.5 MacFarland standard (240 μM BaCl2 in 0.18 M H2SO4). This culture was diluted ten-fold with Iso-sensitest broth before use. Peptoids were initially dissolved in DMSO (5 mM) and diluted further in Iso-sensitest broth to achieve a concentration range of 4–200 μM using 2-fold serial dilutions. 50 μl of inoculum and 50 μl of peptoid solution were added to each test well (final concentration range of 2–100 μM). Experiments were performed in triplicate. A positive control for bacterial growth contained only the inoculum and Iso-sensitest broth. Other controls contained the inoculum and serial dilutions of ampicillin (from 250 μg mL−1 to 2 μg mL−1), serial dilutions of DMSO and the inoculum to confirm no inhibitory effect on bacterial growth, and Iso-sensitest broth alone as a sterile control. The MIC was defined as the lowest concentration which completely inhibited bacterial growth after incubation at 37 °C for 16 h with shaking. Quantitative data was attained from absorbance values using a Biotek Synergy H4 plate reader.
Cytotoxicity assay with HepG2 or HaCaT
Cytotoxicity analyses were performed in 96-well plates (Costar, Fisher Scientific) using alamarBlue® (Invitrogen) for cell viability detection using a modified protocol as described previously. HepG2 or HaCaT cells were subcultured at 37 °C, 5% CO2 in DMEM high glucose supplemented with heat-inactivated foetal bovine sera (FBS, 10%; Biosera Ltd) and penicillin/streptomycin (P/S, 1%). Cells were counted using a Neubauer Improved Haemocytometer. HepG2 cells were seeded 1 day prior to treatment in 96 well plates at a concentration of 2 × 105 cells per mL in 100 μL of medium (2 × 104 cells per well). Then cells were pre-incubated with the compounds in triplicate in a dilution series in triplicate from 2–100 μM (5 mM stock solutions in DMSO diluted from 100 μM to 3 μM; untreated cells with DMSO as a negative control) in 50 μl of the media for 1 hour. Afterwards, 40 μL were removed from each well before the addition of 90 μL of the media, followed by incubation for 24 hours at 37 °C, 5% CO2. Then, 10 μL of alamarBlue® (Invitrogen) was added to each well before a 2 hour incubation prior to assessing cell viability using a fluorescent plate reader (Biotek; Ex 560 nm/Em 600 nm). All data were measured in triplicate on a minimum of two occasions to ensure a robust data set was collected. The ED50 values were calculated from the dose response results achieved from the serial dilutions.Acknowledgements
We thank the Engineering and Physical Sciences Research Council (HLB, Doctoral Training Grant), The Swiss National Foundation (GAE), The Royal Society of Chemistry (HLB, Early Researcher Mobility Grant), Van Mildert College (HLB, Postgraduate Award). In particular, we thank Ronald Zuckermann at the Molecular Foundry for facilitating a research placement. Work at the Molecular Foundry was supported by the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract no DE-AC01-05CH11231.References
- Infectious Diseases Society of America, The 10 × '20 Initiative: pursuing a global commitment to develop 10 new antibacterial drugs by 2020, Clin. Infect. Dis., 2010, 50, 1081 CrossRef PubMed.
- Y. Li, Q. Xiang, Q. Zhang, Y. Huang and Z. Su, Peptides, 2012, 37, 207 CrossRef CAS PubMed.
- K. Fosgerau and T. Hoffmann, Drug Discovery Today, 2015, 20, 122 CrossRef CAS PubMed.
- D. J. Craik, D. P. Fairlie, S. Liras and D. Price, Chem. Biol. Drug Des., 2013, 81, 136 CAS.
- D. K. Mercer and D. A. O'Neil, Future Med. Chem., 2013, 5, 315 CrossRef CAS PubMed.
- R. E. Hancock and H. G. Sahl, Nat. Biotechnol., 2006, 24, 1551 CrossRef CAS PubMed.
- F. T. Lundy, J. Nelson, D. Lockhart, B. Greer, P. Harriott and J. J. Marley, Mol. Immunol., 2008, 45, 190 CrossRef CAS PubMed.
- N. Y. Yount and M. R. Yeaman, Annu. Rev. Pharmacol. Toxicol., 2012, 52, 505 CrossRef PubMed.
- A. T. Y. Yeung, S. L. Gellatly and R. E. W. Hancock, Cell. Mol. Life Sci., 2011, 68, 2161 CrossRef CAS PubMed.
- M. J. Dawson and R. W. Scott, Curr. Opin. Pharmacol., 2012, 12, 545 CrossRef CAS PubMed.
- H. Jenssen, P. Hamill and R. E. Hancock, Clin. Microbiol. Rev., 2006, 19, 491 CrossRef CAS PubMed.
- M. Torrent, D. Pulido, L. Rivas and D. Andreu, Curr. Drug Targets, 2012, 13, 1138 CrossRef CAS PubMed.
- F. L. Chadbourne, C. Raleigh, H. Z. Ali, P. W. Denny and S. L. Cobb, J. Pept. Sci., 2011, 17, 751 CrossRef CAS PubMed.
- G. A. Eggimann, K. Sweeney, H. L. Bolt, N. Rozatian, S. L. Cobb and P. W. Denny, Molecules, 2015, 20, 2775 CrossRef PubMed.
- P. Kosikowska and A. Lesner, Expert Opin. Ther. Pat., 2016, 26, 689 CrossRef CAS PubMed.
- M.-D. Seo, H.-S. Won, J.-H. Kim, T. Mishig-Ochir and B.-J. Lee, Molecules, 2012, 17, 12276 CrossRef CAS PubMed.
- J. L. Fox, Nat. Biotechnol., 2013, 31, 379 CrossRef CAS PubMed.
- Y. J. Gordon, E. G. Romanowski and A. M. McDermott, Curr. Eye Res., 2005, 30, 505 CrossRef CAS PubMed.
- K. Fosgerau and T. Hoffmann, Drug Discovery Today, 2015, 20, 122 CrossRef CAS PubMed.
- S. M. Miller, R. J. Simon, S. Ng, R. N. Zuckermann, J. M. Kerr and W. H. Moos, Drug Dev. Res., 1995, 35, 20 CrossRef CAS.
- J. A. Patch and A. E. Barron, J. Am. Chem. Soc., 2003, 125, 12092 CrossRef CAS PubMed.
- R. Kapoor, P. R. Eimerman, J. W. Hardy, J. D. Cirillo, C. H. Contag and A. E. Barron, Antimicrob. Agents Chemother., 2011, 55, 3058 CrossRef CAS PubMed.
- N. P. Chongsiriwatana, J. A. Patch, A. M. Czyzewski, M. T. Dohm, A. Ivankin, D. Gidalevitz, R. N. Zuckermann and A. E. Barron, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2794 CrossRef CAS PubMed.
- R. Kapoor, M. W. Wadman, M. T. Dohm, A. M. Czyzewski, A. M. Spormann and A. E. Barron, Antimicrob. Agents Chemother., 2011, 55, 3054 CrossRef CAS PubMed.
- T. S. Ryge, N. Frimodt-Moller and P. R. Hansen, Chemotherapy, 2008, 54, 152 CrossRef CAS PubMed.
- C. A. Olsen, H. L. Ziegler, H. M. Nielsen, N. Frimodt-Moller, J. W. Jaroszewski and H. Franzyk, ChemBioChem, 2010, 11, 1356 CrossRef CAS PubMed.
- M. L. Huang, S. B. Y. Shin, M. A. Benson, V. J. Torres and K. Kirshenbaum, ChemMedChem, 2012, 7, 114 CrossRef CAS PubMed.
- M. L. Huang, M. A. Benson, S. B. Y. Shin, V. J. Torres and K. Kirshenbaum, Eur. J. Org. Chem., 2013, 3560 CrossRef CAS.
- B. Mojsoska, R. N. Zuckermann and H. Jenssen, Antimicrob. Agents Chemother., 2015, 59(7), 4112–4120 CrossRef CAS PubMed.
- K. J. Fisher, J. A. Turkett, A. E. Corson and K. L. Bicker, ACS Comb. Sci., 2016, 18, 287 CrossRef CAS PubMed.
- L. Vedel, G. Bonke, C. Foged, H. Ziegler, H. Franzyk, J. W. Jaroszewski and C. A. Olsen, ChemBioChem, 2007, 8, 1781 CrossRef CAS PubMed.
- H. L. Bolt, G. A. Eggimann, P. W. Denny and S. L. Cobb, MedChemComm, 2016, 7, 799 RSC.
- G. A. Eggimann, H. L. Bolt, P. W. Denny and S. L. Cobb, ChemMedChem, 2015, 10, 233 CrossRef CAS PubMed.
- M. Uchida, G. McDermott, M. Wetzler, M. A. Le Gros, M. Myllys, C. Knoechel, A. E. Barron and C. A. Larabell, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 19375 CrossRef CAS PubMed.
- A. E. Corson, S. A. Armstrong, M. E. Wright, E. E. McClelland and K. L. Bicker, ACS Med. Chem. Lett., 2016, 7(12), 1139–1144 CrossRef CAS PubMed.
- Y. Luo, H. L. Bolt, G. A. Eggimann, D. F. Mc Auley, R. Mc Mullan, T. Curran, M. Zhou, C. A. B. Jahoda, S. L. Cobb and F. T. Lundy, ChemBioChem, 2016, 18, 111–118 CrossRef PubMed.
- A. M. Czyzewski, H. Jenssen, C. D. Fjell, M. Waldbrook, N. P. Chongsiriwatana, E. Yuen, R. E. W. Hancock and A. E. Barron, PLoS One, 2016, 11, e0135961 Search PubMed.
- C. W. Wu, K. Kirshenbaum, T. J. Sanborn, J. A. Patch, K. Huang, K. A. Dill, R. N. Zuckermann and A. E. Barron, J. Am. Chem. Soc., 2003, 125, 13525 CrossRef CAS PubMed.
- R. N. Zuckermann, J. M. Kerr, S. B. H. Kent and W. H. Moos, J. Am. Chem. Soc., 1992, 114, 10646 CrossRef CAS.
- H. Tran, S. L. Gael, M. D. Connolly and R. N. Zuckermann, J. Visualized Exp., 2011, e3373 Search PubMed.
- K. Andreev, C. Bianchi, J. S. Laursen, L. Citterio, L. Hein-Kristensen, L. Gram, I. Kuzmenko, C. A. Olsen and D. Gidalevitz, Biochim. Biophys. Acta, 2014, 1838, 2492 CrossRef CAS PubMed.
- H. L. Bolt and S. L. Cobb, Org. Biomol. Chem., 2016, 14, 1211 CAS.
- L. Silvestro, J. N. Weiser and P. H. Axelsen, Antimicrob. Agents Chemother., 2000, 44, 602 CrossRef CAS PubMed.
- M. Zasloff, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 5449 CrossRef CAS.
- N. Ruiz, D. Kahne and T. J. Silhavy, Nat. Rev. Microbiol., 2006, 4, 57 CrossRef PubMed.
- H. Nikaido, in Escherichia coli and Salmonella: cellular and molecular biology, ed. F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter and H. E. Umbarger, ASM Press, Washington, D.C., 2nd edn, 1996, p. 29 Search PubMed.
- F. Guilhelmelli, N. Vilela, P. Albuquerque, L. D. S. Derengowski, I. Silva-Pereira and C. M. Kyaw, Front. Microbiol., 2013, 4, 353 Search PubMed.
- H. C. Neu, Science, 1992, 257, 1064 CAS.
- S. Gruenheid and H. Le Moual, FEMS Microbiol. Lett., 2012, 330, 81 CrossRef CAS PubMed.
- P. A. Wender, D. J. Mitchell, K. Pattabiraman, E. T. Pelkey, L. Steinman and J. B. Rothbard, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 13003 CrossRef CAS PubMed.
- J. B. Rothbard, T. C. Jessop and P. A. Wender, Adv. Drug Delivery Rev., 2005, 57, 495 CrossRef CAS PubMed.
- H. L. Bolt, C. E. J. Williams, R. V. Brooks, R. N. Zuckermann, S. L. Cobb and E. H. C. Bromley, Biopolymers, 2017 DOI:10.1002/bip.23014.
- L. A. Rawlinson, J. P. O'Gara, D. S. Jones and D. J. Brayden, J. Med. Microbiol., 2011, 60, 968 CrossRef PubMed.
- N. Malanovic and K. Lohner, Biochim. Biophys. Acta, 2016, 1858(5), 936–946 CrossRef CAS PubMed.
- J. Turner, Y. Cho, N. N. Dinh, A. J. Waring and R. I. Lehrer, Antimicrob. Agents Chemother., 1998, 42, 2206 CAS.
- B. Mojsoska and H. Jenssen, Pharmaceuticals, 2015, 8, 366 CrossRef CAS PubMed.
- M. R. Yeaman and N. Y. Yount, Pharmacol. Rev., 2003, 55, 27 CrossRef CAS PubMed.
- J. M. Andrews, J. Antimicrob. Chemother., 2001, 48, 5 CrossRef CAS PubMed.
Footnotes |
| † The authors declare no competing interests. |
| ‡ Electronic supplementary information (ESI) available. See DOI: 10.1039/c6md00648e |
| This journal is © The Royal Society of Chemistry 2017 |